Nitric Oxide in Plant Functioning: Metabolism, Signaling, and Responses to Infestation with Ecdysozoa Parasites
Abstract
Simple Summary
Abstract
1. Introduction
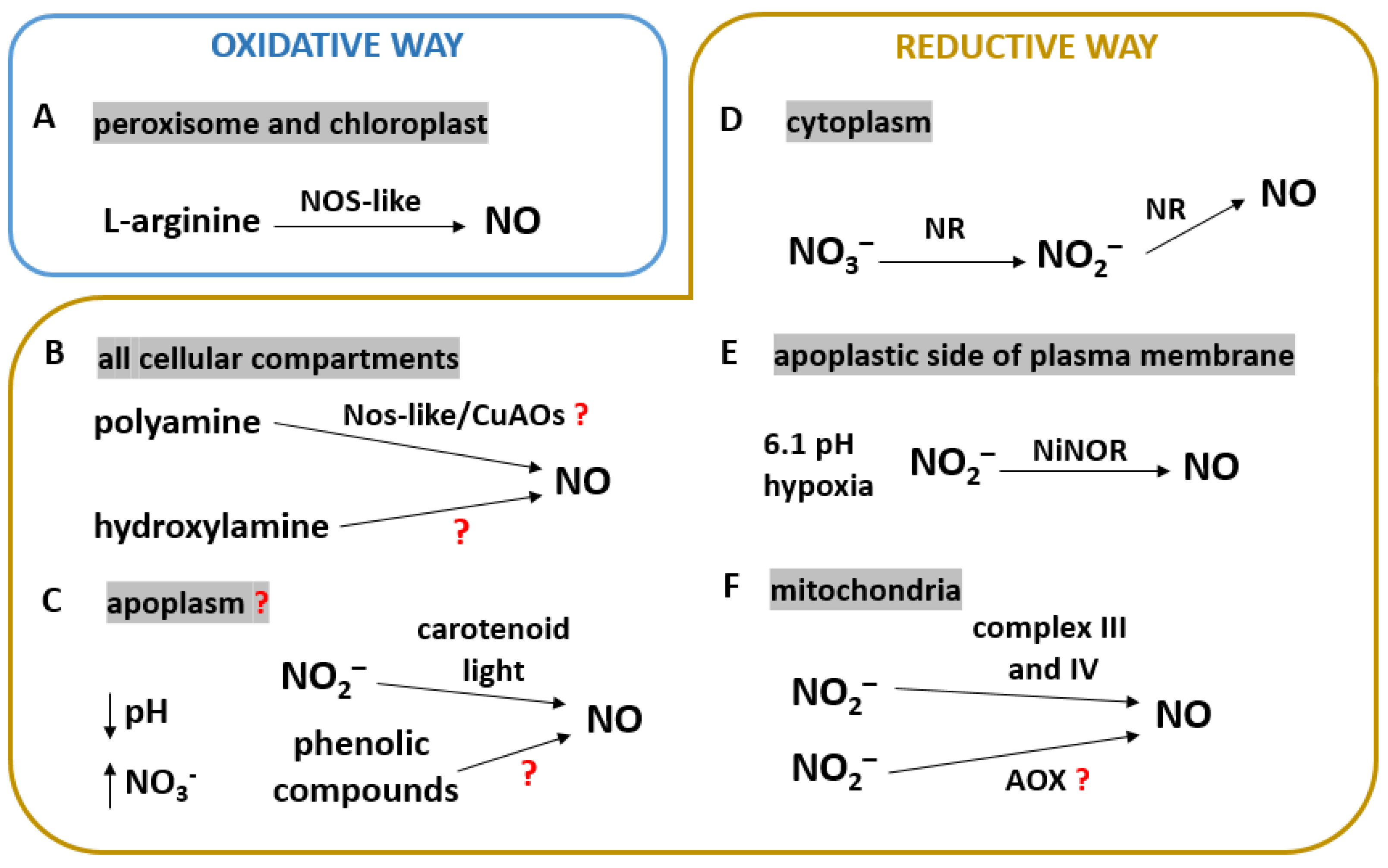
2. Biosynthesis of NO in Plants
2.1. NOS-like Activity
2.2. NO Synthesis via Polyamine and Hydroxylamine Pathways
2.3. Non-Enzymatic Production of NO
2.4. Role of NR in NO Synthesis
2.5. A Plasma Membrane-Bound Nitrite Reductase
2.6. NO Synthesis in Mitochondria
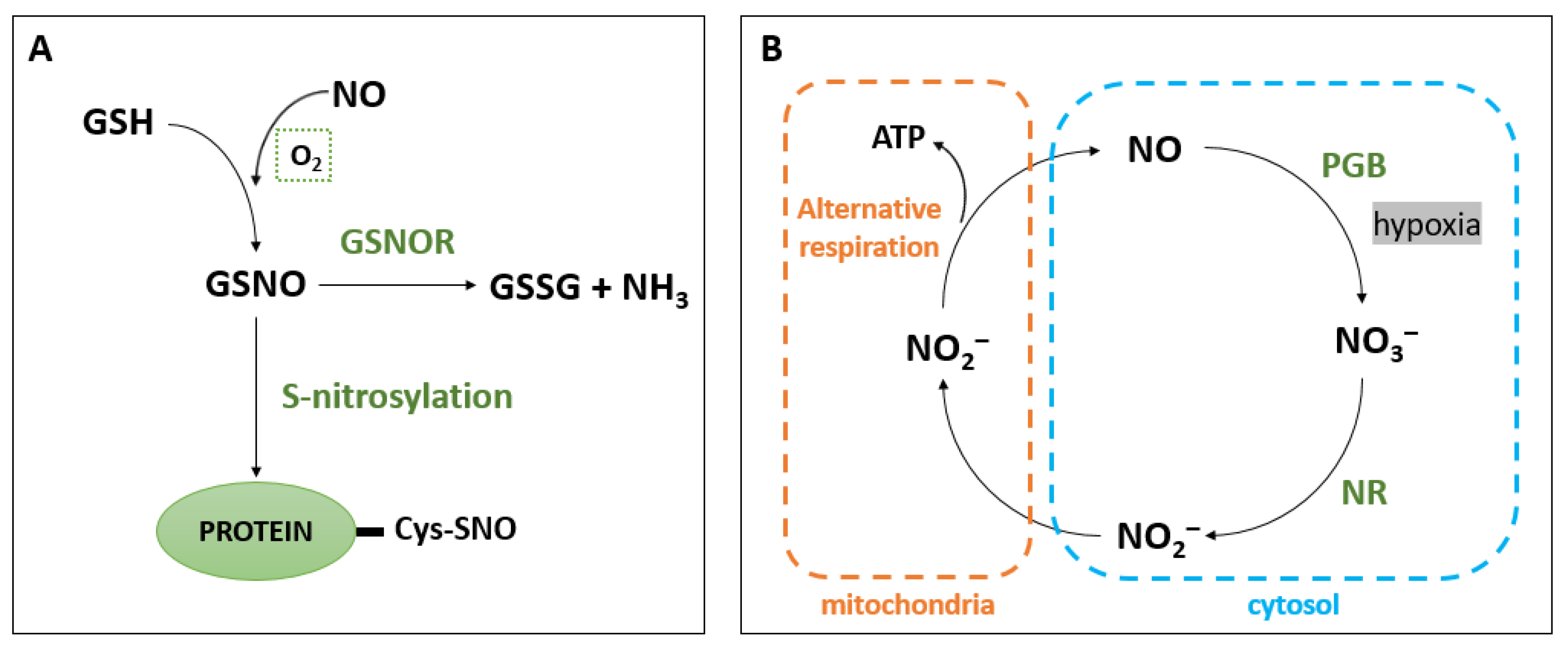
3. NO Scavenging
4. The Role of NO in PTMs
4.1. S-Nitrosylation
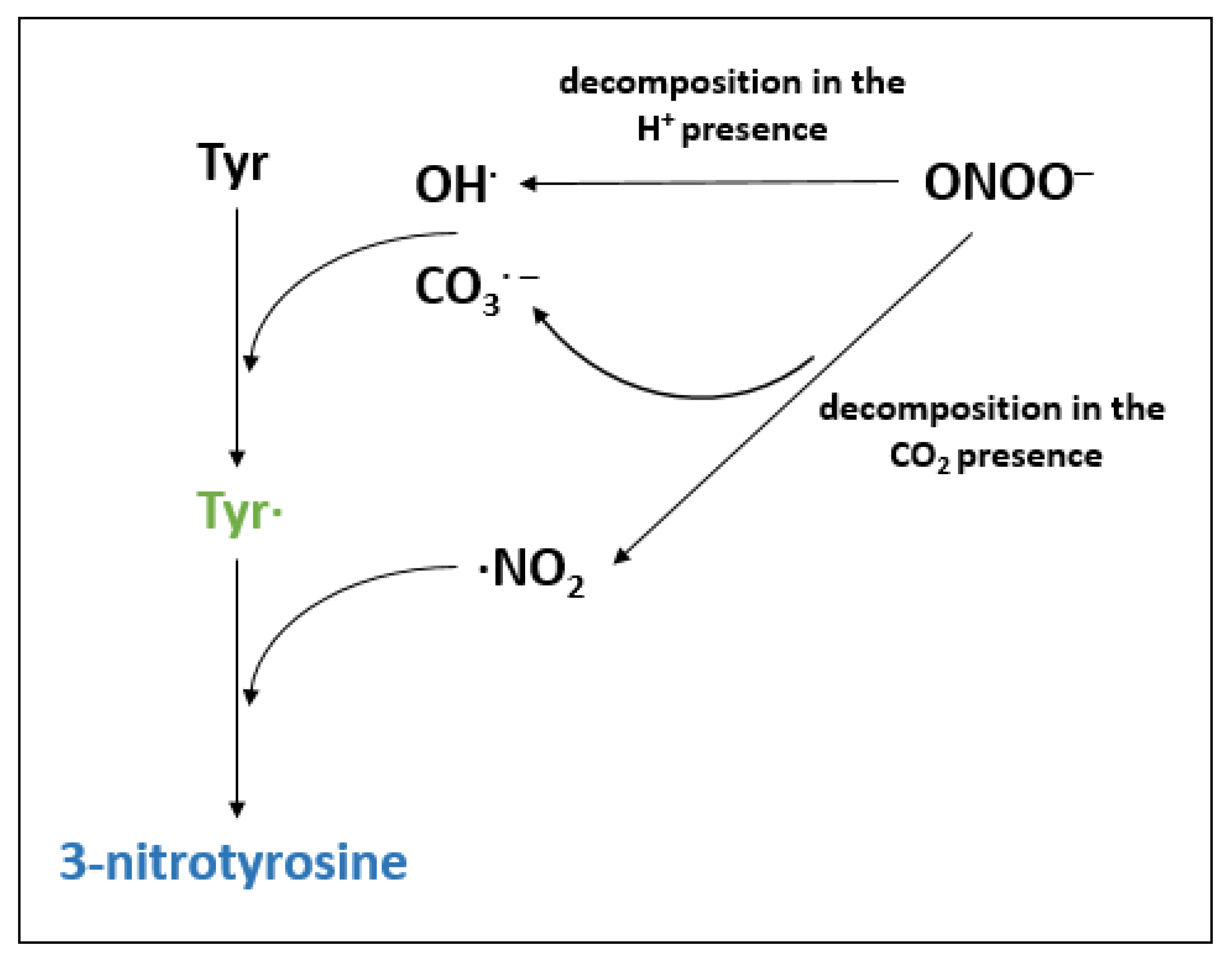
4.2. Protein Nitration
4.3. Phosphorylation
5. Crosstalk between NO, ROS, and Phytohormones
6. In Search of NO-Dependent Defense Mechanisms during Infection with Herbivorous Ecdysozoa Species
6.1. Nematodes
6.2. Insects
6.3. Arachnids
6.4. Are There Common Patterns of NO-Dependent Defensive Responses against Ecdysozoa Species Infestation?
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kolbert, Z.; Barroso, J.B.; Brouquisse, R.; Corpas, F.J.; Gupta, K.J.; Lindermayr, C.; Loake, G.J.; Palma, J.M.; Petřivalský, M.; Wendehenne, D.; et al. A Forty Year Journey: The Generation and Roles of NO in Plants. Nitric Oxide 2019, 93, 53–70. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Tiwari, S.; Singh, V.P.; Prasad, S.M. Nitric Oxide in Plants: An Ancient Molecule with New Tasks. Plant Growth Regul. 2020, 90, 1–13. [Google Scholar] [CrossRef]
- Beligni, M.V.; Lamattina, L. Nitric Oxide in Plants: The History Is Just Beginning: Nitric Oxide in Plants. Plant Cell Environ. 2001, 24, 267–278. [Google Scholar] [CrossRef]
- Sami, F.; Faizan, M.; Faraz, A.; Siddiqui, H.; Yusuf, M.; Hayat, S. Nitric Oxide-Mediated Integrative Alterations in Plant Metabolism to Confer Abiotic Stress Tolerance, NO Crosstalk with Phytohormones and NO-Mediated Post Translational Modifications in Modulating Diverse Plant Stress. Nitric Oxide 2018, 73, 22–38. [Google Scholar] [CrossRef]
- Corpas, F.J. Nitric Oxide and Hydrogen Sulfide in Higher Plants under Physiological and Stress Conditions. Antioxidants 2019, 8, 457. [Google Scholar] [CrossRef]
- Astier, J.; Gross, I.; Durner, J. Nitric Oxide Production in Plants: An Update. J. Exp. Bot. 2018, 69, 3401–3411. [Google Scholar] [CrossRef]
- Ciacka, K.; Staszek, P.; Sobczynska, K.; Krasuska, U.; Gniazdowska, A. Nitric Oxide in Seed Biology. Int. J. Mol. Sci. 2022, 23, 14951. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Chaki, M.; Valderrama, R.; Sánchez-Calvo, B.; Mata-Pérez, C.; Padilla, M.N.; Corpas, F.J.; Barroso, J.B. Nitric Oxide Buffering and Conditional Nitric Oxide Release in Stress Response. J. Exp. Bot. 2018, 69, 3425–3438. [Google Scholar] [CrossRef]
- Asgher, M.; Per, T.S.; Masood, A.; Fatma, M.; Freschi, L.; Corpas, F.J.; Khan, N.A. Nitric Oxide Signaling and Its Crosstalk with Other Plant Growth Regulators in Plant Responses to Abiotic Stress. Environ. Sci. Pollut. Res. Int. 2017, 24, 2273–2285. [Google Scholar] [CrossRef]
- Nykiel, M.; Gietler, M.; Fidler, J.; Prabucka, B.; Rybarczyk-Płońska, A.; Graska, J.; Boguszewska-Mańkowska, D.; Muszyńska, E.; Morkunas, I.; Labudda, M. Signal Transduction in Cereal Plants Struggling with Environmental Stresses: From Perception to Response. Plants 2022, 11, 1009. [Google Scholar] [CrossRef]
- Procházková, D.; Wilhelmová, N. Nitric Oxide, Reactive Nitrogen Species and Associated Enzymes during Plant Senescence. Nitric Oxide 2011, 24, 61–65. [Google Scholar] [CrossRef]
- Bruand, C.; Meilhoc, E. Nitric Oxide in Plants: Pro- or Anti-Senescence. J. Exp. Bot. 2019, 70, 4419–4427. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, Y.; Liu, L.; Liu, X.; Li, B.; Jin, C.; Lin, X. Molecular Functions of Nitric Oxide and Its Potential Applications in Horticultural Crops. Hortic Res. 2021, 8, 71. [Google Scholar] [CrossRef]
- Hendricks, S.B.; Taylorson, R.B. Promotion of Seed Germination by Nitrate, Nitrite, Hydroxylamine, and Ammonium Salts 1. Plant Physiol 1974, 54, 304–309. [Google Scholar] [CrossRef]
- Hayat, S.; Yadav, S.; Wani, D.A.; Irfan, M.; Ahmad, A. Nitric Oxide Effects on Photosynthetic Rate, Growth, and Antioxidant Activity in Tomato. Int. J. Veg. Sci. 2011, 17, 333–348. [Google Scholar] [CrossRef]
