Identification of Gene Markers Associated with COVID-19 Severity and Recovery in Different Immune Cell Subtypes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Preliminary Selection
2.3. Feature-Ranking Algorithms
2.3.1. Last Absolute Shrinkage and Selection Operator
2.3.2. Light Gradient Boosting Machine
2.3.3. Monte Carlo Feature Selection
2.3.4. Max-Relevance and Min-Redundancy
2.4. Incremental Feature Selection
2.5. Synthetic Minority Oversampling Technique
2.6. Classification Algorithm
2.6.1. Decision Tree
2.6.2. K-Nearest Neighbor
2.6.3. Random Forest
2.7. Performance Evaluation
3. Results
3.1. Preliminary Selection and Feature Ranking Result
3.2. Incremental Feature Selection Results
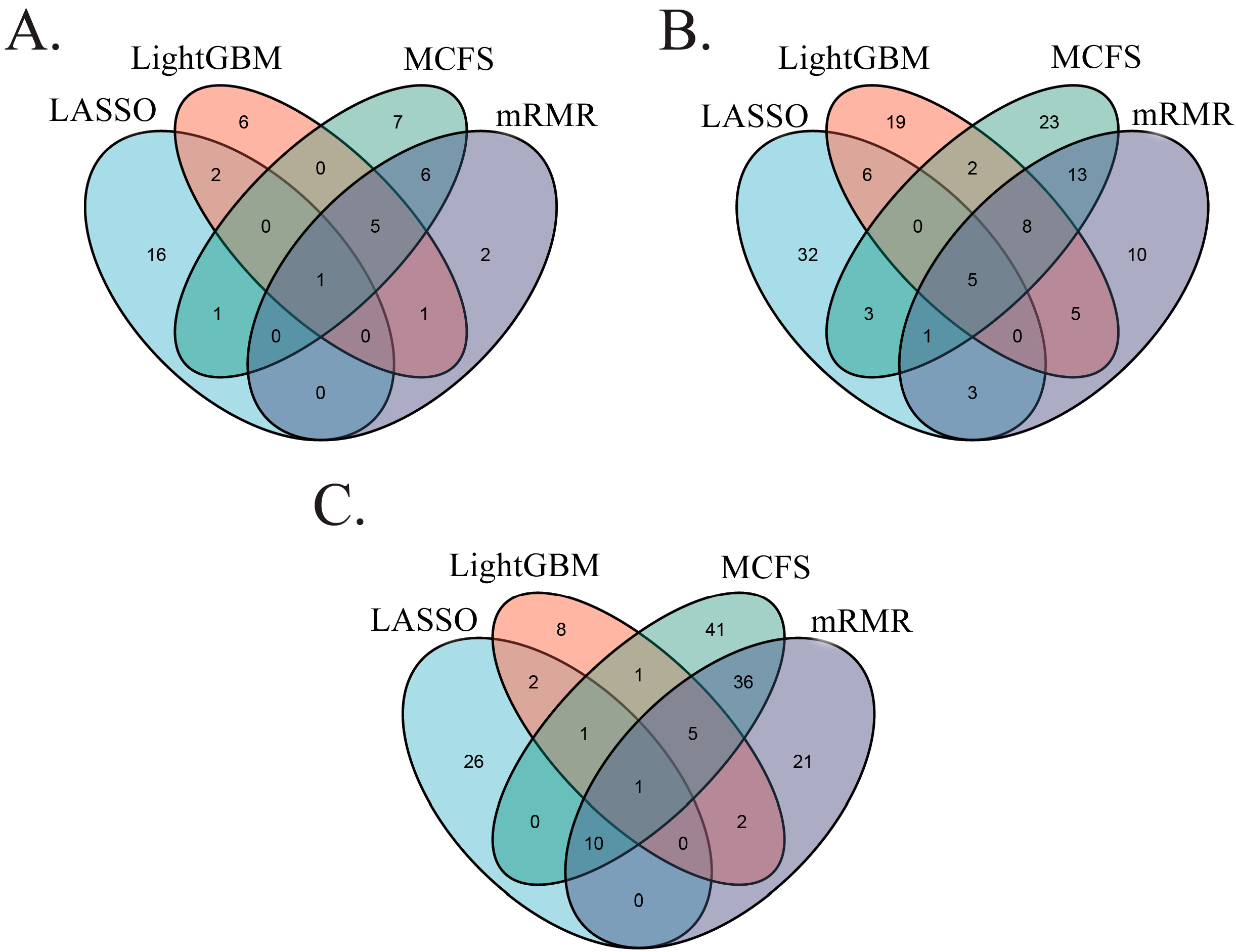
3.3. Intersection of Different Gene Lists
4. Discussion
4.1. Comparative Analysis and Verification with Prior Research
4.2. T-Cell Features Associated with the Severity of COVID-19
4.3. B-Cell Features Associated with the Severity of COVID-19
4.4. Myeloid Cell Features Associated with the Severity of COVID-19
4.5. Limitations of This Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pijls, B.G.; Jolani, S.; Atherley, A.; Derckx, R.T.; Dijkstra, J.I.; Franssen, G.H.; Hendriks, S.; Richters, A.; Venemans-Jellema, A.; Zalpuri, S. Demographic risk factors for COVID-19 infection, severity, icu admission and death: A meta-analysis of 59 studies. BMJ Open 2021, 11, e044640. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.; Shao, C.; Huang, J.; Gan, J.; Huang, X.; Bucci, E.; Piacentini, M.; Ippolito, G.; Melino, G. COVID-19 Infection: The Perspectives on Immune Responses; Nature Publishing Group: Berlin, Germany, 2020; Volume 27, pp. 1451–1454. [Google Scholar]
- Zhang, F.; Gan, R.; Zhen, Z.; Hu, X.; Li, X.; Zhou, F.; Liu, Y.; Chen, C.; Xie, S.; Zhang, B. Adaptive immune responses to sars-cov-2 infection in severe versus mild individuals. Signal Transduct. Target. Ther. 2020, 5, 156. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Wang, X.-M.; Xing, X.; Xu, Z.; Zhang, C.; Song, J.-W.; Fan, X.; Xia, P.; Fu, J.-L.; Wang, S.-Y. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020, 21, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Shuwa, H.A.; Shaw, T.N.; Knight, S.B.; Wemyss, K.; McClure, F.A.; Pearmain, L.; Prise, I.; Jagger, C.; Morgan, D.J.; Khan, S. Alterations in T and B cell function persist in convalescent COVID-19 patients. Med 2021, 2, 720–735.e724. