Exposure to Roundup and Antibiotics Alters Gut Microbial Communities, Growth, and Behavior in Rana berlandieri Tadpoles
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbiome Assay
2.2. Statistical Analysis: Dorsal Body Area, Food Consumption, and Death Rates through Exposure and Recovery Periods
2.3. Behavior Analysis
2.4. Microbiome Analysis
3. Results
3.1. Dorsal Body Area and Food Consumption
3.2. Behavior
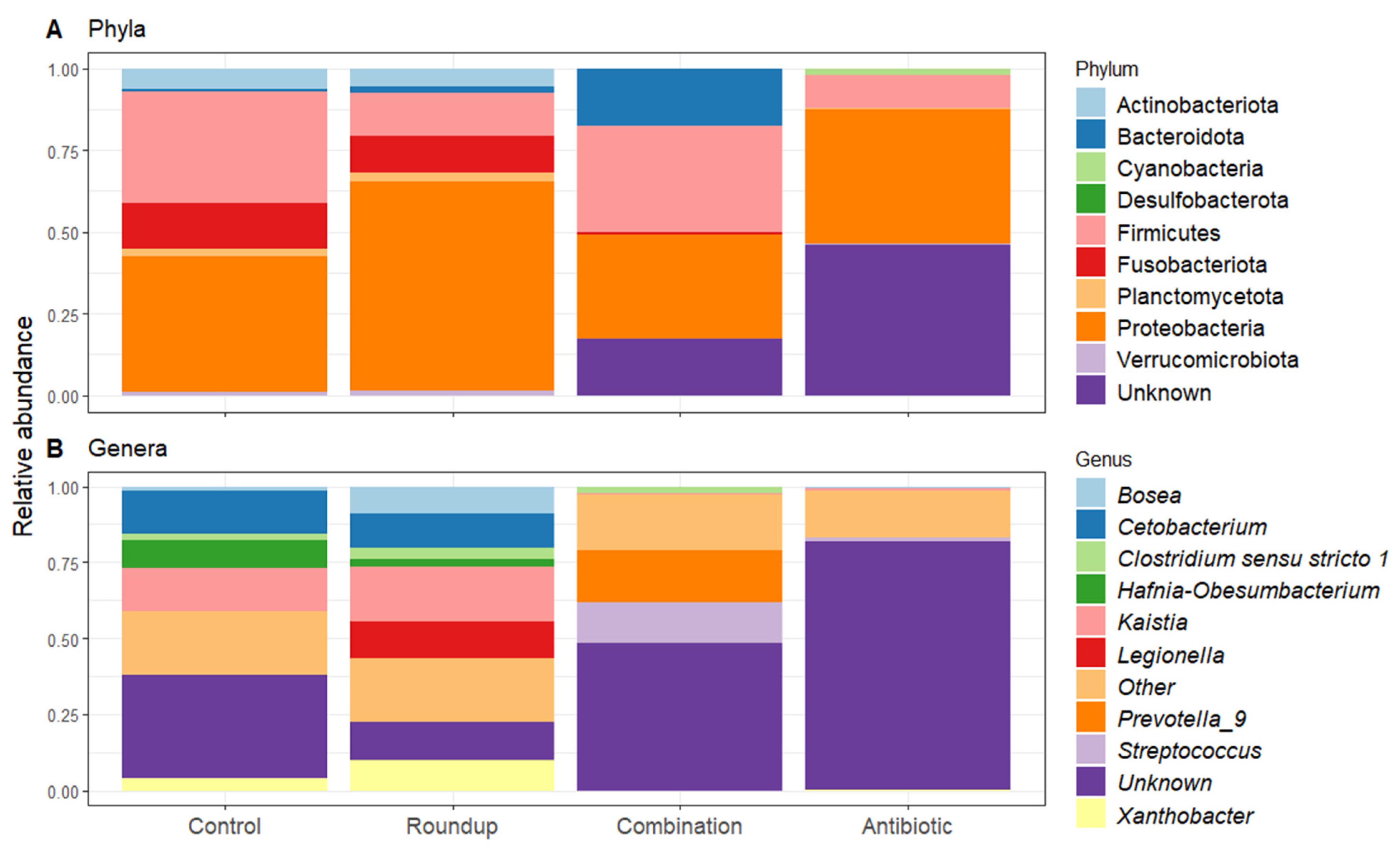
3.3. Gut Microbiome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Douglas, A.E. Fundamentals of Microbiome Science: How Microbes Shape Animal Biology; Princeton University Press: Princeton, NJ, USA, 2015. [Google Scholar]
- McKenzie, V.J.; Bowers, R.M.; Fierer, N.; Knight, R.; Lauber, C.L. Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME J. 2012, 6, 588–596. [Google Scholar] [CrossRef]
- Belden, L.K.; Hughey, M.C.; Rebollar, E.A.; Umile, T.P.; Loftus, S.C.; Burzynski, E.A.; Minbiole, K.P.; House, L.L.; Jensen, R.V.; Becker, M.H.; et al. Panamanian frog species host unique skin bacterial communities. Front. Microbiol. 2015, 6, 1171. [Google Scholar] [CrossRef] [PubMed]
- Assis, A.B.d.; Barreto, C.C.; Navas, C.A. Skin microbiota in frogs from the Brazilian Atlantic Forest: Species, forest type, and potential against pathogens. PLoS ONE 2017, 12, e0179628. [Google Scholar] [CrossRef] [PubMed]
- Hawrelak, J.A.; Myers, S.P. The causes of intestinal dysbiosis: A review. Altern. Med. Rev. 2004, 9, 180–197. [Google Scholar] [PubMed]
- Grenham, S.; Clarke, G.; Cryan, J.F.; Dinan, T.G. Brain-gut-microbe communication in health and disease. Front. Physiol. 2011, 2, 94. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Warne, R.W.; Kirschman, L.; Zeglin, L. Manipulation of gut microbiota during critical developmental windows affects host physiological performance and disease susceptibility across ontogeny. J. Anim. Ecol. 2019, 88, 845–856. [Google Scholar] [CrossRef]
- Johnson, K.V.; Burnet, P.W. Microbiome: Should we diversify from diversity? Gut Microbes 2016, 7, 455–458. [Google Scholar] [CrossRef]
- Sergaki, C.; Lagunas, B.; Lidbury, I.; Gifford, M.L.; Schäfer, P. Challenges and Approaches in Microbiome Research: From Fundamental to Applied. Front. Plant Sci. 2018, 9, 1205. [Google Scholar] [CrossRef]
- Kurenbach, B.; Hill, A.M.; Godsoe, W.; van Hamelsveld, S.; Heinemann, J.A. Agrichemicals and antibiotics in combination increase antibiotic resistance evolution. PeerJ 2018, 6, e5801. [Google Scholar] [CrossRef]