- Wojciechowska, N.; Sobieszczuk-Nowicka, E.; Bagniewska-Zadworna, A. Plant Organ Senescence–Regulation by Manifold Pathways. Plant Biol. 2018, 20, 167–181. [Google Scholar] [CrossRef]
- Zhang, Y.-M.; Guo, P.; Xia, X.; Guo, H.; Li, Z. Multiple Layers of Regulation on Leaf Senescence: New Advances and Perspectives. Front Plant Sci. 2021, 12, 788996. [Google Scholar] [CrossRef]
- Zhou, X.; Joshi, S.; Khare, T.; Patil, S.; Shang, J.; Kumar, V. Nitric Oxide, Crosstalk with Stress Regulators and Plant Abiotic Stress Tolerance. Plant Cell Rep. 2021, 40, 1395–1414. [Google Scholar] [CrossRef]
- Cui, M.; Ok, S.; Yoo, K.; Jung, K.; Yoo, S.; Shin, J. An Arabidopsis Cell Growth Defect Factor-Related Protein, CRS, Promotes Plant Senescence by Increasing the Production of Hydrogen Peroxide. Plant Cell Physiol. 2013, 54, 155–167. [Google Scholar] [CrossRef]
- Dvořák, P.; Krasylenko, Y.; Zeiner, A.; Šamaj, J.; Takáč, T. Signaling Toward Reactive Oxygen Species-Scavenging Enzymes in Plants. Front Plant Sci. 2020, 11, 618835. [Google Scholar] [CrossRef]
- Del Castello, F.; Nejamkin, A.; Cassia, R.; Correa-Aragunde, N.; Fernández, B.; Foresi, N.; Lombardo, C.; Ramirez, L.; Lamattina, L. The Era of Nitric Oxide in Plant Biology: Twenty Years Tying up Loose Ends. Nitric Oxide 2019, 85, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Pande, A.; Mun, B.-G.; Khan, M.; Rahim, W.; Lee, D.-S.; Lee, G.-M.; Al Azawi, T.N.I.; Hussain, A.; Yun, B.-W. Nitric Oxide Signaling and Its Association with Ubiquitin-Mediated Proteasomal Degradation in Plants. Int. J. Mol. Sci. 2022, 23, 1657. [Google Scholar] [CrossRef] [PubMed]
- Gietler, M.; Nykiel, M.; Orzechowski, S.; Fettke, J.; Zagdańska, B. Proteomic Analysis of S-Nitrosylated and S-Glutathionylated Proteins in Wheat Seedlings with Different Dehydration Tolerances. Plant Physiol. Biochem. 2016, 108, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Gietler, M.; Nykiel, M.; Orzechowski, S.; Fettke, J.; Zagdańska, B. Protein Carbonylation Linked to Wheat Seedling Tolerance to Water Deficiency. Environ. Exp. Bot. 2017, 137, 84–95. [Google Scholar] [CrossRef]
- Kudoyarova, G. Phytohormones 2020. Biomolecules 2022, 12, 1305. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galvan, A.; Fernandez, E. Nitrate Reductase Regulates Plant Nitric Oxide Homeostasis. Trends Plant Sci. 2017, 22, 163–174. [Google Scholar] [CrossRef]
- Gupta, K.J.; Fernie, A.R.; Kaiser, W.M.; van Dongen, J.T. On the Origins of Nitric Oxide. Trends Plant Sci. 2011, 16, 160–168. [Google Scholar] [CrossRef]
- Gupta, K.J.; Kaladhar, V.C.; Fitzpatrick, T.B.; Fernie, A.R.; Møller, I.M.; Loake, G.J. Nitric Oxide Regulation of Plant Metabolism. Mol. Plant 2022, 15, 228–242. [Google Scholar] [CrossRef]
- Bredt, D.S.; Snyder, S.H. Isolation of Nitric Oxide Synthetase, a Calmodulin-Requiring Enzyme. Proc. Natl. Acad. Sci. USA 1990, 87, 682–685. [Google Scholar] [CrossRef]
- Stuehr, D.J.; Santolini, J.; Wang, Z.-Q.; Wei, C.-C.; Adak, S. Update on Mechanism and Catalytic Regulation in the NO Synthases. J. Biol. Chem. 2004, 279, 36167–36170. [Google Scholar] [CrossRef]
- Alderton, W.K.; Cooper, C.E.; Knowles, R.G. Nitric Oxide Synthases: Structure, Function and Inhibition. Biochem. J. 2001, 357, 593–615. [Google Scholar] [CrossRef]
- Salgado, I.; Oliveira, H.C.; Gaspar, M. Plant Nitric Oxide Signaling Under Environmental Stresses. In Mechanism of Plant Hormone Signaling under Stress; Pandey, G.K., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 345–370. ISBN 978-1-118-88902-2. [Google Scholar]
- Förstermann, U.; Sessa, W.C. Nitric Oxide Synthases: Regulation and Function. Eur. Heart J. 2012, 33, 829–837. [Google Scholar] [CrossRef]
- Durner, J.; Wendehenne, D.; Klessig, D.F. Defense Gene Induction in Tobacco by Nitric Oxide, Cyclic GMP, and Cyclic ADP-Ribose. Proc. Natl. Acad. Sci. USA 1998, 95, 10328–10333. [Google Scholar] [CrossRef]
- Delledonne, M.; Xia, Y.; Dixon, R.A.; Lamb, C. Nitric Oxide Functions as a Signal in Plant Disease Resistance. Nature 1998, 394, 585–588. [Google Scholar] [CrossRef]