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-W.; Zhang, C.; Fan, X.; Meng, F.-P.; Xu, Z.; Xia, P.; Cao, W.-J.; Yang, T.; Dai, X.-P.; Wang, S.-Y. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020, 11, 3410. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Strålin, K.; Gorin, J.-B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020, 183, 158–168.e114. [Google Scholar] [CrossRef] [PubMed]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Liu, S.; Yang, L.; Yu, W.; Zhang, Y. Myeloid cells in COVID-19 microenvironment. Signal Transduct. Target. Ther. 2021, 6, 372. [Google Scholar] [CrossRef]
- Liu, H.; Setiono, R. Incremental feature selection. Appl. Intell. 1998, 9, 217–230. [Google Scholar] [CrossRef]
- Ren, X.; Wen, W.; Fan, X.; Hou, W.; Su, B.; Cai, P.; Li, J.; Liu, Y.; Tang, F.; Zhang, F.; et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell 2021, 184, 1895–1913.e1819. [Google Scholar] [CrossRef]
- Ranstam, J.; Cook, J. Lasso regression. J. Br. Surg. 2018, 105, 1348. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3146–3154. [Google Scholar]
- Dramiński, M.; Koronacki, J. Rmcfs: An R package for monte Carlo feature selection and interdependency discovery. J. Stat. Softw. 2018, 85, 1–28. [Google Scholar] [CrossRef]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Li, H.; Zhang, S.; Chen, L.; Pan, X.; Li, Z.; Huang, T.; Cai, Y.D. Identifying functions of proteins in mice with functional embedding features. Front. Genet. 2022, 13, 909040. [Google Scholar] [CrossRef]
- Li, H.; Huang, F.; Liao, H.; Li, Z.; Feng, K.; Huang, T.; Cai, Y.D. Identification of COVID-19-specific immune markers using a machine learning method. Front. Mol. Biosci. 2022, 9, 952626. [Google Scholar] [CrossRef]
- Li, Z.; Guo, W.; Ding, S.; Chen, L.; Feng, K.; Huang, T.; Cai, Y.D. Identifying key microrna signatures for neurodegenerative diseases with machine learning methods. Front. Genet. 2022, 13, 880997. [Google Scholar] [CrossRef]
- Lu, J.; Meng, M.; Zhou, X.; Ding, S.; Feng, K.; Zeng, Z.; Huang, T.; Cai, Y.D. Identification of COVID-19 severity biomarkers based on feature selection on single-cell RNA-Seq data of CD8+ T cells. Front. Genet. 2022, 13, 1053772. [Google Scholar] [CrossRef]
