Learning to Fuse Multiple Brain Functional Networks for Automated Autism Identification
Abstract
Simple Summary
Abstract
1. Introduction
- An adaptive multi-FCNs fusion strategy is proposed for ASD diagnosis based on rs-fMRI by utilizing label information and diverse component FCNs, resulting in a more flexible and highly discriminative fused FCN.
- The fusion weights of component FCNs and the classifier are simultaneously optimized in a unified framework, making the model straightforward to implement and enhancing its generalization ability. This differs from the traditional FCN fusion methods which generally involve numerous hyperparameters and can easily lead to the overfitting problem on the limited medical data.
- Extensive experiments on the ABIDE datasets demonstrate the comparative performance of our method against several state-of-the-art FCNs fusion approaches.
2. Materials and Methods
2.1. Data Acquisition and Preprocessing
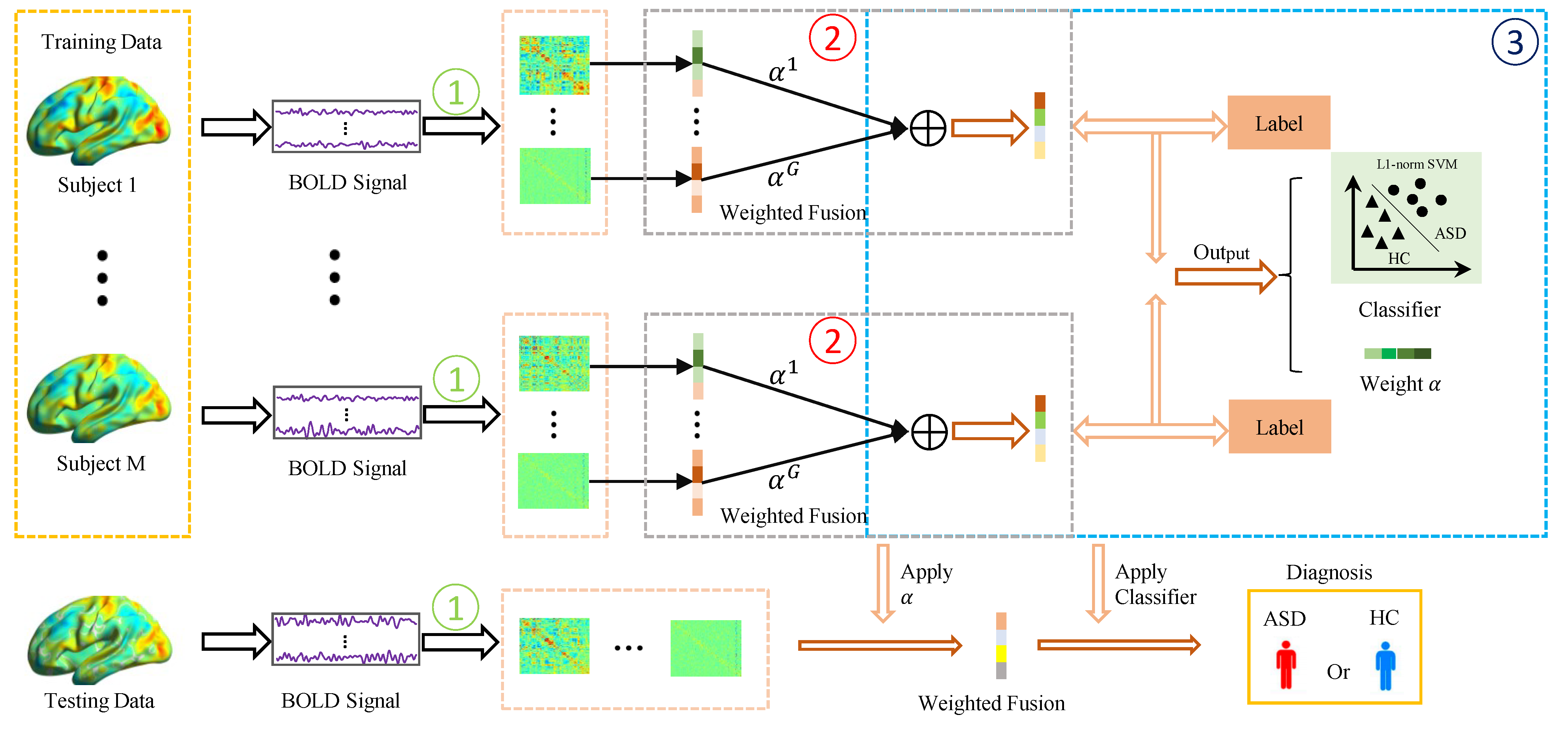
2.2. Proposed Method
2.2.1. Joint Multi-FCN Fusion and Disease Classification
2.2.2. Optimization Algorithm
| Algorithm 1 Algorithm of the proposed model. |
| Input: Input data , y; parameter ; maximum iteration number: ; and the iteration stopping threshold . Output: Weight vector C, fusion weight vector . Initialize:, choose . For Step 1: Fixed , updated C By solving Equation (3) Step 2: Fixed C, updated By solving Equation (5) Calculate Equation (1) as . If Break and Return . End End |
3. Experiments
3.1. Estimated Multi-FCN
3.2. Methods for Comparison
- Average: It fuses multiple FCNs by distributing the same weight for each FCN.