- Guo, F.; Okamoto, M.; Crawford, N. Identification of a Plant Nitric Oxide Synthase Gene Involved in Hormonal Signaling. Science 2003, 302, 103. [Google Scholar] [CrossRef]
- Zemojtel, T.; Fröhlich, A.; Palmieri, M.C.; Kolanczyk, M.; Mikula, I.; Wyrwicz, L.S.; Wanker, E.E.; Mundlos, S.; Vingron, M.; Martasek, P.; et al. Plant Nitric Oxide Synthase: A Never-Ending Story? Trends Plant Sci. 2006, 11, 524–525; author reply 526–528. [Google Scholar] [CrossRef]
- Moreau, M.; Lee, G.I.; Wang, Y.; Crane, B.R.; Klessig, D.F. AtNOS/AtNOA1 Is a Functional Arabidopsis Thaliana CGTPase and Not a Nitric-Oxide Synthase. J. Biol. Chem. 2008, 283, 32957–32967. [Google Scholar] [CrossRef]
- Liu, H.; Lau, E.; Lam, M.P.Y.; Chu, H.; Li, S.; Huang, G.; Guo, P.; Wang, J.; Jiang, L.; Chu, I.K.; et al. OsNOA1/RIF1 Is a Functional Homolog of AtNOA1/RIF1: Implication for a Highly Conserved Plant CGTPase Essential for Chloroplast Function. New Phytol. 2010, 187, 83–105. [Google Scholar] [CrossRef]
- Jeandroz, S.; Wipf, D.; Stuehr, D.J.; Lamattina, L.; Melkonian, M.; Tian, Z.; Zhu, Y.; Carpenter, E.J.; Wong, G.K.-S.; Wendehenne, D. Occurrence, Structure, and Evolution of Nitric Oxide Synthase-like Proteins in the Plant Kingdom. Sci. Signal 2016, 9, re2. [Google Scholar] [CrossRef]
- Choudhary, S.; Wani, K.I.; Naeem, M.; Khan, M.M.A.; Aftab, T. Cellular Responses, Osmotic Adjustments, and Role of Osmolytes in Providing Salt Stress Resilience in Higher Plants: Polyamines and Nitric Oxide Crosstalk. J. Plant Growth Regul. 2022. [Google Scholar] [CrossRef]
- Kumar, N.; Gautam, A.; Dubey, A.K. 15–Polyamines Metabolism and NO Signaling in Plants. In Nitric Oxide in Plant Biology; Pratap Singh, V., Singh, S., Tripathi, D.K., Romero-Puertas, M.C., Sandalio, L.M., Eds.; Academic Press: Cambridge, MS, USA, 2022; pp. 345–372. ISBN 978-0-12-818797-5. [Google Scholar]
- Krasuska, U.; Ciacka, K.; Gniazdowska, A. Nitric Oxide-Polyamines Cross-Talk during Dormancy Release and Germination of Apple Embryos. Nitric Oxide 2017, 68, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Recalde, L.; Gómez Mansur, N.M.; Cabrera, A.V.; Matayoshi, C.L.; Gallego, S.M.; Groppa, M.D.; Benavides, M.P. Unravelling Ties in the Nitrogen Network: Polyamines and Nitric Oxide Emerging as Essential Players in Signalling Roadway. Ann. Appl. Biol. 2021, 178, 192–208. [Google Scholar] [CrossRef]
- Tun, N.N.; Santa-Catarina, C.; Begum, T.; Silveira, V.; Handro, W.; Floh, E.I.S.; Scherer, G.F.E. Polyamines Induce Rapid Biosynthesis of Nitric Oxide (NO) in Arabidopsis Thaliana Seedlings. Plant Cell Physiol. 2006, 47, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Liu, Z.; Zhu, L.; Ma, Z.; Wang, J.; Zhu, J. Exogenous Melatonin Improves Plant Iron Deficiency Tolerance via Increased Accumulation of Polyamine-Mediated Nitric Oxide. Int. J. Mol. Sci. 2016, 17, 1777. [Google Scholar] [CrossRef]
- Siddiqui, M.H.; Alamri, S.A.; Al-Khaishany, M.Y.; Al-Qutami, M.A.; Ali, H.M.; AL-Rabiah, H.; Kalaji, H.M. Exogenous Application of Nitric Oxide and Spermidine Reduces the Negative Effects of Salt Stress on Tomato. Hortic. Environ. Biotechnol. 2017, 58, 537–547. [Google Scholar] [CrossRef]
- Cai, W.; Liu, W.; Wang, W.-S.; Fu, Z.-W.; Han, T.-T.; Lu, Y.-T. Overexpression of Rat Neurons Nitric Oxide Synthase in Rice Enhances Drought and Salt Tolerance. PLoS ONE 2015, 10, e0131599. [Google Scholar] [CrossRef]
- Rai, K.K.; Pandey, N.; Rai, N.; Rai, S.K.; Pandey-Rai, S. Salicylic Acid and Nitric Oxide: Insight Into the Transcriptional Regulation of Their Metabolism and Regulatory Functions in Plants. Front. Agron. 2021, 3, 781027. [Google Scholar] [CrossRef]
- Tiwari, R.K.; Kumar, R.; Lal, M.K.; Kumar, A.; Altaf, M.A.; Devi, R.; Mangal, V.; Naz, S.; Altaf, M.M.; Dey, A.; et al. Melatonin-Polyamine Interplay in the Regulation of Stress Responses in Plants. J. Plant Growth Regul. 2022. [Google Scholar] [CrossRef]
- Nandy, S.; Mandal, S.; Gupta, S.K.; Anand, U.; Ghorai, M.; Mundhra, A.; Rahman, M.H.; Ray, P.; Mitra, S.; Ray, D.; et al. Role of Polyamines in Molecular Regulation and Cross-Talks Against Drought Tolerance in Plants. J. Plant Growth Regul. 2022. [Google Scholar] [CrossRef]