- Huang, F.; Fu, M.; Li, J.; Chen, L.; Feng, K.; Huang, T.; Cai, Y.-D. Analysis and prediction of protein stability based on interaction network, gene ontology, and KEGG pathway enrichment scores. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2023, 1871, 140889. [Google Scholar] [CrossRef]
- Huang, F.; Ma, Q.; Ren, J.; Li, J.; Wang, F.; Huang, T.; Cai, Y.-D. Identification of smoking associated transcriptome aberration in blood with machine learning methods. BioMed Res. Int. 2023, 2023, 5333361. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhang, Y.; Guo, W.; Feng, K.; Yuan, Y.; Huang, T.; Cai, Y.-D. Identification of genes associated with the impairment of olfactory and gustatory functions in COVID-19 via machine-learning methods. Life 2023, 13, 798. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, L.; Lu, J. A similarity-based method for prediction of drug side effects with heterogeneous information. Math. Biosci. 2018, 306, 136–144. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the International Joint Conference on Artificial Intelligence, Montreal, QC, Canada, 20–25 August 1995; Lawrence Erlbaum Associates Ltd.: Mahwah, NJ, USA, 1995; pp. 1137–1145. [Google Scholar]
- Safavian, S.R.; Landgrebe, D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 1991, 21, 660–674. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Wang, H.; Chen, L. Pmptce-hnea: Predicting metabolic pathway types of chemicals and enzymes with a heterogeneous network embedding algorithm. Curr. Bioinform. 2023. [Google Scholar] [CrossRef]
- Li, X.; Lu, L.; Chen, L. Identification of protein functions in mouse with a label space partition method. Math. Biosci. Eng. 2022, 19, 3820–3842. [Google Scholar] [CrossRef]
- Powers, D. Evaluation: From precision, recall and F-measure to ROC., informedness, markedness & correlation. J. Mach. Learn. Technol. 2011, 2, 37–63. [Google Scholar]
- Tang, S.; Chen, L. Iatc-nfmlp: Identifying classes of anatomical therapeutic chemicals based on drug networks, fingerprints and multilayer perceptron. Curr. Bioinform. 2022, 17, 814–824. [Google Scholar]