- MVJB: This method superimposes multiple FCNs into a tensor and uses tensor decomposition to learn a joint embedding representation of each ROI. Then PC is used to calculate the correlation between the embedding representations of ROIs to obtain the fused FCN [14].
- FC-kNN: It uses the criterion of intraclass compactness and interclass separability to fuse the commonality and specificity of two FCNs [15].
- MVS-GCN: MVS-GCN first generates dense FCNs and then binarizes them into multiple FCNs by different thresholds. Rather than fusing these FCNs directly, it uses multitask embedding learning to extract potential correlation features from different FCNs [17].
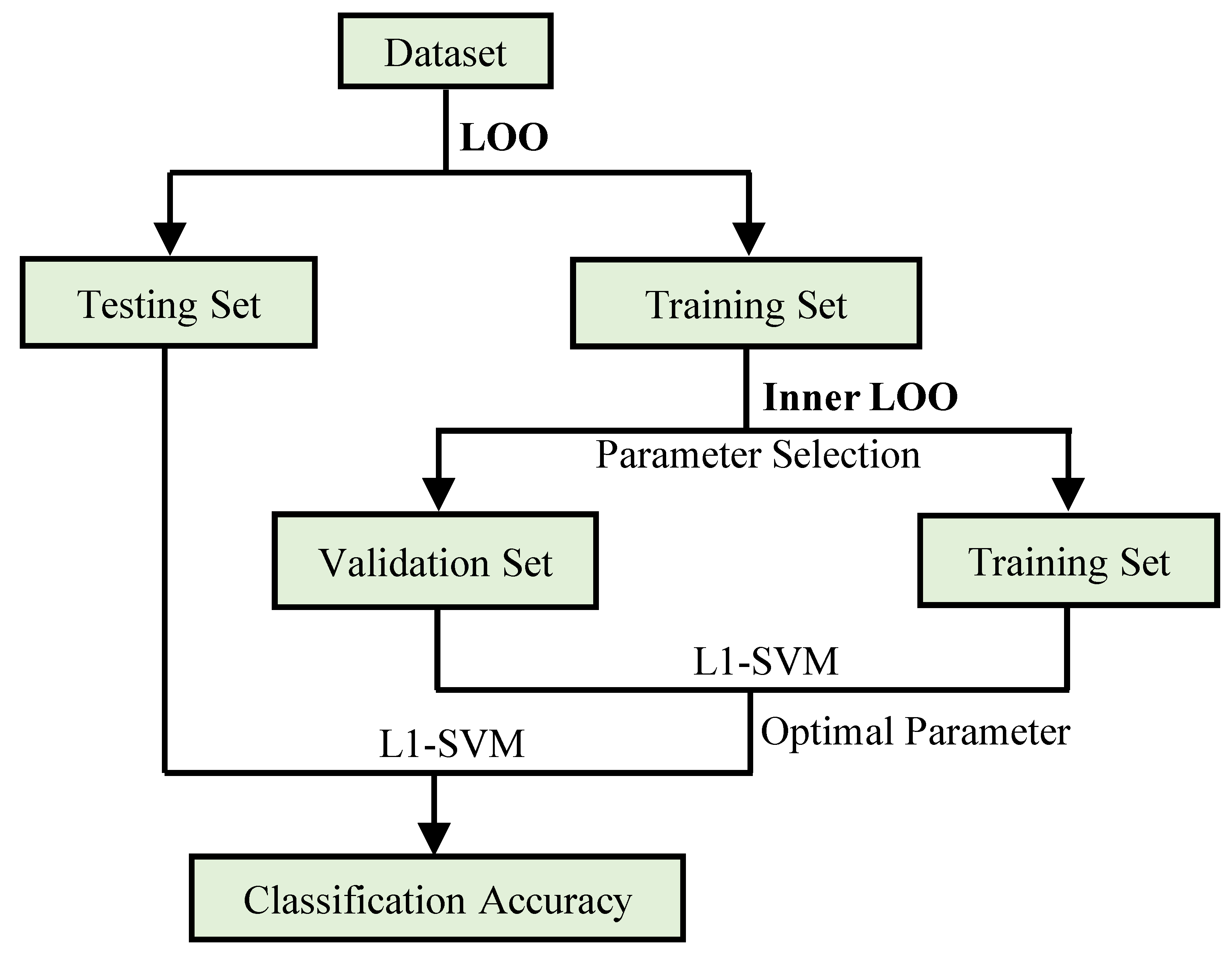
3.3. Experimental Setting
3.4. Classification Performance
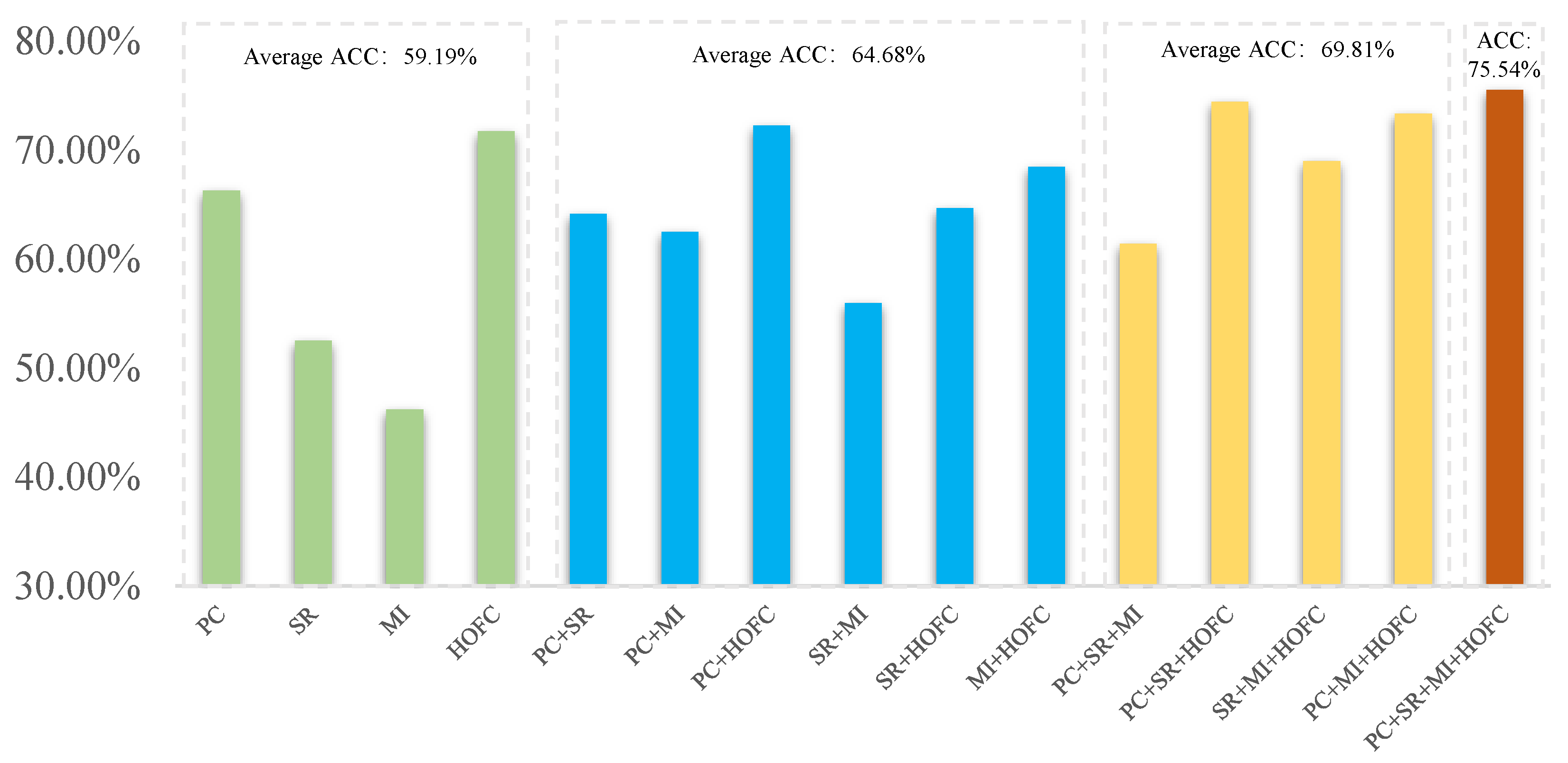
- Most of the muti-FCN fusion methods generally achieve better recognition performance than single FCN method. This further illustrates that it is not easy to acquire a good representation of brain only using a single type of FCN since the interaction between different ROIs in the real brain is extremely complex.