- Wimalasekera, R.; Villar, C.; Begum, T.; Scherer, G.F.E. COPPER AMINE OXIDASE1 (CuAO1) of Arabidopsis Thaliana Contributes to Abscisic Acid- and Polyamine-Induced Nitric Oxide Biosynthesis and Abscisic Acid Signal Transduction. Mol. Plant 2011, 4, 663–678. [Google Scholar] [CrossRef]
- Groß, F.; Rudolf, E.-E.; Thiele, B.; Durner, J.; Astier, J. Copper Amine Oxidase 8 Regulates Arginine-Dependent Nitric Oxide Production in Arabidopsis Thaliana. J. Exp. Bot. 2017, 68, 2149–2162. [Google Scholar] [CrossRef]
- Rümer, S.; Gupta, K.; Kaiser, W. Plant Cells Oxidize Hydroxylamines to NO. J. Exp. Bot. 2009, 60, 2065–2072. [Google Scholar] [CrossRef]
- Bethke, P.C.; Badger, M.R.; Jones, R.L. Apoplastic Synthesis of Nitric Oxide by Plant Tissues. Plant Cell 2004, 16, 332–341. [Google Scholar] [CrossRef]
- Treffon, P.; Vierling, E. Focus on Nitric Oxide Homeostasis: Direct and Indirect Enzymatic Regulation of Protein Denitrosation Reactions in Plants. Antioxidants 2022, 11, 1411. [Google Scholar] [CrossRef]
- Cooney, R.V.; Harwood, P.J.; Custer, L.J.; Franke, A.A. Light-Mediated Conversion of Nitrogen Dioxide to Nitric Oxide by Carotenoids. Env. Health Perspect. 1994, 102, 460–462. [Google Scholar] [CrossRef]
- Yamasaki, H.; Sakihama, Y. Simultaneous Production of Nitric Oxide and Peroxynitrite by Plant Nitrate Reductase: In Vitro Evidence for the NR-Dependent Formation of Active Nitrogen Species. FEBS Lett. 2000, 468, 89–92. [Google Scholar] [CrossRef]
- Campbell, W.H. Structure and Function of Eukaryotic NAD(P)H:Nitrate Reductase. Cell Mol. Life Sci. 2001, 58, 194–204. [Google Scholar] [CrossRef]
- Allagulova, C.R.; Avalbaev, A.M.; Lubyanova, A.R.; Lastochkina, O.V.; Shakirova, F.M. Current Concepts of the Mechanisms of Nitric Oxide Formation in Plants. Russ. J. Plant Physiol. 2022, 69, 61. [Google Scholar] [CrossRef]
- Rai, K.K. Revisiting the Critical Role of ROS and RNS in Plant Defense. J. Plant Growth Regul. 2022. [Google Scholar] [CrossRef]
- Labudda, M.; Różańska, E.; Muszyńska, E.; Marecka, D.; Głowienka, M.; Roliński, P.; Prabucka, B. Heterodera schachtii Infection Affects Nitrogen Metabolism in Arabidopsis Thaliana. Plant Pathol. 2020, 69, 794–803. [Google Scholar] [CrossRef]
- Zhao, C.; Cai, S.; Wang, Y.; Chen, Z.-H. Loss of Nitrate Reductases NIA1 and NIA2 Impairs Stomatal Closure by Altering Genes of Core ABA Signaling Components in Arabidopsis. Plant Signal. Behav. 2016, 11, e1183088. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Zhao, S.; Dong, H.; Zhang, H.; Sun, L.; Miao, C. Nia1 and Nia2 Are Involved in Exogenous Salicylic Acid-Induced Nitric Oxide Generation and Stomatal Closure in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Olas, J.J.; Wahl, V. Tissue-Specific NIA1 and NIA2 Expression in Arabidopsis thaliana. Plant Signal. Behav. 2019, 14, 1656035. [Google Scholar] [CrossRef] [PubMed]
- Kolbert, Z.; Bartha, B.; Erdei, L. Exogenous Auxin-Induced NO Synthesis Is Nitrate Reductase-Associated in Arabidopsis Thaliana Root Primordia. J. Plant Physiol. 2008, 165, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.-F.; Zhang, Z.-W.; Yang, X.-Y.; Wang, C.-Q.; Lan, T.; Tang, X.-Y.; Chen, G.-D.; Zeng, J.; Yuan, S. Nitrate Reductase Is a Key Enzyme Responsible for Nitrogen-Regulated Auxin Accumulation in Arabidopsis Roots. Biochem. Biophys. Res. Commun. 2020, 532, 633–639. [Google Scholar] [CrossRef]
- Tang, X.; Peng, Y.; Li, Z.; Guo, H.; Xia, X.; Li, B.; Yin, W. The Regulation of Nitrate Reductases in Response to Abiotic Stress in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 1202. [Google Scholar] [CrossRef]
- Costa-Broseta, Á.; Perea-Resa, C.; Castillo, M.-C.; Ruíz, M.F.; Salinas, J.; León, J. Nitric Oxide Controls Constitutive Freezing Tolerance in Arabidopsis by Attenuating the Levels of Osmoprotectants, Stress-Related Hormones and Anthocyanins. Sci. Rep. 2018, 8, 9268. [Google Scholar] [CrossRef]
- Tewari, R.K.; Horemans, N.; Nauts, R.; Wannijn, J.; Van Hees, M.; Vandenhove, H. The Nitric Oxide Suppressed Arabidopsis Mutants- Atnoa1 and Atnia1nia2noa1-2 Produce Nitric Oxide in MS Growth Medium and on Uranium Exposure. Plant Physiol. Biochem. 2019, 140, 9–17. [Google Scholar] [CrossRef]