- Clarke, G.; Stilling, R.M.; Kennedy, P.J.; Stanton, C.; Cryan, J.F.; Dinan, T.G. Minireview: Gut microbiota: The neglected endocrine organ. Mol. Endocrinol. 2014, 28, 1221–1238. [Google Scholar] [CrossRef] [PubMed]
- Neuman, H.; Debelius, J.W.; Knight, R.; Koren, O. Microbial endocrinology: The interplay between the microbiota and the endocrine system. FEMS Microbiol. Rev. 2015, 39, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Bauer, K.C.; Huus, K.E.; Finlay, B.B. Microbes and the mind: Emerging hallmarks of the gut microbiota-brain axis. Cell. Microbiol. 2016, 18, 632–644. [Google Scholar] [CrossRef]
- Schwarzer, M.; Strigini, M.; Leulier, F. Gut Microbiota and Host Juvenile Growth. Calcif. Tissue Int. 2018, 102, 387–405. [Google Scholar] [CrossRef]
- Moran, N.A.; McCutcheon, J.P.; Nakabachi, A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 2008, 42, 165–190. [Google Scholar] [CrossRef]
- Falcinelli, S.; Picchietti, S.; Rodiles, A.; Cossignani, L.; Merrifield, D.L.; Taddei, A.R.; Maradonna, F.; Olivotto, I.; Gioacchini, G.; Carnevali, O. Lactobacillus rhamnosus lowers zebrafish lipid content by changing gut microbiota and host transcription of genes involved in lipid metabolism. Sci. Rep. 2015, 5, 9336. [Google Scholar] [CrossRef] [PubMed]
- Elsabagh, M.; Mohamed, R.; Moustafa, E.M.; Hamza, A.; Farrag, F.; Decamp, O.; Dawood, M.A.O.; Eltholth, M. Assessing the impact of Bacillus strains mixture probiotic on water quality, growth performance, blood profile and intestinal morphology of Nile tilapia, Oreochromis niloticus. Aquacult. Nutr. 2018, 24, 1613–1622. [Google Scholar] [CrossRef]
- Davidson, G.L.; Raulo, A.; Knowles, S.C.L. Identifying Microbiome-Mediated Behaviour in Wild Vertebrates. Trends Ecol. Evol. 2020, 35, 972–980. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Raes, J.; van den Bogert, B.; Arumugam, M.; Booijink, C.C.; Troost, F.J.; Bork, P.; Wels, M.; de Vos, W.M.; Kleerebezem, M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012, 6, 1415–1426. [Google Scholar] [CrossRef]
- Shanks, O.C.; Kelty, C.A.; Archibeque, S.; Jenkins, M.; Newton, R.J.; McLellan, S.L.; Huse, S.M.; Sogin, M.L. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl. Environ. Microbiol. 2011, 77, 2992–3001. [Google Scholar] [CrossRef]
- Ley, R.E.; Lozupone, C.A.; Hamady, M.; Knight, R.; Gordon, J.I. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008, 6, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Glasl, B.; Herndl, G.J.; Frade, P.R. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 2016, 10, 2280–2292. [Google Scholar] [CrossRef] [PubMed]