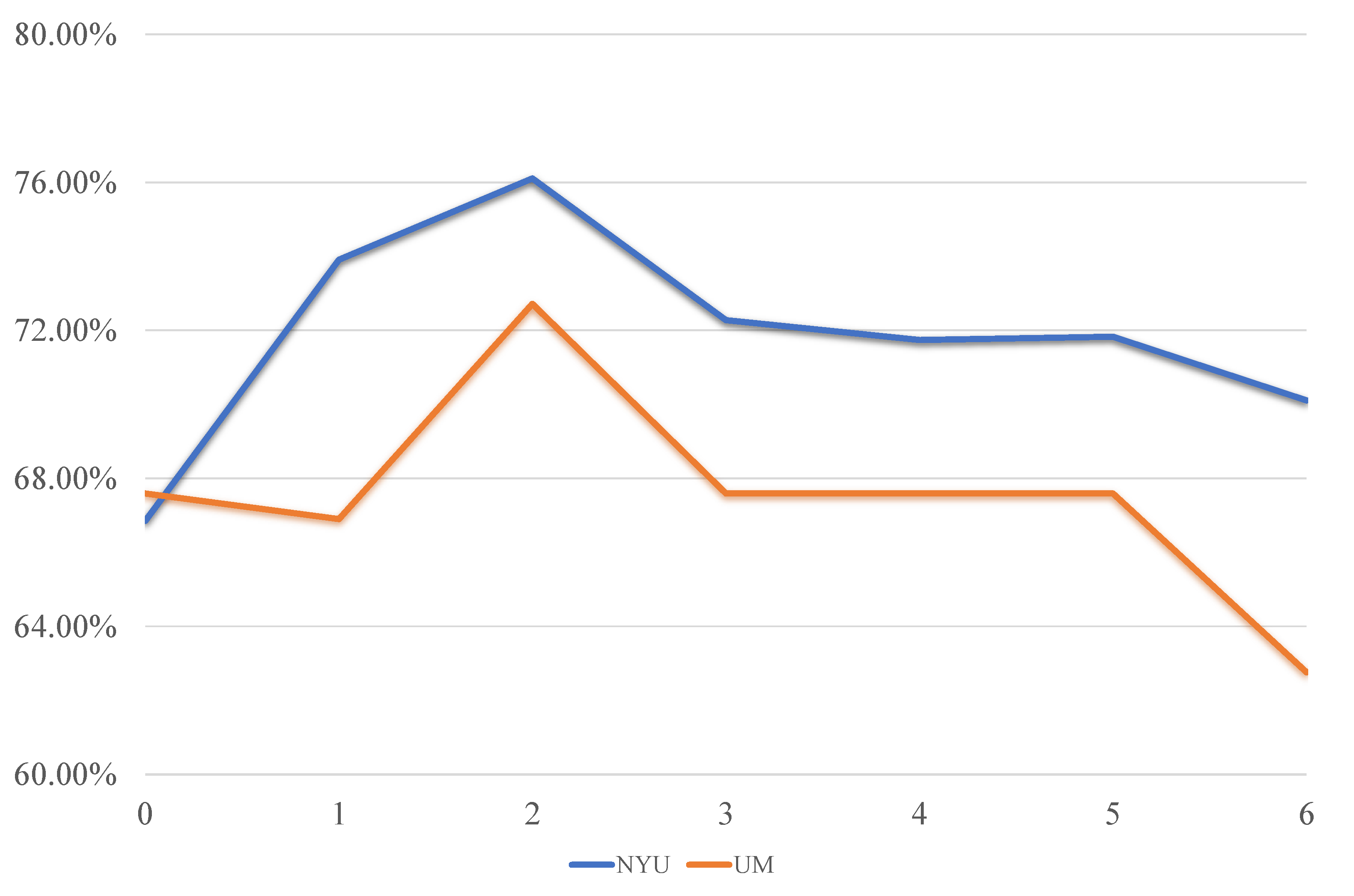
- The simple average-weighted approach cannot work well on the used two datasets. In contrast, our proposed method improves the ASD classification performance by and at NYU and UM sites, respectively. This may benefit from the adaptively optimized fusion weights combined with the label information for each type of FCN, as shown in Equation (1).
- Compared to MVJB [14] and FC-kNN [15], our method also contributes significantly to the improvement in accuracy. On the one hand, this is due to the fact that we incorporate the fused weight learning into the classification task, which may help to improve the discriminative ability of the final fused FCN. On the other hand, our method is not limited by the number of fused FCNs, and thus can obtain information from more FCNs.
- Despite its simplicity, our method can outperform MVS-GCN [17], a deep-learning-based multiview learning scheme. The possible reason is that the MVS-GCN framework needs to determine a lot of hyperparameters, which easily incurs the difficulty in parameter selection and may cause the overfitting problem, since the amount of training data is limited in our experiments.