- Tejada-Jimenez, M.; Llamas, A.; Galván, A.; Fernández, E. Role of Nitrate Reductase in NO Production in Photosynthetic Eukaryotes. Plants 2019, 8, 56. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, Á.; Ocaña-Calahorro, F.; Mariscal, V.; Carreras, A.; Barroso, J.B.; Galván, A.; Fernández, E. A Dual System Formed by the ARC and NR Molybdoenzymes Mediates Nitrite-Dependent NO Production in Chlamydomonas. Plant Cell Environ. 2016, 39, 2097–2107. [Google Scholar] [CrossRef]
- Sanz-Luque, E.; Ocaña-Calahorro, F.; de Montaigu, A.; Chamizo-Ampudia, A.; Llamas, Á.; Galván, A.; Fernández, E. THB1, a Truncated Hemoglobin, Modulates Nitric Oxide Levels and Nitrate Reductase Activity. Plant J. 2015, 81, 467–479. [Google Scholar] [CrossRef]
- Minaeva, E.; Zalutskaya, Z.; Filina, V.; Ermilova, E. Truncated Hemoglobin 1 Is a New Player in Chlamydomonas Reinhardtii Acclimation to Sulfur Deprivation. PLoS ONE 2017, 12, e0186851. [Google Scholar] [CrossRef]
- Stöhr, C.; Strube, F.; Marx, G.; Ullrich, W.R.; Rockel, P. A Plasma Membrane-Bound Enzyme of Tobacco Roots Catalyses the Formation of Nitric Oxide from Nitrite. Planta 2001, 212, 835–841. [Google Scholar] [CrossRef]
- Meyer, C.; Stöhr, C. Soluble and Plasma Membrane-Bound Enzymes Involved in Nitrate and Nitrite Metabolism. In Photosynthetic Nitrogen Assimilation and Associated Carbon and Respiratory Metabolism; Foyer, C.H., Noctor, G., Eds.; Advances in Photosynthesis and Respiration; Springer: Netherlands, Dordrecht, 2002; pp. 49–62. ISBN 978-0-306-48138-3. [Google Scholar]
- Moche, M.; Stremlau, S.; Hecht, L.; Göbel, C.; Feussner, I.; Stöhr, C. Effect of Nitrate Supply and Mycorrhizal Inoculation on Characteristics of Tobacco Root Plasma Membrane Vesicles. Planta 2010, 231, 425–436. [Google Scholar] [CrossRef]
- Gupta, K.J.; Kaiser, W.M. Production and Scavenging of Nitric Oxide by Barley Root Mitochondria. Plant Cell Physiol. 2010, 51, 576–584. [Google Scholar] [CrossRef]
- Cvetkovska, M.; Vanlerberghe, G.C. Alternative Oxidase Modulates Leaf Mitochondrial Concentrations of Superoxide and Nitric Oxide. New Phytol. 2012, 195, 32–39. [Google Scholar] [CrossRef]
- Cvetkovska, M.; Vanlerberghe, G.C. Alternative Oxidase Impacts the Plant Response to Biotic Stress by Influencing the Mitochondrial Generation of Reactive Oxygen Species. Plant Cell Environ. 2013, 36, 721–732. [Google Scholar] [CrossRef]
- Vishwakarma, A.; Kumari, A.; Mur, L.A.J.; Gupta, K.J. A Discrete Role for Alternative Oxidase under Hypoxia to Increase Nitric Oxide and Drive Energy Production. Free Radic. Biol. Med. 2018, 122, 40–51. [Google Scholar] [CrossRef]
- Zafari, S.; Vanlerberghe, G.C.; Igamberdiev, A.U. Nitric Oxide Turnover Under Hypoxia Results in the Rapid Increased Expression of the Plastid-Localized Phosphorylated Pathway of Serine Biosynthesis. Front. Plant Sci. 2021, 12, 780842. [Google Scholar] [CrossRef]
- Zafari, S.; Vanlerberghe, G.C.; Igamberdiev, A.U. The Role of Alternative Oxidase in the Interplay between Nitric Oxide, Reactive Oxygen Species, and Ethylene in Tobacco (Nicotiana Tabacum L.) Plants Incubated under Normoxic and Hypoxic Conditions. Int. J. Mol. Sci. 2022, 23, 7153. [Google Scholar] [CrossRef]
- Kumari, A.; Pathak, P.K.; Bulle, M.; Igamberdiev, A.U.; Gupta, K.J. Alternative Oxidase Is an Important Player in the Regulation of Nitric Oxide Levels under Normoxic and Hypoxic Conditions in Plants. J. Exp. Bot. 2019, 70, 4345–4354. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Mur, L.A.J.; Wany, A.; Kumari, A.; Fernie, A.R.; Ratcliffe, R.G. The Role of Nitrite and Nitric Oxide under Low Oxygen Conditions in Plants. New Phytol. 2020, 225, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Sun, C.; Lin, X.; Busch, W. The Emerging Role of GSNOR in Oxidative Stress Regulation. Trends Plant Sci. 2021, 26, 156–168. [Google Scholar] [CrossRef] [PubMed]
- León, J.; Costa-Broseta, Á. Present Knowledge and Controversies, Deficiencies, and Misconceptions on Nitric Oxide Synthesis, Sensing, and Signaling in Plants. Plant Cell Environ. 2020, 43, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jahnová, J.; Luhová, L.; Petřivalský, M. S-Nitrosoglutathione Reductase-The Master Regulator of Protein S-Nitrosation in Plant NO Signaling. Plants 2019, 8, 48. [Google Scholar] [CrossRef]