- Legrand, T.; Catalano, S.R.; Wos-Oxley, M.L.; Stephens, F.; Landos, M.; Bansemer, M.S.; Stone, D.A.J.; Qin, J.G.; Oxley, A.P.A. The Inner Workings of the Outer Surface: Skin and Gill Microbiota as Indicators of Changing Gut Health in Yellowtail Kingfish. Front. Microbiol. 2017, 8, 2664. [Google Scholar] [CrossRef] [PubMed]
- Wuerthner, V.P.; Hernández-Gómez, O.; Hua, J. Amphibian Skin Microbiota Response to Variable Housing Conditions and Experimental Treatment across Space and Time. J. Herpetol. 2019, 53, 324–335, 312. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Hernández-Gómez, O.; Wuerthner, V.; Hua, J. Amphibian Host and Skin Microbiota Response to a Common Agricultural Antimicrobial and Internal Parasite. Microb. Ecol. 2020, 79, 175–191. [Google Scholar] [CrossRef]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef]
- Koprivnikar, J.; Forbes, M.R.; Baker, R.L. Contaminant effects on host-parasite interactions: Atrazine, frogs, and trematodes. Environ. Toxicol. Chem. 2007, 26, 2166–2170. [Google Scholar] [CrossRef]
- Pochini, K.M.; Hoverman, J.T. Immediate and lag effects of pesticide exposure on parasite resistance in larval amphibians. Parasitology 2017, 144, 817–822. [Google Scholar] [CrossRef]
- Buss, N.; Hua, J. Parasite susceptibility in an amphibian host is modified by salinization and predators. Environ. Pollut. 2018, 236, 754–763. [Google Scholar] [CrossRef]
- Benbrook, C.M. Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 2016, 28, 3. [Google Scholar] [CrossRef] [PubMed]
- Stecher, B.; Robbiani, R.; Walker, A.W.; Westendorf, A.M.; Barthel, M.; Kremer, M.; Chaffron, S.; Macpherson, A.J.; Buer, J.; Parkhill, J.; et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007, 5, 2177–2189. [Google Scholar] [CrossRef] [PubMed]
- Jani, A.J.; Briggs, C.J. The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proc. Natl. Acad. Sci. USA 2014, 111, E5049–E5058. [Google Scholar] [CrossRef] [PubMed]
- Soen, Y. Environmental disruption of host-microbe co-adaptation as a potential driving force in evolution. Front. Genet. 2014, 5, 168. [Google Scholar] [CrossRef]
- Huang, B.H.; Chang, C.W.; Huang, C.W.; Gao, J.; Liao, P.C. Composition and Functional Specialists of the Gut Microbiota of Frogs Reflect Habitat Differences and Agricultural Activity. Front. Microbiol. 2017, 8, 2670. [Google Scholar] [CrossRef]
- Blair, W.F. Mating call and stage of speciation in the Microhyla olivacea-M. carolinensis complex. Evolution 1955, 9, 469–480. [Google Scholar]