4. Discussion
4.1. Hyperparameter Analysis
4.2. Classification Performance with Different Numbers of Fused FCNs
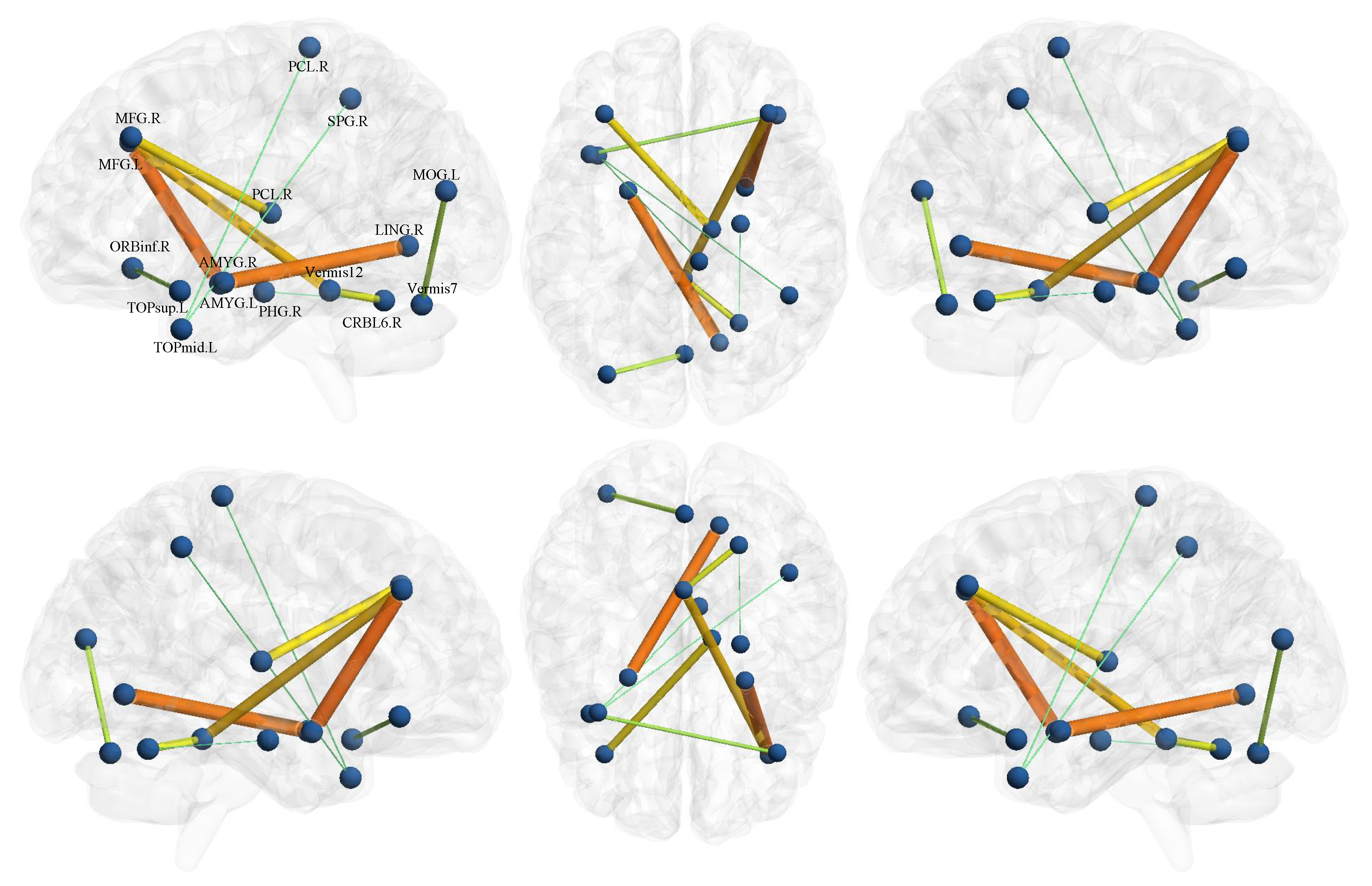
4.3. Discriminative Features
4.4. Fusion Weight Analysis
4.5. Time Complexity Analysis
4.6. Comparison with State-of-the-Art Methods
4.7. Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FCN | Functional connectivity network |
| ASD | Autism spectrum disorder |
| ROI | Regions of interest |
| HC | Healthy control |
| ABIDE | Autism Brain Imaging Data Exchange |
| BOLD | Blood oxygen level-dependent |
| rs-fMRI | Resting-state functional magnetic resonance imaging |
| PC | Pearson’s correlation |
| SR | Sparse representation |
| MI | Mutual information |
| HOFC | Higher-order functional connectivity |
| NYU | New York University |
| UM | University of Michigan |
| AO | Alternating optimization |
| SVM | Support vector machine |
| LOO | Leave-one-out |
References
- Hyman, S.L.; Levy, S.E.; Myers, S.M.; Kuo, D.Z.; Apkon, S.; Davidson, L.F.; Ellerbeck, K.A.; Foster, J.E.; Noritz, G.H.; Leppert, M.O.; et al. Identification, evaluation, and management of children with autism spectrum disorder. Pediatrics 2020, 145, e20193447. [Google Scholar] [CrossRef]
- Talantseva, O.I.; Romanova, R.S.; Shurdova, E.M.; Dolgorukova, T.A.; Sologub, P.S.; Titova, O.S.; Kleeva, D.F.; Grigorenko, E.L. The global prevalence of autism spectrum disorder: A three-level meta-analysis. Front. Psychiatry 2023, 14, 1071181. [Google Scholar] [CrossRef] [PubMed]
- Duvall, S.; Armstrong, K.; Shahabuddin, A.; Grantz, C.; Fein, D.; Lord, C. A road map for identifying autism spectrum disorder: Recognizing and evaluating characteristics that should raise red or “pink” flags to guide accurate differential diagnosis. Clin. Neuropsychol. 2022, 36, 1172–1207. [Google Scholar] [CrossRef]
- Chu, Y.; Wang, G.; Cao, L.; Qiao, L.; Liu, M. Multi-Scale Graph Representation Learning for Autism Identification With Functional MRI. Front. Neuroinform. 2022, 15, 802305. [Google Scholar] [CrossRef]
- Eslami, T.; Mirjalili, V.; Fong, A.; Laird, A.R.; Saeed, F. ASD-DiagNet: A hybrid learning approach for detection of autism spectrum disorder using fMRI data. Front. Neuroinform. 2019, 13, 70. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J.; Tang, Z.; Lei, B. Deep spatial-temporal feature fusion from adaptive dynamic functional connectivity for MCI identification. IEEE Trans. Med. Imaging 2020, 39, 2818–2830. [Google Scholar] [CrossRef] [PubMed]
- Korisky, A.; Gordon, I.; Goldstein, A. Oxytocin impacts top-down and bottom-up social perception in adolescents with ASD: A MEG study of neural connectivity. Mol. Autism 2022, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Vidaurre, D.; Beckmann, C.F.; Glasser, M.F.; Jenkinson, M.; Miller, K.L.; Nichols, T.E.; Robinson, E.C.; Salimi-Khorshidi, G.; Woolrich, M.W.; et al. Functional connectomics from resting-state fMRI. Trends Cogn. Sci. 2013, 17, 666–682. [Google Scholar] [CrossRef]
- Lee, H.; Lee, D.S.; Kang, H.; Kim, B.N.; Chung, M.K. Sparse brain network recovery under compressed sensing. IEEE Trans. Med. Imaging 2011, 30, 1154–1165. [Google Scholar]
- Wang, Z.; Alahmadi, A.; Zhu, D.; Li, T. Brain functional connectivity analysis using mutual information. In Proceedings of the 2015 IEEE Global Conference on Signal and Information Processing (GlobalSIP), Orlando, FL, USA, 14–16 December 2015; pp. 542–546. [Google Scholar]