- Frungillo, L.; Skelly, M.J.; Loake, G.J.; Spoel, S.H.; Salgado, I. S-Nitrosothiols Regulate Nitric Oxide Production and Storage in Plants through the Nitrogen Assimilation Pathway. Nat. Commun. 2014, 5, 5401. [Google Scholar] [CrossRef]
- Zhan, N.; Wang, C.; Chen, L.; Yang, H.; Feng, J.; Gong, X.; Ren, B.; Wu, R.; Mu, J.; Li, Y.; et al. S-Nitrosylation Targets GSNO Reductase for Selective Autophagy during Hypoxia Responses in Plants. Mol. Cell 2018, 71, 143–154.e6. [Google Scholar] [CrossRef]
- Timilsina, A.; Dong, W.; Hasanuzzaman, M.; Liu, B.; Hu, C. Nitrate–Nitrite–Nitric Oxide Pathway: A Mechanism of Hypoxia and Anoxia Tolerance in Plants. Int. J. Mol. Sci. 2022, 23, 11522. [Google Scholar] [CrossRef]
- Gupta, K.J.; Hancock, J.T.; Petrivalsky, M.; Kolbert, Z.; Lindermayr, C.; Durner, J.; Barroso, J.B.; Palma, J.M.; Brouquisse, R.; Wendehenne, D.; et al. Recommendations on Terminology and Experimental Best Practice Associated with Plant Nitric Oxide Research. New Phytol. 2020, 225, 1828–1834. [Google Scholar] [CrossRef]
- Hill, R.; Hargrove, M.; Arredondo-Peter, R. Phytoglobin: A Novel Nomenclature for Plant Globins Accepted by the Globin Community at the 2014 XVIII Conference on Oxygen-Binding and Sensing Proteins. F1000Res 2016, 5, 212. [Google Scholar] [CrossRef]
- Smagghe, B.J.; Hoy, J.A.; Percifield, R.; Kundu, S.; Hargrove, M.S.; Sarath, G.; Hilbert, J.-L.; Watts, R.A.; Dennis, E.S.; Peacock, W.J.; et al. Review: Correlations between Oxygen Affinity and Sequence Classifications of Plant Hemoglobins. Biopolymers 2009, 91, 1083–1096. [Google Scholar] [CrossRef]
- Hoy, J.A.; Hargrove, M.S. The Structure and Function of Plant Hemoglobins. Plant Physiol. Biochem. 2008, 46, 371–379. [Google Scholar] [CrossRef]
- Gupta, K.J.; Hebelstrup, K.H.; Mur, L.A.J.; Igamberdiev, A.U. Plant Hemoglobins: Important Players at the Crossroads between Oxygen and Nitric Oxide. FEBS Lett. 2011, 585, 3843–3849. [Google Scholar] [CrossRef]
- Gupta, K.J.; Igamberdiev, A.U. Reactive Nitrogen Species in Mitochondria and Their Implications in Plant Energy Status and Hypoxic Stress Tolerance. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Hill, R.D. Plant Mitochondrial Function during Anaerobiosis. Ann. Bot. 2009, 103, 259–268. [Google Scholar] [CrossRef]
- Gupta, K.J.; Igamberdiev, A.U. The Anoxic Plant Mitochondrion as a Nitrite: NO Reductase. Mitochondrion 2011, 11, 537–543. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Baron, K.; Manac’H-Little, N.; Stoimenova, M.; Hill, R.D. The Haemoglobin/Nitric Oxide Cycle: Involvement in Flooding Stress and Effects on Hormone Signalling. Ann. Bot. 2005, 96, 557–564. [Google Scholar] [CrossRef]
- Becana, M.; Yruela, I.; Sarath, G.; Catalán, P.; Hargrove, M.S. Plant Hemoglobins: A Journey from Unicellular Green Algae to Vascular Plants. New Phytol. 2020, 227, 1618–1635. [Google Scholar] [CrossRef]
- Hebelstrup, K.H.; Møller, I.M. Mitochondrial Signaling in Plants Under Hypoxia: Use of Reactive Oxygen Species (ROS) and Reactive Nitrogen Species (RNS). In Reactive Oxygen and Nitrogen Species Signaling and Communication in Plants; Gupta, K.J., Igamberdiev, A.U., Eds.; Signaling and Communication in Plants; Springer International Publishing: Cham, Switzerland, 2015; Volume 23, pp. 63–77. ISBN 978-3-319-10078-4. [Google Scholar]
- Igamberdiev, A.U.; Bykova, N.V.; Shah, J.K.; Hill, R.D. Anoxic Nitric Oxide Cycling in Plants: Participating Reactions and Possible Mechanisms. Physiol. Plant. 2010, 138, 393–404. [Google Scholar] [CrossRef]
- Berger, A.; Brouquisse, R.; Pathak, P.K.; Hichri, I.; Inderjit; Bhatia, S.; Boscari, A.; Igamberdiev, A.U.; Gupta, K.J. Pathways of Nitric Oxide Metabolism and Operation of Phytoglobins in Legume Nodules: Missing Links and Future Directions. Plant Cell Environ. 2018, 41, 2057–2068. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Hill, R.D. Nitrate, NO and Haemoglobin in Plant Adaptation to Hypoxia: An Alternative to Classic Fermentation Pathways. J. Exp. Bot. 2004, 55, 2473–2482. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, W.; Beckett, P.M.; Colmer, T.D.; Setter, T.L.; Greenway, H. Tolerance of Roots to Low Oxygen: ‘Anoxic’ Cores, the Phytoglobin-Nitric Oxide Cycle, and Energy or Oxygen Sensing. J. Plant Physiol. 2019, 239, 92–108. [Google Scholar] [CrossRef] [PubMed]