- Samsel, A.; Seneff, S. Glyphosate’s Suppression of Cytochrome P450 Enzymes and Amino Acid Biosynthesis by the Gut Microbiome: Pathways to Modern Diseases. Entropy 2013, 15, 1416–1463. [Google Scholar] [CrossRef]
- Tang, Q.; Tang, J.; Ren, X.; Li, C. Glyphosate exposure induces inflammatory responses in the small intestine and alters gut microbial composition in rats. Environ. Pollut. 2020, 261, 114129. [Google Scholar] [CrossRef]
- Ruuskanen, S.; Fuchs, B.; Nissinen, R.; Puigbò, P.; Rainio, M.; Saikkonen, K.; Helander, M. Ecosystem consequences of herbicides: The role of microbiome. Trends Ecol. Evol. 2023, 38, 35–43. [Google Scholar] [CrossRef]
- Krynak, K.L.; Burke, D.J.; Benard, M.F. Rodeo™ Herbicide Negatively Affects Blanchard’s Cricket Frogs (Acris blanchardi) Survival and Alters the Skin-Associated Bacterial Community. J. Herpetol. 2017, 51, 402–410, 409. [Google Scholar] [CrossRef]
- Cuzziol Boccioni, A.P.; García-Effron, G.; Peltzer, P.M.; Lajmanovich, R.C. Effect of glyphosate and ciprofloxacin exposure on enteric bacteria of tadpoles. Rev. Argent. Microbiol. 2023, 55, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Zaneveld, J.R.; McMinds, R.; Vega Thurber, R. Stress and stability: Applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2017, 2, 17121. [Google Scholar] [CrossRef] [PubMed]
- Kohl, K.D.; Cary, T.L.; Karasov, W.H.; Dearing, M.D. Restructuring of the amphibian gut microbiota through metamorphosis. Environ. Microbiol. Rep. 2013, 5, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Relyea, R.A. The Lethal Impact of Roundup on Aquatic and Terrestrial Amphibians. Ecol. Appl. 2005, 15, 1118–1124. [Google Scholar] [CrossRef]
- Kamath, A. Fluoroquinolone induced neurotoxicity: A review. J. Adv. Pharm. Educ. Res. 2013, 3, 16–19. [Google Scholar]
- Relyea, R.A. The lethal impacts of roundup and predatory stress on six species of North American tadpoles. Arch. Environ. Contam. Toxicol. 2005, 48, 351–357. [Google Scholar] [CrossRef]
- Relyea, R.A. New effects of Roundup on amphibians: Predators reduce herbicide mortality; herbicides induce antipredator morphology. Ecol. Appl. 2012, 22, 634–647. [Google Scholar] [CrossRef]
- Relyea, R.A. Growth and survival of five amphibian species exposed to combinations of pesticides. Environ. Toxicol. Chem. Int. J. 2004, 23, 1737–1742. [Google Scholar] [CrossRef]
- Gabor, C.R.; Perkins, H.R.; Heitmann, A.T.; Forsburg, Z.R.; Aspbury, A.S. Roundup™ with corticosterone functions as an infodisruptor to antipredator response in tadpoles. Front. Ecol. Evol. 2019, 7, 114. [Google Scholar] [CrossRef]