- Gómez-Verdejo, V.; Martínez-Ramón, M.; Florensa-Vila, J.; Oliviero, A. Analysis of fMRI time series with mutual information. Med. Image Anal. 2012, 16, 451–458. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, X.; Shi, F.; Li, G.; Kim, M.; Giannakopoulos, P.; Haller, S.; Shen, D. Topographical information-based high-order functional connectivity and its application in abnormality detection for mild cognitive impairment. J. Alzheimer’s Dis. 2016, 54, 1095–1112. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, K.; Hao, Q.; Duan, P.; Kang, X. Hyperspectral anomaly detection with multiscale attribute and edge-preserving filters. IEEE Geosci. Remote Sens. Lett. 2018, 15, 1605–1609. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, L.; Zhang, J.; Qiao, L.; Liu, M. Fusing Multiview Functional Brain Networks by Joint Embedding for Brain Disease Identification. J. Pers. Med. 2023, 13, 251. [Google Scholar] [CrossRef] [PubMed]
- Gan, J.; Peng, Z.; Zhu, X.; Hu, R.; Ma, J.; Wu, G. Brain functional connectivity analysis based on multi-graph fusion. Med. Image Anal. 2021, 71, 102057. [Google Scholar] [CrossRef]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Wen, G.; Cao, P.; Bao, H.; Yang, W.; Zheng, T.; Zaiane, O. MVS-GCN: A prior brain structure learning-guided multi-view graph convolution network for autism spectrum disorder diagnosis. Comput. Biol. Med. 2022, 142, 105239. [Google Scholar] [CrossRef]
- Di Martino, A.; Yan, C.G.; Li, Q.; Denio, E.; Castellanos, F.X.; Alaerts, K.; Anderson, J.S.; Assaf, M.; Bookheimer, S.Y.; Dapretto, M.; et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry 2014, 19, 659–667. [Google Scholar] [CrossRef]
- Yan, C.G.; Wang, X.D.; Zuo, X.N.; Zang, Y.F. DPABI: Data processing & analysis for (resting-state) brain imaging. Neuroinformatics 2016, 14, 339–351. [Google Scholar]
- Friston, K.J.; Williams, S.; Howard, R.; Frackowiak, R.S.; Turner, R. Movement-related effects in fMRI time-series. Magn. Reson. Med. 1996, 35, 346–355. [Google Scholar] [CrossRef]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, O.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 2002, 15, 273–289. [Google Scholar] [CrossRef]
- Al-Sharoa, E.; Al-Khassaweneh, M.; Aviyente, S. Tensor based temporal and multilayer community detection for studying brain dynamics during resting state fMRI. IEEE Trans. Biomed. Eng. 2018, 66, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.E.; Chang, K.W.; Hsieh, C.J.; Wang, X.R.; Lin, C.J. LIBLINEAR: A library for large linear classification. J. Mach. Learn. Res. 2008, 9, 1871–1874. [Google Scholar]
- Schmidt, M.; Fung, G.; Rosales, R. Optimization Methods for l1-Regularization; Technical Report TR-2009-19; University of British Columbia: Vancouver, BC, Canada, 2009. [Google Scholar]
- Yuan, G.X.; Chang, K.W.; Hsieh, C.J.; Lin, C.J. A comparison of optimization methods and software for large-scale l1-regularized linear classification. J. Mach. Learn. Res. 2010, 11, 3183–3234. [Google Scholar]
- Li, W.; Tian, X. Numerical solution method for general interval quadratic programming. Appl. Math. Comput. 2008, 202, 589–595. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, L.; Qiao, L.; Shen, D. Estimating functional connectivity networks via low-rank tensor approximation with applications to MCI identification. IEEE Trans. Biomed. Eng. 2019, 67, 1912–1920. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Phadikar, S.; Deb, N.; Sinha, N.; Das, P.; Ghaderpour, E. Automatic Eyeblink and Muscular Artifact Detection and Removal From EEG Signals Using k-Nearest Neighbor Classifier and Long Short-Term Memory Networks. IEEE Sens. J. 2023, 23, 5422–5436. [Google Scholar] [CrossRef]