- Wu, C.; Chen, L. A model with deep analysis on a large drug network for drug classification. Math. Biosci. Eng. 2023, 20, 383–401. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, L. Identification of drug–disease associations by using multiple drug and disease networks. Curr. Bioinform. 2022, 17, 48–59. [Google Scholar] [CrossRef]
- Gorodkin, J. Comparing two k-category assignments by a k-category correlation coefficient. Comput. Biol. Chem. 2004, 28, 367–374. [Google Scholar] [CrossRef]
- Kang, S.; Narazaki, M.; Metwally, H.; Kishimoto, T. Historical overview of the interleukin-6 family cytokine. J. Exp. Med. 2020, 217, e2019034. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Bost, P.; Giladi, A.; Liu, Y.; Bendjelal, Y.; Xu, G.; David, E.; Blecher-Gonen, R.; Cohen, M.; Medaglia, C.; Li, H. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell 2020, 181, 1475–1488.e12. [Google Scholar] [CrossRef]
- Daamen, A.R.; Bachali, P.; Owen, K.A.; Kingsmore, K.M.; Hubbard, E.L.; Labonte, A.C.; Robl, R.; Shrotri, S.; Grammer, A.C.; Lipsky, P.E. Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. Sci. Rep. 2021, 11, 7052. [Google Scholar] [CrossRef]
- Antica, M.; Scollay, R. Development of t lymphocytes at extrathymic sites. J. Immunol. 1999, 163, 206–211. [Google Scholar] [CrossRef]
- Pimm, M.L.; Liu, X.; Tuli, F.; Heritz, J.; Lojko, A.; Henty-Ridilla, J.L. Visualizing molecules of functional human profilin. eLife 2022, 11, e76485. [Google Scholar] [CrossRef]
- Mazucanti, C.H.; Egan, J.M. Sars-cov-2 disease severity and diabetes: Why the connection and what is to be done? Immun. Ageing 2020, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- DeLoach, A.; Cozart, M.; Kiaei, A.; Kiaei, M. A retrospective review of the progress in amyotrophic lateral sclerosis drug discovery over the last decade and a look at the latest strategies. Expert Opin. Drug Discov. 2015, 10, 1099–1118. [Google Scholar] [CrossRef]
- Cheriyath, V.; Kaur, J.; Davenport, A.; Khalel, A.; Chowdhury, N.; Gaddipati, L. G1P3 (IFI6), a mitochondrial localised antiapoptotic protein, promotes metastatic potential of breast cancer cells through mtROS. Br. J. Cancer 2018, 119, 52–64. [Google Scholar] [CrossRef]