- Hibbitts, T.J.; LaDuc, T.J.V.; Hibbitts, T.L.; Hibbitts, T.D. Texas Amphibians: A Field Guide; University of Texas Press: Austin, TX, USA, 2012. [Google Scholar]
- Gosner, K. A simplified table for staging anuran embryos and larvae with notes on identification. Herpetologica 1960, 16, 183–190. [Google Scholar]
- Knutie, S.A.; Wilkinson, C.L.; Kohl, K.D.; Rohr, J.R. Early-life disruption of amphibian microbiota decreases later-life resistance to parasites. Nat. Commun. 2017, 8, 86. [Google Scholar] [CrossRef]
- Davis, A.K.; Connell, L.L.; Grosse, A.; Maerz, J.C. A fast, non-invasive method of measuring growth in tadpoles using image analysis. Herpetol. Rev. 2008, 39, 56–57. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Gontang, E.A.; Aylward, F.O.; Carlos, C.; Glavina del Rio, T.; Chovatia, M.; Fern, A.; Lo, C.-C.; Malfatti, S.A.; Tringe, S.G.; Currie, C.R. Major changes in microbial diversity and community composition across gut sections of a juvenile Panchlora cockroach. PLoS ONE 2017, 12, e0177189. [Google Scholar] [CrossRef]
- Gabor, C.R.; Villatoro-Castañeda, M.; Carlos-Shanley, C.; Ujhegyi, N.; Bókony, V. Gut Bacterial Communities Vary across Habitats and Their Diversity Increases with Increasing Glucocorticoids in Toad Tadpoles. Diversity 2022, 15, 23. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High resolution sample inference from amplicon data. Nat. Methods 2015, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Charney, N.; Record, S. Jost diversity measures for community data. Package ‘vegetarian’. R Package 2015, 23, 3. [Google Scholar]
- Hill, M.O. Diversity and evenness: A unifying notation and its consequences. Ecology 1973, 54, 427–432. [Google Scholar] [CrossRef]
- Jost, L. Entropy and diversity. Oikos 2006, 113, 363–375. [Google Scholar] [CrossRef]
- Borcard, D.; Gillet, F.; Legendre, P. Numerical Ecology with R; Springer: New York, NY, USA, 2011; Volume 2. [Google Scholar]
- Jost, L. Partitioning diversity into independent alpha and beta components. Ecology 2007, 88, 2427–2439. [Google Scholar] [CrossRef]
- Ma, Z.S. Measuring microbiome diversity and similarity with Hill numbers. In Metagenomics; Elsevier: Amsterdam, The Netherlands, 2018; pp. 157–178. [Google Scholar]
- Lahti, L.; Shetty, S. Introduction to the Microbiome R Package. 2018. Available online: https://microbiome.github.io/tutorials (accessed on 15 May 2023).