- De Castro, J.; Hill, R.D.; Stasolla, C.; Badea, A. Waterlogging Stress Physiology in Barley. Agronomy 2022, 12, 780. [Google Scholar] [CrossRef]
- Chammakhi, C.; Boscari, A.; Pacoud, M.; Aubert, G.; Mhadhbi, H.; Brouquisse, R. Nitric Oxide Metabolic Pathway in Drought-Stressed Nodules of Faba Bean (Vicia faba L.). Int. J. Mol. Sci. 2022, 23, 13057. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Jardinaud, M.-F.; Gao, J.; Pecrix, Y.; Wen, J.; Mysore, K.; Xu, P.; Sanchez-Canizares, C.; Ruan, Y.; Li, Q.; et al. NIN-like Protein Transcription Factors Regulate Leghemoglobin Genes in Legume Nodules. Science 2021, 374, 625–628. [Google Scholar] [CrossRef]
- Kumari, A.; Pathak, P.K.; Loake, G.J.; Gupta, K.J. The PHYTOGLOBIN-NO Cycle Regulates Plant Mycorrhizal Symbiosis. Trends Plant Sci. 2019, 24, 981–983. [Google Scholar] [CrossRef]
- Cochrane, D.W.; Shah, J.K.; Hebelstrup, K.H.; Igamberdiev, A.U. Expression of Phytoglobin Affects Nitric Oxide Metabolism and Energy State of Barley Plants Exposed to Anoxia. Plant Sci. 2017, 265, 124–130. [Google Scholar] [CrossRef]
- Kumari, A.; Singh, P.; Kaladhar, V.C.; Manbir; Paul, D.; Pathak, P.K.; Gupta, K.J. Phytoglobin-NO Cycle and AOX Pathway Play a Role in Anaerobic Germination and Growth of Deepwater Rice. Plant Cell Environ. 2022, 45, 178–190. [Google Scholar] [CrossRef]
- Ageeva-Kieferle, A.; Georgii, E.; Winkler, B.; Ghirardo, A.; Albert, A.; Hüther, P.; Mengel, A.; Becker, C.; Schnitzler, J.-P.; Durner, J.; et al. Nitric Oxide Coordinates Growth, Development, and Stress Response via Histone Modification and Gene Expression. Plant Physiol. 2021, 187, 336–360. [Google Scholar] [CrossRef]
- Pande, A.; Mun, B.-G.; Rahim, W.; Khan, M.; Lee, D.; Lee, G.; Al Azzawi, T.; Hussain, A.; Kim, C.; Wook, B. Phytohormonal Regulation Through Protein S-Nitrosylation Under Stress. Front. Plant Sci. 2022, 13, 865542. [Google Scholar] [CrossRef]
- Zhang, J.; Liao, W. Protein S-Nitrosylation in Plant Abiotic Stresses. Funct. Plant Biol. 2020, 47, 1. [Google Scholar] [CrossRef]
- Wang, C.; Wei, L.; Zhang, J.; Hu, D.; Gao, R.; Liu, Y.; Feng, L.; Gong, W.; Liao, W. Nitric Oxide Enhances Salt Tolerance in Tomato Seedlings by Regulating Endogenous S-Nitrosylation Levels. J Plant Growth Regul 2022, 42, 275–329. [Google Scholar] [CrossRef]
- Lubega, J.; Umbreen, S.; Loake, G.J. Recent Advances in the Regulation of Plant Immunity by S-Nitrosylation. J. Exp. Bot. 2021, 72, 864–872. [Google Scholar] [CrossRef]
- Chen, L.; Sun, S.; Song, C.-P.; Zhou, J.-M.; Li, J.; Zuo, J. Nitric Oxide Negatively Regulates Gibberellin Signaling to Coordinate Growth and Salt Tolerance in Arabidopsis. J. Genet. Genom. 2022, 49, 756–765. [Google Scholar] [CrossRef]
- Jing, H.; Yang, X.; Feng, J.; Zhang, J.; Strader, L.C.; Zuo, J. S-Nitrosylation of Aux/IAA Protein Represses Auxin Signaling. bioRxiv 2022. preprint. [Google Scholar]
- Wang, Y.; Yun, B.-W.; Kwon, E.; Hong, J.K.; Yoon, J.; Loake, G.J. S-Nitrosylation: An Emerging Redox-Based Post-Translational Modification in Plants. J. Exp. Bot. 2006, 57, 1777–1784. [Google Scholar] [CrossRef]
- Smith, B.C.; Marletta, M.A. Mechanisms of S-Nitrosothiol Formation and Selectivity in Nitric Oxide Signaling. Curr. Opin. Chem. Biol. 2012, 16, 498–506. [Google Scholar] [CrossRef]
- Broniowska, K.A.; Hogg, N. The Chemical Biology of S-Nitrosothiols. Antioxid. Redox. Signal 2012, 17, 969–980. [Google Scholar] [CrossRef]
- Basu, S.; Keszler, A.; Azarova, N.A.; Nwanze, N.; Perlegas, A.; Shiva, S.; Broniowska, K.A.; Hogg, N.; Kim-Shapiro, D.B. A Novel Role for Cytochrome c: Efficient Catalysis of S-Nitrosothiol Formation. Free Radic. Biol. Med. 2010, 48, 255–263. [Google Scholar] [CrossRef]
- Corpas, F.; Alché, J.; Barroso, J. Current Overview of S-Nitrosoglutathione (GSNO) in Higher Plants. Front. Plant Sci. 2013, 4, 126. [Google Scholar] [CrossRef]