- Sajid, M.; Ullah, H.; Yan, K.; He, M.; Feng, J.; Shereen, M.A.; Hao, R.; Li, Q.; Guo, D.; Chen, Y. The functional and antiviral activity of interferon alpha-inducible ifi6 against hepatitis b virus replication and gene expression. Front. Immunol. 2021, 12, 634937. [Google Scholar] [CrossRef]
- Kimura, T.; Kambe, T. The functions of metallothionein and zip and znt transporters: An overview and perspective. Int. J. Mol. Sci. 2016, 17, 336. [Google Scholar] [CrossRef]
- Gao, F.; Wang, H.; Li, X.; Guo, F.; Yuan, Y.; Wang, X.; Zhang, Y.; Bai, G. Alteration of the immune microenvironment in HBsAg and HBeAg dual-positive pregnant women presenting a high HBV viral load. J. Inflamm. Res. 2021, 14, 5619–5632. [Google Scholar] [CrossRef]
- Choi, Y.; Jeon, H.; Akin, J.W.; Curry Jr, T.E.; Jo, M. The FOS/AP-1 regulates metabolic changes and cholesterol synthesis in human periovulatory granulosa cells. Endocrinology 2021, 162, bqab127. [Google Scholar] [CrossRef] [PubMed]
- Jain, J.; McCaffrey, P.G.; Valge-Archer, V.E.; Rao, A. Nuclear factor of activated t cells contains FOS and jun. Nature 1992, 356, 801–804. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Gopanenko, A.V.; Malygin, A.A.; Karpova, G.G. The es26 protein is involved in the formation of a nucleophosmin binding site on the human 40s ribosomal subunit. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2018, 1866, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Kasela, S.; Kisand, K.; Tserel, L.; Kaleviste, E.; Remm, A.; Fischer, K.; Esko, T.; Westra, H.-J.; Fairfax, B.P.; Makino, S. Pathogenic implications for autoimmune mechanisms derived by comparative eQTL analysis of CD4+ versus CD8+ T cells. PLoS Genet. 2017, 13, e1006643. [Google Scholar] [CrossRef]
- Miller, R.; Phillips, R. Development of b lymphocytes. In Biology of Aging and Development; Springer: Berlin/Heidelberg, Germany, 1975; pp. 241–251. [Google Scholar]
- Kuri-Cervantes, L.; Pampena, M.B.; Meng, W.; Rosenfeld, A.M.; Ittner, C.A.; Weisman, A.R.; Agyekum, R.S.; Mathew, D.; Baxter, A.E.; Vella, L.A. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020, 5, eabd7114. [Google Scholar] [CrossRef] [PubMed]
- Fathi, N.; Rezaei, N. Lymphopenia in COVID-19: Therapeutic opportunities. Cell Biol. Int. 2020, 44, 1792–1797. [Google Scholar] [CrossRef]