- Kohl, K.D.; Cary, T.L.; Karasov, W.H.; Dearing, M.D. Larval exposure to polychlorinated biphenyl 126 (PCB-126) causes persistent alteration of the amphibian gut microbiota. Environ. Toxicol. Chem. 2015, 34, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Krynak, K.L. Environmental Influences Onamphibian Innate Immune Defense Traits; Case Western Reserve University: Cleveland, OH, USA, 2015. [Google Scholar]
- Hughey, M.C.; Walke, J.B.; Becker, M.H.; Umile, T.P.; Burzynski, E.A.; Minbiole, K.P.; Iannetta, A.A.; Santiago, C.N.; Hopkins, W.A.; Belden, L.K. Short-term exposure to coal combustion waste has little impact on the skin microbiome of adult spring peepers (Pseudacris crucifer). Appl. Environ. Microbiol. 2016, 82, 3493–3502. [Google Scholar] [CrossRef]
- Zhang, W.; Guo, R.; Yang, Y.; Ding, J.; Zhang, Y. Long-term effect of heavy-metal pollution on diversity of gastrointestinal microbial community of Bufo raddei. Toxicol. Lett. 2016, 258, 192–197. [Google Scholar] [CrossRef]
- Chang, C.W.; Huang, B.H.; Lin, S.M.; Huang, C.L.; Liao, P.C. Changes of diet and dominant intestinal microbes in farmland frogs. BMC Microbiol. 2016, 16, 33. [Google Scholar] [CrossRef] [PubMed]
- Chai, L.; Dong, Z.; Chen, A.; Wang, H. Changes in intestinal microbiota of Bufo gargarizans and its association with body weight during metamorphosis. Arch. Microbiol. 2018, 200, 1087–1099. [Google Scholar] [CrossRef]
- Scalvenzi, T.; Clavereau, I.; Bourge, M.; Pollet, N. Gut microbial ecology of Xenopus tadpoles across life stages. Peer Community J. 2021, 1, e41. [Google Scholar] [CrossRef]
- Earl, J.E.; Whiteman, H.H. Are commonly used fitness predictors accurate? A meta-analysis of amphibian size and age at metamorphosis. Copeia 2015, 103, 297–309. [Google Scholar] [CrossRef]
- Wojtaszek, B.F.; Buscarini, T.M.; Chartrand, D.T.; Stephenson, G.R.; Thompson, D.G. Effect of Release® herbicide on mortality, avoidance response, and growth of amphibian larvae in two forest wetlands. Environ. Toxicol. Chem. Int. J. 2005, 24, 2533–2544. [Google Scholar] [CrossRef]
- Edge, C.B.; Gahl, M.K.; Thompson, D.G.; Houlahan, J.E. Laboratory and field exposure of two species of juvenile amphibians to a glyphosate-based herbicide and Batrachochytrium dendrobatidis. Sci. Total Environ. 2013, 444, 145–152. [Google Scholar] [CrossRef]
- Bach, N.C.; Natale, G.S.; Somoza, G.M.; Ronco, A.E. Effect on the growth and development and induction of abnormalities by a glyphosate commercial formulation and its active ingredient during two developmental stages of the South-American Creole frog, Leptodactylus latrans. Environ. Sci. Pollut. Res. 2016, 23, 23959–23971. [Google Scholar] [CrossRef]