- Chandrasekhar, N.; Peddakrishna, S. Enhancing Heart Disease Prediction Accuracy through Machine Learning Techniques and Optimization. Processes 2023, 11, 1210. [Google Scholar] [CrossRef]
- Rizk, Y.; Hajj, N.; Mitri, N.; Awad, M. Deep belief networks and cortical algorithms: A comparative study for supervised classification. Appl. Comput. Inform. 2019, 15, 81–93. [Google Scholar] [CrossRef]
- Montemurro, N.; Aliaga, N.; Graff, P.; Escribano, A.; Lizana, J. New Targets and New Technologies in the Treatment of Parkinson’s Disease: A Narrative Review. Int. J. Environ. Res. Public Health 2022, 19, 8799. [Google Scholar] [CrossRef]
- Chen, Y.; Dai, J.; Tang, L.; Mikhailova, T.; Liang, Q.; Li, M.; Zhou, J.; Kopp, R.F.; Weickert, C.; Chen, C.; et al. Neuroimmune transcriptome changes in patient brains of psychiatric and neurological disorders. Mol. Psychiatry 2023, 28, 710–721. [Google Scholar] [CrossRef]
- Xia, M.; Wang, J.; He, Y. BrainNet Viewer: A network visualization tool for human brain connectomics. PLoS ONE 2013, 8, e68910. [Google Scholar] [CrossRef] [PubMed]
- Menon, V. The triple network model, insight, and large-scale brain organization in autism. Biol. Psychiatry 2018, 84, 236–238. [Google Scholar] [CrossRef]
- Bi, X.a.; Liu, Y.; Jiang, Q.; Shu, Q.; Sun, Q.; Dai, J. The diagnosis of autism spectrum disorder based on the random neural network cluster. Front. Hum. Neurosci. 2018, 12, 257. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Q.; Zhang, H.; Chen, J.; Wang, S.; Shen, D. Sparse multiview task-centralized ensemble learning for ASD diagnosis based on age-and sex-related functional connectivity patterns. IEEE Trans. Cybern. 2018, 49, 3141–3154. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Fu, C.; You, Y.; Fu, D. Classification of ASD based on fMRI data with deep learning. Cogn. Neurodyn. 2021, 15, 961–974. [Google Scholar] [CrossRef]
- Kang, L.; Chen, J.; Huang, J.; Jiang, J. Autism spectrum disorder recognition based on multi-view ensemble learning with multi-site fMRI. Cogn. Neurodyn. 2023, 17, 345–355. [Google Scholar] [CrossRef]
- Kam, T.E.; Suk, H.I.; Lee, S.W. Multiple functional networks modeling for autism spectrum disorder diagnosis. Hum. Brain Mapp. 2017, 38, 5804–5821. [Google Scholar] [CrossRef]
- Niu, K.; Guo, J.; Pan, Y.; Gao, X.; Peng, X.; Li, N.; Li, H. Multichannel deep attention neural networks for the classification of autism spectrum disorder using neuroimaging and personal characteristic data. Complexity 2020, 2020, 1357853. [Google Scholar] [CrossRef]
- Li, W.; Qiao, L.; Zhang, L.; Wang, Z.; Shen, D. Functional brain network estimation with time series self-scrubbing. IEEE J. Biomed. Health Inform. 2019, 23, 2494–2504. [Google Scholar] [CrossRef]
| Datasets | Class | Gender (M/F) | Age (Years) | FIQ |
|---|---|---|---|---|
| NYU | ASD (N = 79) | |||
| HC (N = 105) | ||||
| UM | ASD (N = 68) | |||
| HC (N = 77) |
| Method | Definition |
|---|---|
| Pearson’s correlation (PC) | |
| Sparse representation (SR) | |
| Mutual Information (MI) | |
| Higher Order Functional Connections (HOFC) |