- Ferretti, M.B.; Ghalei, H.; Ward, E.A.; Potts, E.L.; Karbstein, K. Rps26 directs mrna-specific translation by recognition of kozak sequence elements. Nat. Struct. Mol. Biol. 2017, 24, 700–707. [Google Scholar] [CrossRef]
- Vázquez-Jiménez, A.; Avila-Ponce De León, U.E.; Matadamas-Guzman, M.; Muciño-Olmos, E.A.; Martínez-López, Y.E.; Escobedo-Tapia, T.; Resendis-Antonio, O. On deep landscape exploration of COVID-19 patients cells and severity markers. Front. Immunol. 2021, 12, 705646. [Google Scholar] [CrossRef] [PubMed]
- Anzurez, A.; Naka, I.; Miki, S.; Nakayama-Hosoya, K.; Isshiki, M.; Watanabe, Y.; Nakamura-Hoshi, M.; Seki, S.; Matsumura, T.; Takano, T. Association of hla-drb1* 09: 01 with severe COVID-19. Hla 2021, 98, 37–42. [Google Scholar] [CrossRef]
- Ma, D.; Liu, S.; Hu, L.; He, Q.; Shi, W.; Yan, D.; Cao, Y.; Zhang, G.; Wang, Z.; Wu, J. Single-cell RNA sequencing identify SDCBP in ACE2-positive bronchial epithelial cells negatively correlates with COVID-19 severity. J. Cell. Mol. Med. 2021, 25, 7001–7012. [Google Scholar] [CrossRef]
- Pozzi, C.; Di Pisa, F.; Bernacchioni, C.; Ciambellotti, S.; Turano, P.; Mangani, S. Iron binding to human heavy-chain ferritin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1909–1920. [Google Scholar] [CrossRef]
- Michalski, J.E.; Kurche, J.S.; Schwartz, D.A. From ards to pulmonary fibrosis: The next phase of the COVID-19 pandemic? Transl. Res. 2022, 241, 13–24. [Google Scholar] [CrossRef]
- Xu, S.; Sun, Z.; Li, L.; Liu, J.; He, J.; Song, D.; Shan, G.; Liu, H.; Wu, X. Induction of t cells suppression by dendritic cells transfected with vsig4 recombinant adenovirus. Immunol. Lett. 2010, 128, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, L. Bioinformatics approach to identify the influences of sars-cov2 infections on atherosclerosis. Front. Cardiovasc. Med. 2022, 9, 907665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Mears, J.R.; Shakib, L.; Beynor, J.I.; Shanaj, S.; Korsunsky, I.; Nathan, A.; Donlin, L.T.; Raychaudhuri, S. IFN-γ and TNF-α drive a CXCL10+ CCL2+ macrophage phenotype expanded in severe COVID-19 lungs and inflammatory diseases with tissue inflammation. Genome Med. 2021, 13, 64. [Google Scholar] [CrossRef] [PubMed]

| Immune Cell Subtypes | COVID-19 Process | Sample Size (Cells/Patients/Sample) |
|---|---|---|
| B cells | Control | 44,788/25/28 |