- Wilbur, H.M.; Collins, J.P. Ecological Aspects of Amphibian Metamorphosis: Nonnormal distributions of competitive ability reflect selection for facultative metamorphosis. Science 1973, 182, 1305–1314. [Google Scholar] [CrossRef]
- Semlitsch, R.D.; Scott, D.E.; Pechmann, J.H. Time and size at metamorphosis related to adult fitness in Ambystoma talpoideum. Ecology 1988, 69, 184–192. [Google Scholar] [CrossRef]
- Berven, K.A. Factors affecting population fluctuations in larval and adult stages of the wood frog (Rana sylvatica). Ecology 1990, 71, 1599–1608. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, L.; Xu, Y.; Deng, Y.; Zhang, L.; Qin, Y.; Wang, Z.; Liu, R.; Zhou, Z.; Diao, J. Amphibian (Rana nigromaculata) exposed to cyproconazole: Changes in growth index, behavioral endpoints, antioxidant biomarkers, thyroid and gonad development. Aquat. Toxicol. 2019, 208, 62–70. [Google Scholar] [CrossRef]
- Bridges, C.M. Tadpole swimming performance and activity affected by acute exposure to sublethal levels of carbaryl. Environ. Toxicol. Chem. Int. J. 1997, 16, 1935–1939. [Google Scholar] [CrossRef]
- Klavarioti, M.; Mantzavinos, D.; Kassinos, D. Removal of residual pharmaceuticals from aqueous systems by advanced oxidation processes. Environ. Int. 2009, 35, 402–417. [Google Scholar] [CrossRef]
- Martini, F.; Tarazona, J.V.; Pablos, M. Are fish and standardized FETAX assays protective enough for amphibians? A case study on Xenopus laevis larvae assay with biologically active substances present in livestock wastes. Sci. World J. 2012, 2012, 605804. [Google Scholar] [CrossRef]
- Rico, A.; Oliveira, R.; McDonough, S.; Matser, A.; Khatikarn, J.; Satapornvanit, K.; Nogueira, A.J.; Soares, A.M.; Domingues, I.; Van den Brink, P.J. Use, fate and ecological risks of antibiotics applied in tilapia cage farming in Thailand. Environ. Pollut. 2014, 191, 8–16. [Google Scholar] [CrossRef]
- Lopez-Cadenas, C.; Sierra-Vega, M.; Garcia-Vieitez, J.J.; Diez-Liébana, M.J.; Sahagun-Prieto, A.; Fernandez-Martinez, N. Enrofloxacin: Pharmacokinetics and metabolism in domestic animal species. Curr. Drug Metab. 2013, 14, 1042–1058. [Google Scholar] [CrossRef]