| Datasets | Method | ACC | SEN | SPE | BAC | PPV | NPV | F1 | AUC |
|---|---|---|---|---|---|---|---|---|---|
| NYU | PC | 66.30% | 58.23% | 72.38% | 65.30% | 61.33% | 69.72% | 59.74% | 70.56% |
| SR | 62.50% | 43.03% | 77.14% | 60.09% | 58.62% | 64.29% | 49.64% | 66.74% | |
| MI | 46.20% | 40.51% | 50.48% | 45.49% | 38.10% | 53.00% | 39.26% | 41.29% | |
| HOFC | 71.74% | 62.03% | 79.05% | 70.54% | 69.01% | 73.45% | 65.33% | 77.20% | |
| Average | 69.02% | 59.49% | 76.19% | 67.84% | 65.28% | 71.43% | 62.25% | 73.66% | |
| MVJB | 74.46% | 64.56% | 81.90% | 73.23% | 72.86% | 75.44% | 68.46% | 78.72% | |
| FC-kNN | 70.65% | 64.56% | 75.24% | 69.90% | 66.23% | 73.83% | 65.38% | 75.86% | |
| MVS-GCN | 72.28% | 63.29% | 79.05% | 71.17% | 69.44% | 74.11% | 66.23% | 77.53% | |
| Proposed | 75.54% | 65.82% | 82.86% | 74.34% | 74.29% | 76.32% | 69.80% | 79.07% |
| Datasets | Method | ACC | SEN | SPE | BAC | PPV | NPV | F1 | AUC |
|---|---|---|---|---|---|---|---|---|---|
| UM | PC | 56.55% | 55.88% | 57.14% | 56.51% | 53.52% | 59.46% | 54.68% | 62.18% |
| SR | 51.72% | 50.00% | 53.25% | 51.62% | 48.57% | 54.67% | 49.28% | 52.06% | |
| MI | 49.66% | 22.06% | 74.03% | 48.04% | 42.86% | 51.82% | 29.13% | 41.75% | |
| HOFC | 62.07% | 55.88% | 67.53% | 61.71% | 60.32% | 63.41% | 58.02% | 66.12% | |
| Average | 60.69% | 51.47% | 68.83% | 60.15% | 59.32% | 61.13% | 55.12% | 65.09% | |
| MVJB | 63.45% | 63.24% | 63.64% | 63.44% | 60.56% | 66.22% | 61.87% | 65.16% | |
| FC-kNN | 65.52% | 63.24% | 67.53% | 65.38% | 63.24% | 67.53% | 63.25% | 66.14% | |
| MVS-GCN | 68.27% | 70.59% | 66.23% | 68.41% | 64.86% | 71.83% | 67.60% | 71.56% | |
| Proposed | 71.72% | 70.59% | 72.73% | 71.66% | 69.57% | 73.68% | 70.07% | 77.35% |
| Pairwise Comparison | p-Value | |
|---|---|---|
| Proposed vs. PC | Yes | |
| Proposed vs. SR | Yes | |
| Proposed vs. MI | Yes | |
| Proposed vs. HOFC | Yes | |
| Proposed vs. Average | Yes | |
| Proposed vs. MVJB | Yes | |
| Proposed vs. FC-kNN | Yes | |
| Proposed vs. MVS-GCN | Yes |
| Methods | Proposed | MVJB | FC-kNN | MVS-GCN |
|---|---|---|---|---|
| Time | 1480 s | 1832 s | 2125 s | 2658 s |
| Method | ACC | SEN | SPE | BAC | PPV | NPV | F1 | AUC |
|---|---|---|---|---|---|---|---|---|
| MEL | 74.60% | - | - | - | - | - | 72.20% | 74.30% |
| DRBM | 75.24% | 61.33% | 85.71% | - | 82.10% | - | - | 73.73% |
| Multichannel DANN | 70.91% | 72.02% | 69.92% | - | 75.82% | - | 73.83% | - |
| Proposed | 75.54% | 65.82% | 82.86% | 74.34% | 74.29% | 76.32% | 69.80% | 79.07% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Ma, Y.; Qiao, L.; Zhang, L.; Liu, M. Learning to Fuse Multiple Brain Functional Networks for Automated Autism Identification. Biology 2023, 12, 971. https://doi.org/10.3390/biology12070971
Zhang C, Ma Y, Qiao L, Zhang L, Liu M. Learning to Fuse Multiple Brain Functional Networks for Automated Autism Identification. Biology. 2023; 12(7):971. https://doi.org/10.3390/biology12070971
Chicago/Turabian StyleZhang, Chaojun, Yunling Ma, Lishan Qiao, Limei Zhang, and Mingxia Liu. 2023. "Learning to Fuse Multiple Brain Functional Networks for Automated Autism Identification" Biology 12, no. 7: 971. https://doi.org/10.3390/biology12070971
APA StyleZhang, C., Ma, Y., Qiao, L., Zhang, L., & Liu, M. (2023). Learning to Fuse Multiple Brain Functional Networks for Automated Autism Identification. Biology, 12(7), 971. https://doi.org/10.3390/biology12070971