| Convalescence | 227,795/95/140 | |
| Progression mild/moderate | 58,338/22/33 | |
| Progression severe/critical | 72,779/54/83 | |
| T cells | Control | 42,514/25/28 |
| Convalescence | 261,704/95/140 | |
| Progression mild/moderate | 85,287/22/33 | |
| Progression severe/critical | 245,090/54/83 | |
| Myeloid cells | Control | 21,910/25/28 |
| Convalescence | 115,311/95/140 | |
| Progression mild/moderate | 48,467/22/33 | |
| Progression severe/critical | 160,859/54/83 |
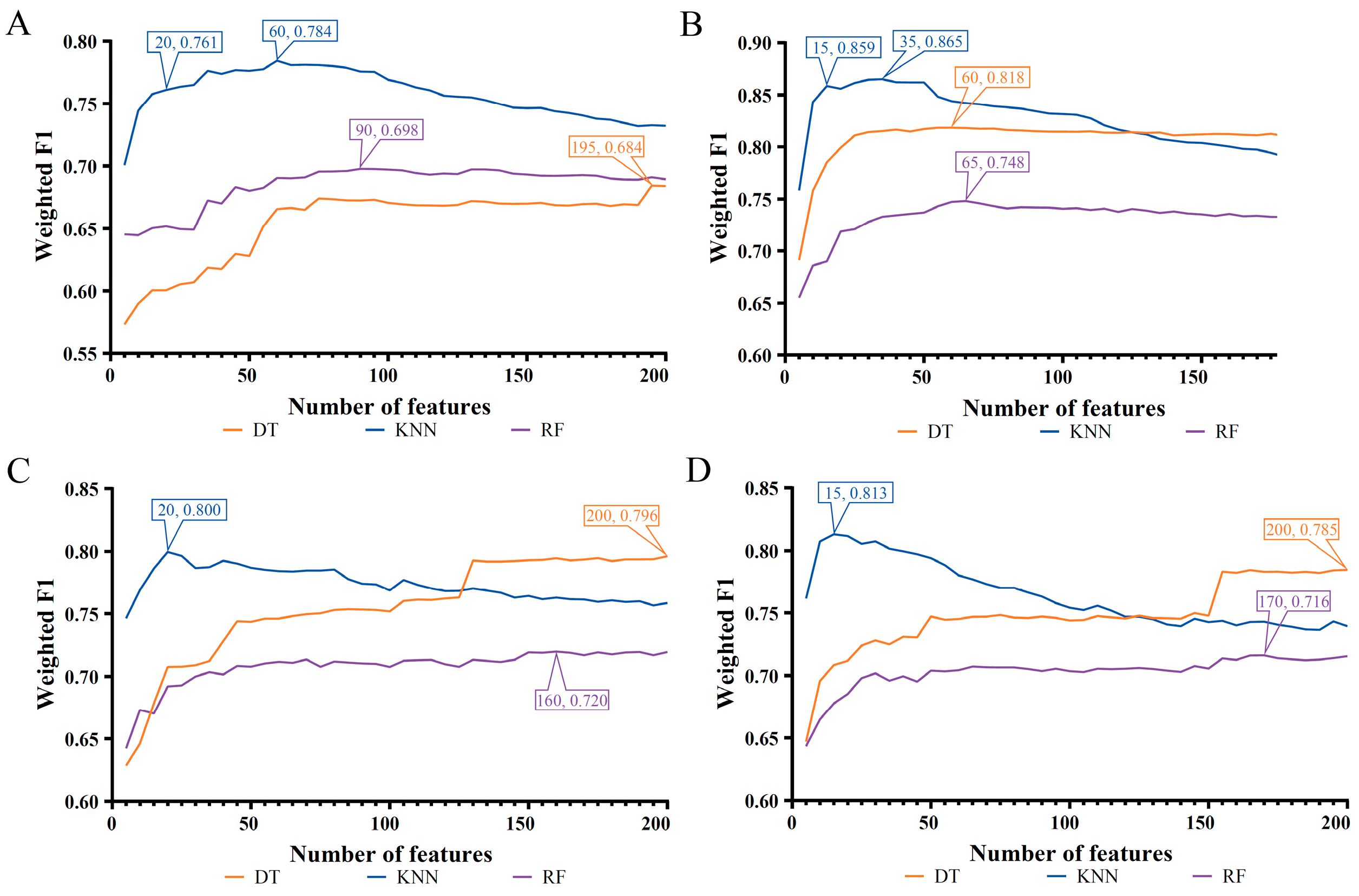
| Feature List | Classification Algorithm | Number of Features | ACC | MCC | Macro F1 | Weighted F1 |
|---|---|---|---|---|---|---|
| LASSO feature list | Decision tree | 195 | 0.676 | 0.513 | 0.638 | 0.684 |
| K-nearest neighbor | 60 | 0.784 | 0.721 | 0.786 | 0.784 | |
| Random forest | 90 | 0.694 | 0.527 | 0.667 | 0.698 | |
| LightGBM feature list | Decision tree | 60 | 0.815 | 0.714 | 0.788 | 0.818 |
| K-nearest neighbor | 35 | 0.863 | 0.810 | 0.859 | 0.865 | |
| Random forest | 65 | 0.745 | 0.645 | 0.746 | 0.748 | |
| MCFS feature list | Decision tree | 200 | 0.792 | 0.679 | 0.762 | 0.796 |
| K-nearest neighbor | 20 | 0.799 | 0.735 | 0.800 | 0.800 | |
| Random forest | 160 | 0.715 | 0.578 | 0.706 | 0.720 | |
| mRMR feature list | Decision tree | 200 | 0.780 | 0.661 | 0.748 | 0.785 |
| K-nearest neighbor | 15 | 0.812 | 0.750 | 0.813 | 0.813 | |
| Random forest | 170 | 0.712 | 0.562 | 0.698 | 0.716 |
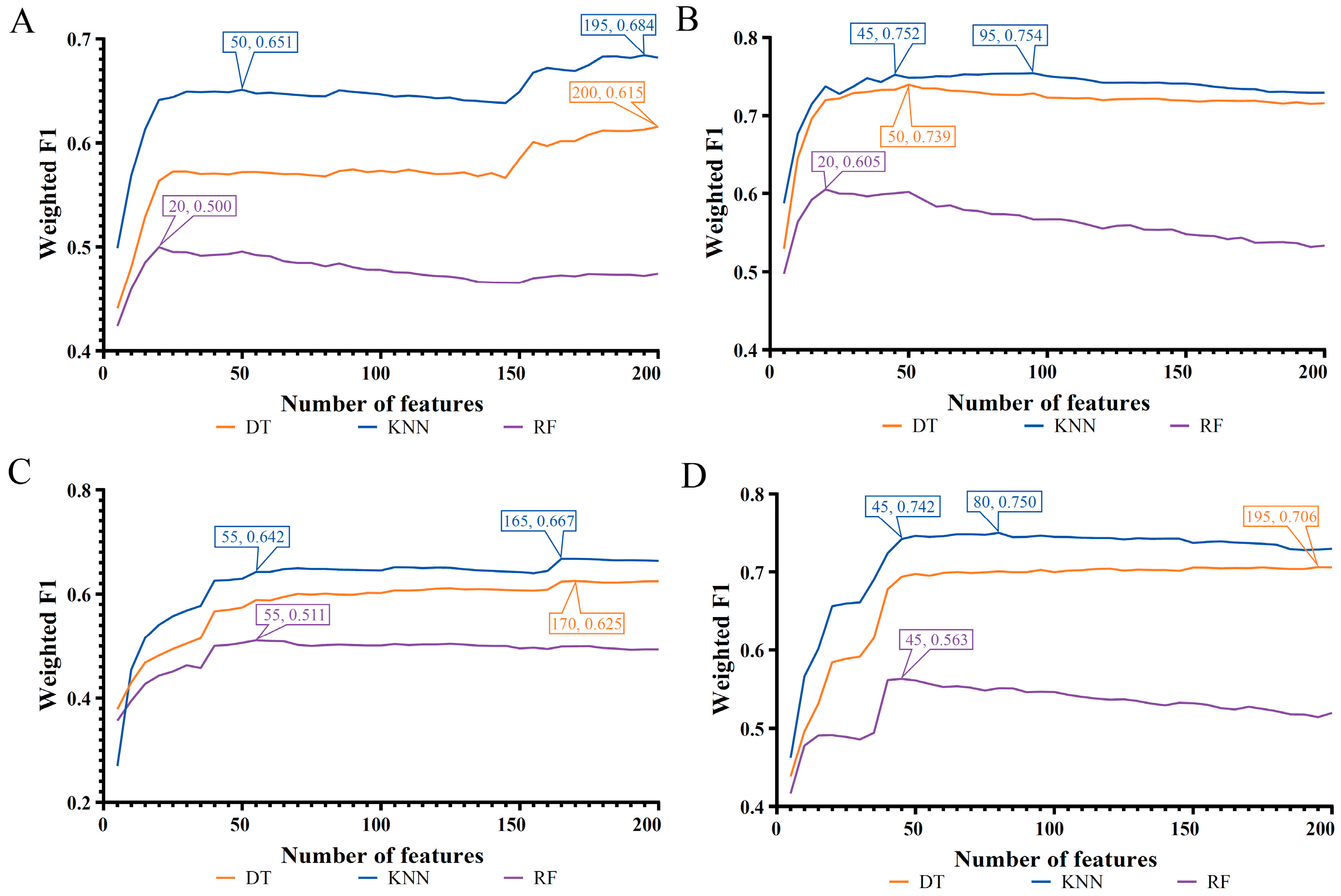
| Feature List | Classification Algorithm | Number of Features | ACC | MCC | Macro F1 | Weighted F1 |
|---|---|---|---|---|---|---|
| LASSO feature list | Decision tree | 200 | 0.611 | 0.428 | 0.582 | 0.615 |
| K-nearest neighbor | 195 | 0.677 | 0.573 | 0.666 | 0.684 | |
| Random forest | 20 | 0.499 | 0.275 | 0.496 | 0.500 | |
| LightGBM feature list | Decision tree | 50 | 0.737 | 0.610 | 0.716 | 0.739 |
| K-nearest neighbor | 95 | 0.751 | 0.655 | 0.744 | 0.754 | |
| Random forest | 20 | 0.608 | 0.435 | 0.620 | 0.605 | |
| MCFS feature list | Decision tree | 170 | 0.622 | 0.442 | 0.599 | 0.625 |
| K-nearest neighbor | 165 | 0.667 | 0.549 | 0.670 | 0.667 | |
| Random forest | 55 | 0.512 | 0.289 | 0.521 | 0.511 | |
| mRMR feature list | Decision tree | 195 | 0.703 | 0.560 | 0.677 | 0.706 |