- Bernier, S.P.; Surette, M.G. Concentration-dependent activity of antibiotics in natural environments. Front. Microbiol. 2013, 4, 20. [Google Scholar] [CrossRef]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Tong, Q.; Du, X.-p.; Hu, Z.-f.; Cui, L.-y.; Bie, J.; Zhang, Q.-z.; Xiao, J.-h.; Lin, Y.; Wang, H.-b. Comparison of the gut microbiota of Rana amurensis and Rana dybowskii under natural winter fasting conditions. FEMS Microbiol. Lett. 2019, 366, fnz241. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, Y.; Gao, J.; Li, X. Effects of octylphenol exposure on the lipid metabolism and microbiome of the intestinal tract of Rana chensinensis tadpole by RNAseq and 16s amplicon sequencing. Ecotoxicol. Environ. Saf. 2020, 197, 110650. [Google Scholar] [CrossRef]
- Kaplan, J.L.; Shi, H.N.; Walker, W.A. The role of microbes in developmental immunologic programming. Pediatr. Res. 2011, 69, 465–472. [Google Scholar] [CrossRef]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef]
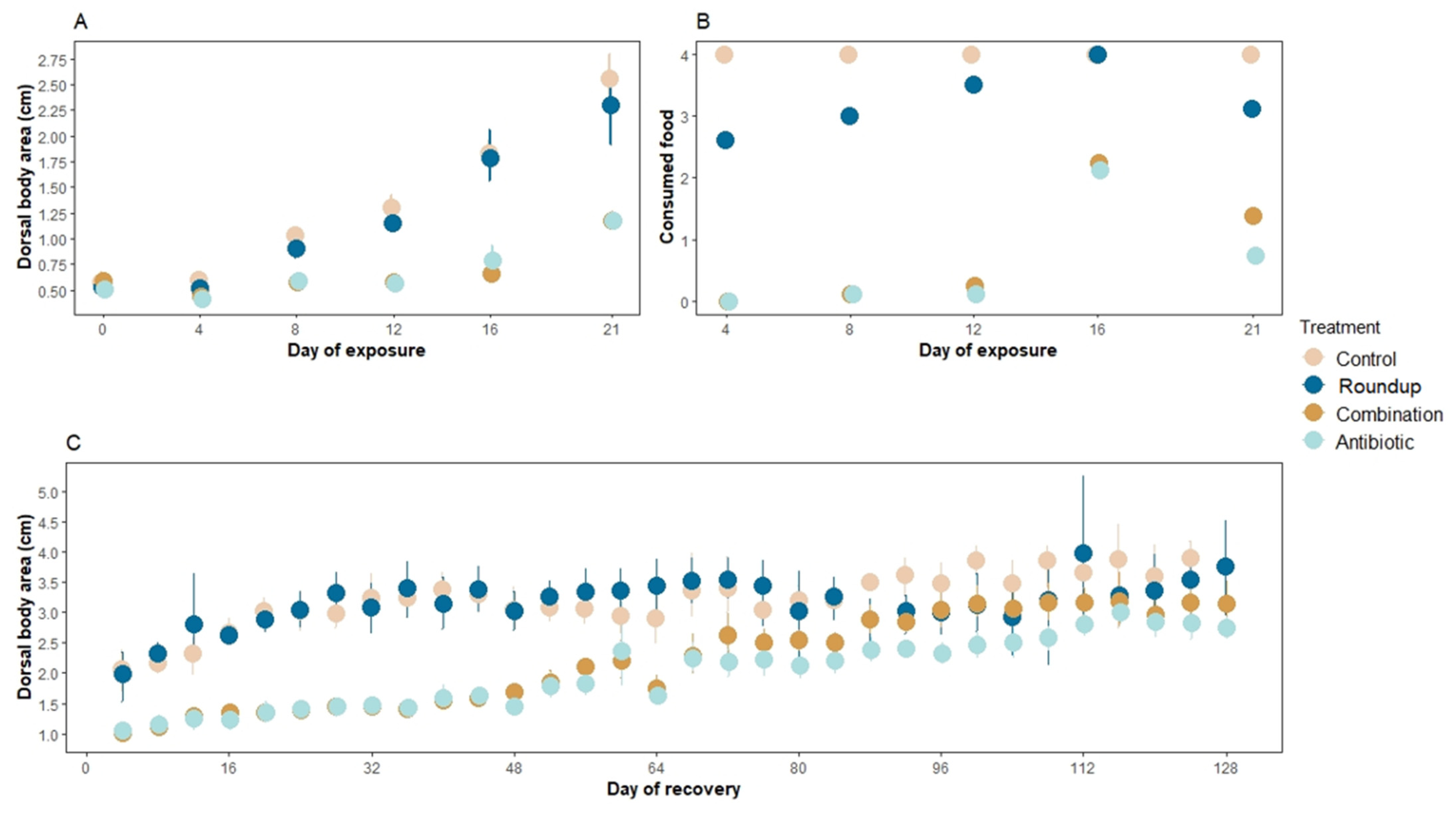
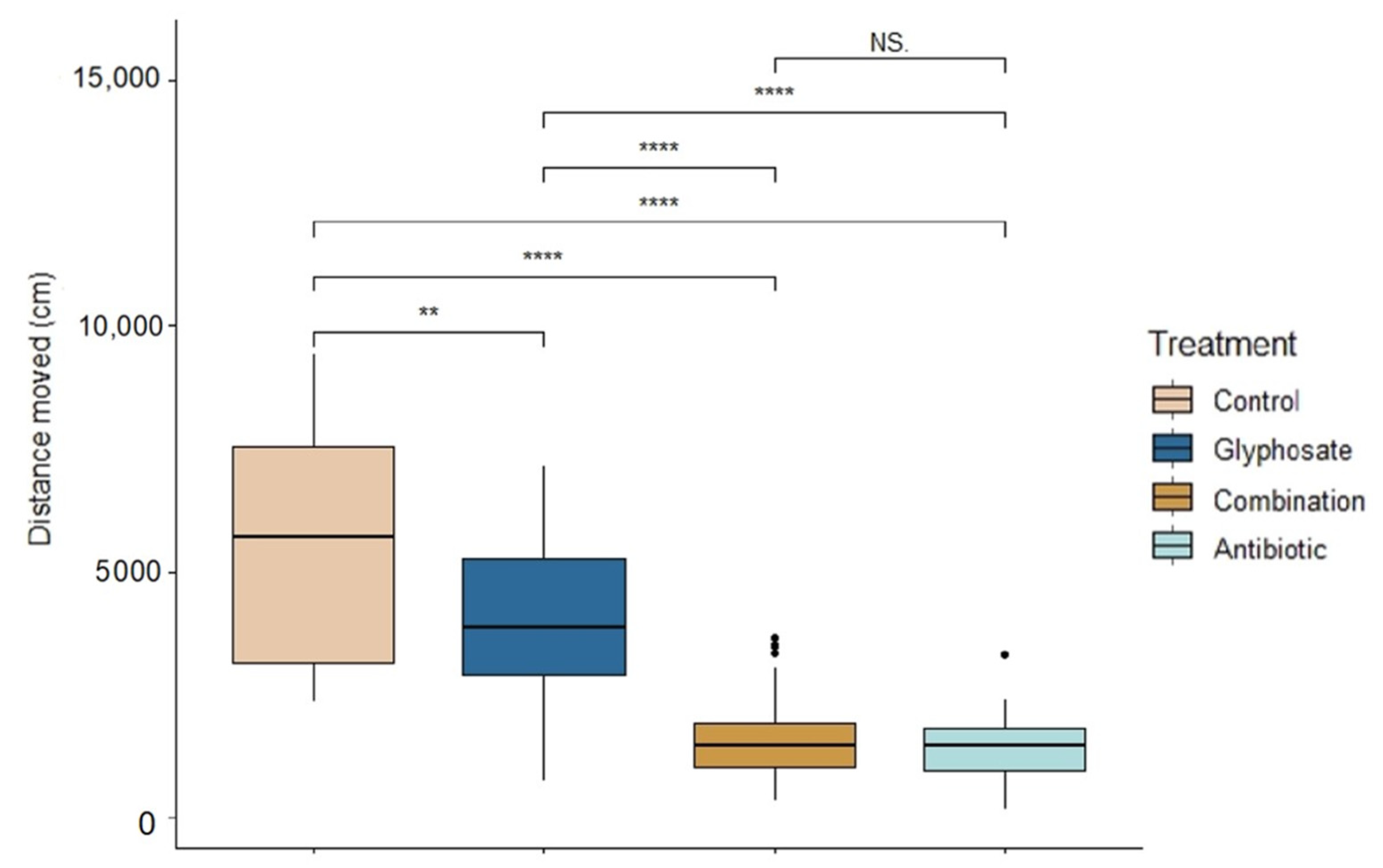
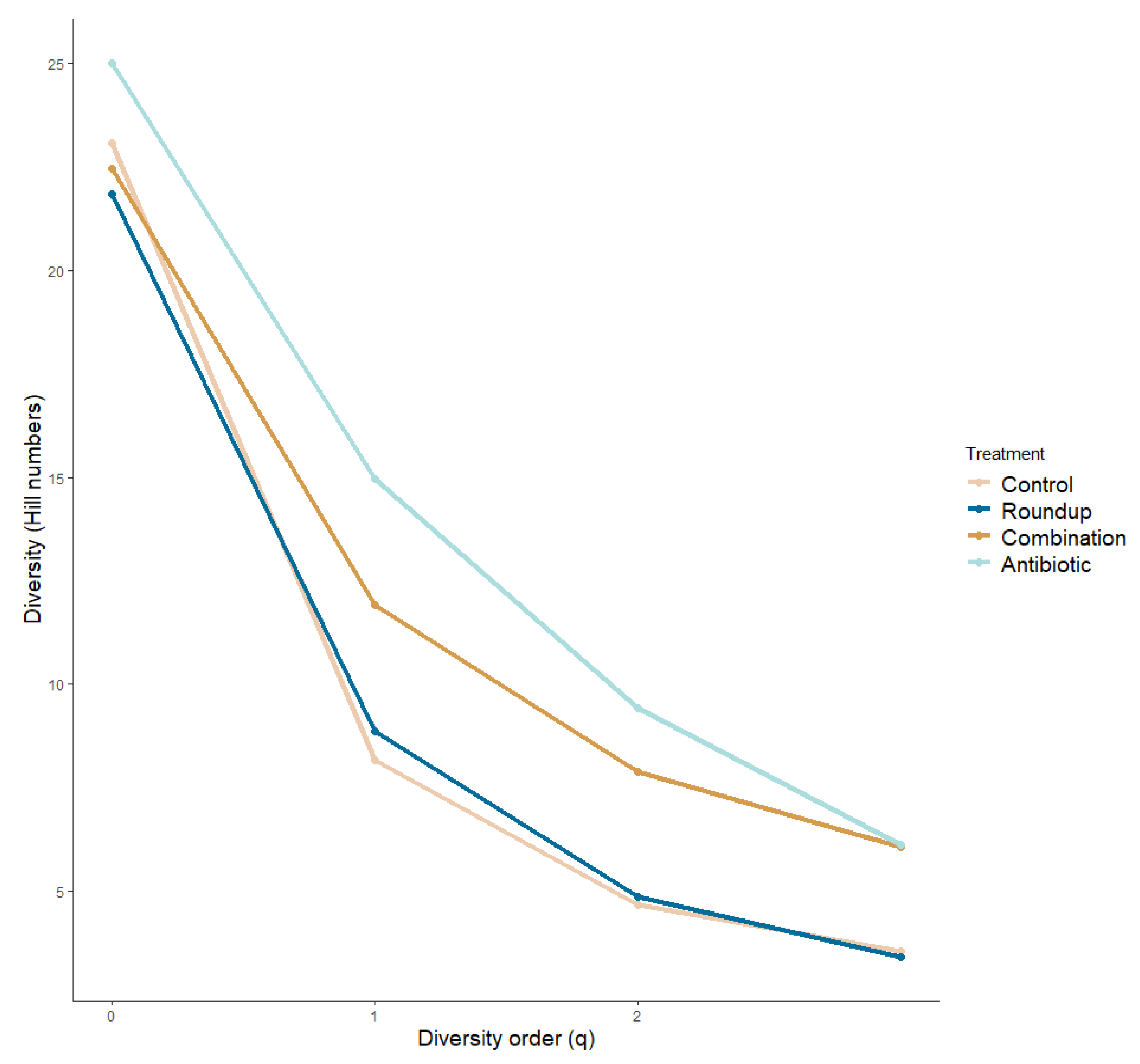
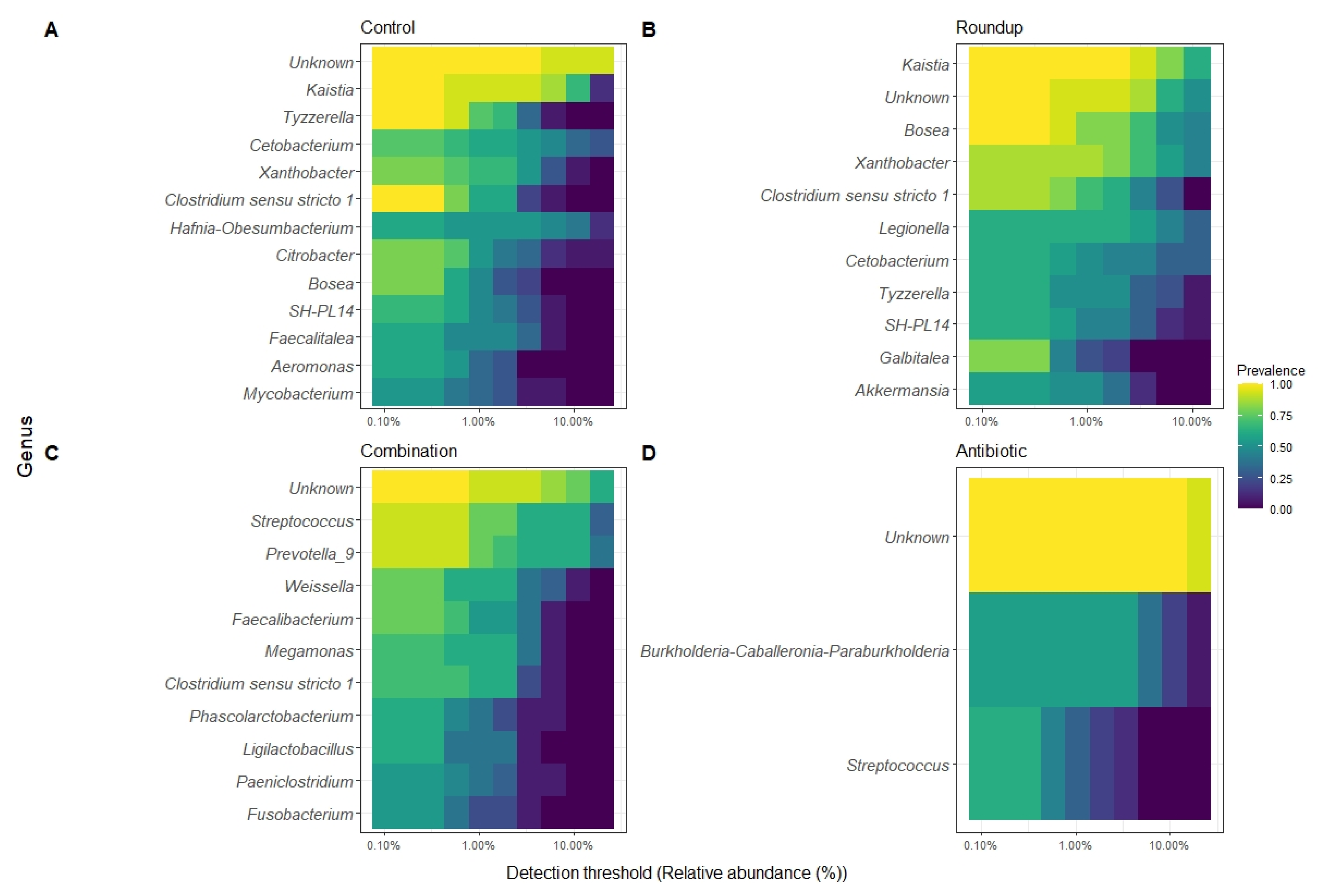
| Treatment Comparison | F | p | Adjusted R2 |
|---|---|---|---|
| Control–Roundup | 5.725 | 0.001 | 0.14 |
| Control–Antibiotic | 9.986 | 0.001 | 0.23 |
| Control–Combination | 12.49 | 0.001 | 0.30 |
| Roundup–Antibiotic | 4.643 | 0.001 | 0.13 |
| Roundup–Combination | 7.896 | 0.001 | 0.20 |
| Antibiotic–Combination | 4.114 | 0.001 | 0.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villatoro-Castañeda, M.; Forsburg, Z.R.; Ortiz, W.; Fritts, S.R.; Gabor, C.R.; Carlos-Shanley, C. Exposure to Roundup and Antibiotics Alters Gut Microbial Communities, Growth, and Behavior in Rana berlandieri Tadpoles. Biology 2023, 12, 1171. https://doi.org/10.3390/biology12091171
Villatoro-Castañeda M, Forsburg ZR, Ortiz W, Fritts SR, Gabor CR, Carlos-Shanley C. Exposure to Roundup and Antibiotics Alters Gut Microbial Communities, Growth, and Behavior in Rana berlandieri Tadpoles. Biology. 2023; 12(9):1171. https://doi.org/10.3390/biology12091171
Chicago/Turabian StyleVillatoro-Castañeda, Melissa, Zachery R. Forsburg, Whitney Ortiz, Sarah R. Fritts, Caitlin R. Gabor, and Camila Carlos-Shanley. 2023. "Exposure to Roundup and Antibiotics Alters Gut Microbial Communities, Growth, and Behavior in Rana berlandieri Tadpoles" Biology 12, no. 9: 1171. https://doi.org/10.3390/biology12091171
APA StyleVillatoro-Castañeda, M., Forsburg, Z. R., Ortiz, W., Fritts, S. R., Gabor, C. R., & Carlos-Shanley, C. (2023). Exposure to Roundup and Antibiotics Alters Gut Microbial Communities, Growth, and Behavior in Rana berlandieri Tadpoles. Biology, 12(9), 1171. https://doi.org/10.3390/biology12091171