| K-nearest neighbor | 80 | 0.748 | 0.648 | 0.747 | 0.750 | |
| Random forest | 45 | 0.565 | 0.363 | 0.583 | 0.563 |
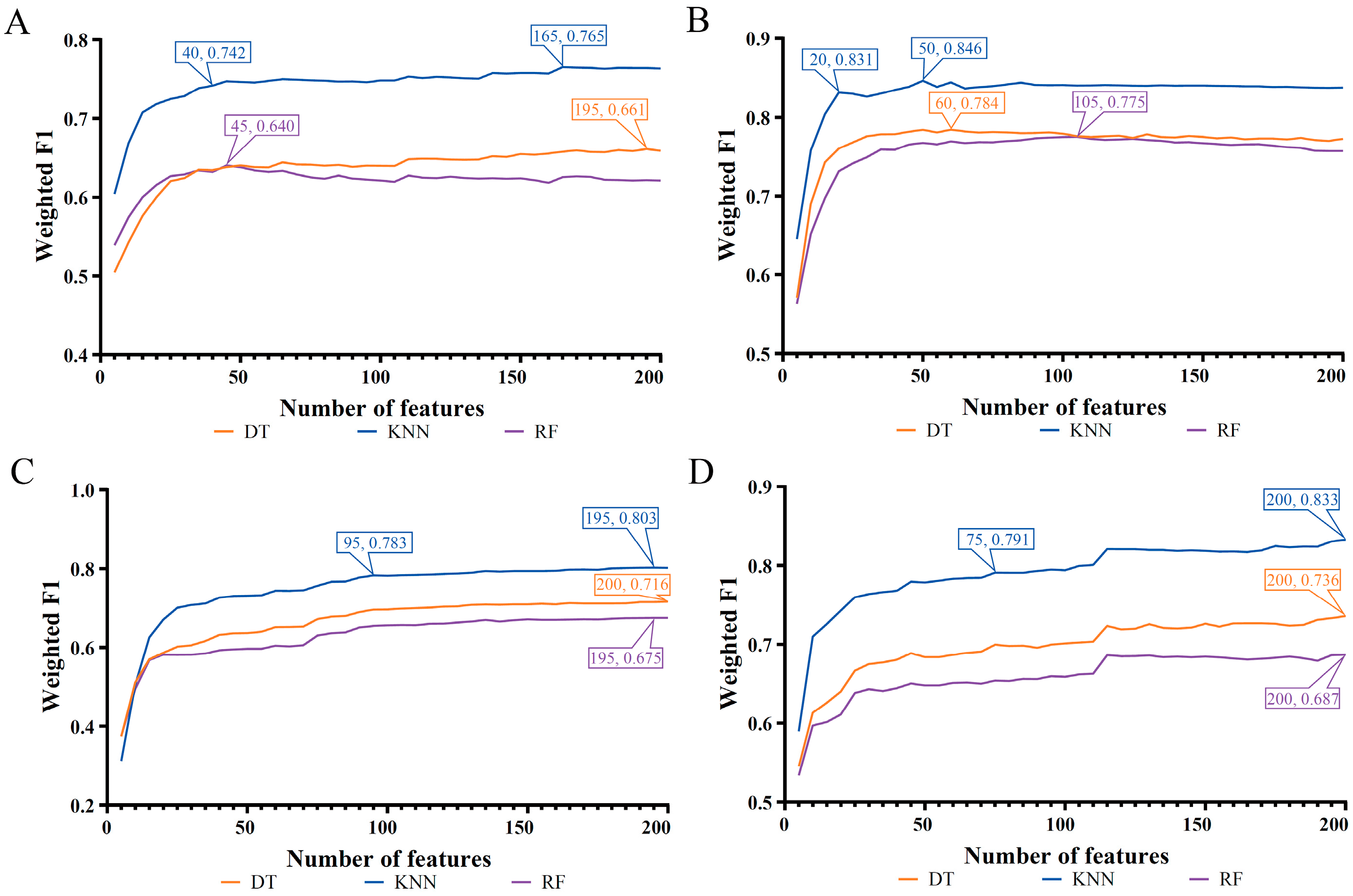
| Feature List | Classification Algorithm | Number of Features | ACC | MCC | Macro F1 | Weighted F1 |
|---|---|---|---|---|---|---|
| LASSO feature list | Decision tree | 195 | 0.655 | 0.491 | 0.610 | 0.661 |
| K-nearest neighbor | 165 | 0.761 | 0.679 | 0.746 | 0.765 | |
| Random forest | 45 | 0.637 | 0.463 | 0.618 | 0.640 | |
| LightGBM feature list | Decision tree | 60 | 0.782 | 0.673 | 0.751 | 0.784 |
| K-nearest neighbor | 50 | 0.844 | 0.778 | 0.829 | 0.846 | |
| Random forest | 105 | 0.775 | 0.669 | 0.776 | 0.775 | |
| MCFS feature list | Decision tree | 200 | 0.711 | 0.572 | 0.669 | 0.716 |
| K-nearest neighbor | 195 | 0.799 | 0.721 | 0.788 | 0.803 | |
| Random forest | 195 | 0.672 | 0.518 | 0.657 | 0.675 | |
| mRMR feature list | Decision tree | 200 | 0.731 | 0.601 | 0.692 | 0.736 |
| K-nearest neighbor | 200 | 0.828 | 0.762 | 0.812 | 0.833 | |
| Random forest | 200 | 0.685 | 0.532 | 0.669 | 0.687 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, J.-X.; Gao, Q.; Zhou, X.-C.; Chen, L.; Guo, W.; Feng, K.-Y.; Lu, L.; Huang, T.; Cai, Y.-D. Identification of Gene Markers Associated with COVID-19 Severity and Recovery in Different Immune Cell Subtypes. Biology 2023, 12, 947. https://doi.org/10.3390/biology12070947
Ren J-X, Gao Q, Zhou X-C, Chen L, Guo W, Feng K-Y, Lu L, Huang T, Cai Y-D. Identification of Gene Markers Associated with COVID-19 Severity and Recovery in Different Immune Cell Subtypes. Biology. 2023; 12(7):947. https://doi.org/10.3390/biology12070947
Chicago/Turabian StyleRen, Jing-Xin, Qian Gao, Xiao-Chao Zhou, Lei Chen, Wei Guo, Kai-Yan Feng, Lin Lu, Tao Huang, and Yu-Dong Cai. 2023. "Identification of Gene Markers Associated with COVID-19 Severity and Recovery in Different Immune Cell Subtypes" Biology 12, no. 7: 947. https://doi.org/10.3390/biology12070947
APA StyleRen, J.-X., Gao, Q., Zhou, X.-C., Chen, L., Guo, W., Feng, K.-Y., Lu, L., Huang, T., & Cai, Y.-D. (2023). Identification of Gene Markers Associated with COVID-19 Severity and Recovery in Different Immune Cell Subtypes. Biology, 12(7), 947. https://doi.org/10.3390/biology12070947