- Wynia-Smith, S.L.; Smith, B.C. Nitrosothiol Formation and S-Nitrosation Signaling through Nitric Oxide Synthases. Nitric Oxide 2017, 63, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Perissinotti, L.L.; Turjanski, A.G.; Estrin, D.A.; Doctorovich, F. Transnitrosation of Nitrosothiols: Characterization of an Elusive Intermediate. J. Am. Chem. Soc. 2005, 127, 486–487. [Google Scholar] [CrossRef] [PubMed]
- Houk, K.N.; Hietbrink, B.N.; Bartberger, M.D.; McCarren, P.R.; Choi, B.Y.; Voyksner, R.D.; Stamler, J.S.; Toone, E.J. Nitroxyl Disulfides, Novel Intermediates in Transnitrosation Reactions. J. Am. Chem. Soc. 2003, 125, 6972–6976. [Google Scholar] [CrossRef] [PubMed]
- Kolbert, Z.; Feigl, G.; Bordé, Á.; Molnár, Á.; Erdei, L. Protein Tyrosine Nitration in Plants: Present Knowledge, Computational Prediction and Future Perspectives. Plant Physiol. Biochem. 2017, 113, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Feeney, M.B.; Schöneich, C. Tyrosine Modifications in Aging. Antioxid Redox Signal 2012, 17, 1571–1579. [Google Scholar] [CrossRef]
- Jones, L.H. Chemistry and Biology of Biomolecule Nitration. Chem. Biol. 2012, 19, 1086–1092. [Google Scholar] [CrossRef]
- Souza, J.M.; Peluffo, G.; Radi, R. Protein Tyrosine Nitration--Functional Alteration or Just a Biomarker? Free Radic. Biol. Med. 2008, 45, 357–366. [Google Scholar] [CrossRef]
- Radi, R. Protein Tyrosine Nitration: Biochemical Mechanisms and Structural Basis of Functional Effects. Acc. Chem. Res. 2013, 46, 550–559. [Google Scholar] [CrossRef]
- Heijnen, H.F.G.; van Donselaar, E.; Slot, J.W.; Fries, D.M.; Blachard-Fillion, B.; Hodara, R.; Lightfoot, R.; Polydoro, M.; Spielberg, D.; Thomson, L.; et al. Subcellular Localization of Tyrosine-Nitrated Proteins Is Dictated by Reactive Oxygen Species Generating Enzymes and by Proximity to Nitric Oxide Synthase. Free Radic. Biol. Med. 2006, 40, 1903–1913. [Google Scholar] [CrossRef]
- Ferrer-Sueta, G.; Radi, R. Chemical Biology of Peroxynitrite: Kinetics, Diffusion, and Radicals. ACS Chem. Biol. 2009, 4, 161–177. [Google Scholar] [CrossRef]
- Sánchez-Vicente, I.; Fernández-Espinosa, M.G.; Lorenzo, O. Nitric Oxide Molecular Targets: Reprogramming Plant Development upon Stress. J. Exp. Bot. 2019, 70, 4441–4460. [Google Scholar] [CrossRef]
- Bayden, A.S.; Yakovlev, V.A.; Graves, P.R.; Mikkelsen, R.B.; Kellogg, G.E. Factors Influencing Protein Tyrosine Nitration—Structure-Based Predictive Models. Free Radic. Biol. Med. 2011, 50, 749–762. [Google Scholar] [CrossRef]
- Corpas, F.; Palma, J.; del Río, L.; Barroso, J. Protein Tyrosine Nitration in Higher Plants Grown under Natural and Stress Conditions. Front. Plant Sci. 2013, 4, 29. [Google Scholar] [CrossRef]
- Abello, N.; Kerstjens, H.A.M.; Postma, D.S.; Bischoff, R. Protein Tyrosine Nitration: Selectivity, Physicochemical and Biological Consequences, Denitration, and Proteomics Methods for the Identification of Tyrosine-Nitrated Proteins. J. Proteome Res. 2009, 8, 3222–3238. [Google Scholar] [CrossRef]
- van der Vliet, A.; Eiserich, J.P.; Shigenaga, M.K.; Cross, C.E. Reactive Nitrogen Species and Tyrosine Nitration in the Respiratory Tract: Epiphenomena or a Pathobiologic Mechanism of Disease? Am. J. Respir. Crit. Care Med 1999, 160, 1–9. [Google Scholar] [CrossRef]
- Radi, R. Nitric Oxide, Oxidants, and Protein Tyrosine Nitration. Proc. Natl. Acad. Sci. USA 2004, 101, 4003–4008. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Protein Nitration: A Connecting Bridge between Nitric Oxide (NO) and Plant Stress. Plant Stress 2021, 2, 100026. [Google Scholar] [CrossRef]
- Shi, W.-Q.; Cai, H.; Xu, D.-D.; Su, X.-Y.; Lei, P.; Zhao, Y.-F.; Li, Y.-M. Tyrosine Phosphorylation/Dephosphorylation Regulates Peroxynitrite-Mediated Peptide Nitration. Regul. Pept. 2007, 144, 1–5. [Google Scholar] [CrossRef]
- Galetskiy, D.; Lohscheider, J.; Kononikhin, A.; Popov, I.; Nikolaev, E.; Adamska, I. Phosphorylation and Nitration Levels of Photosynthetic Proteins Are Conversely Regulated by Light Stress. Plant Mol. Biol. 2011, 77, 461–473. [Google Scholar] [CrossRef]
- Mata-Pérez, C.; Begara-Morales, J.C.; Chaki, M.; Sánchez-Calvo, B.; Valderrama, R.; Padilla, M.N.; Corpas, F.J.; Barroso, J.B. Protein Tyrosine Nitration during Development and Abiotic Stress Response in Plants. Front. Plant Sci. 2016, 7, 1699. [Google Scholar] [CrossRef]
- Wu, X.; Gong, F.; Cao, D.; Hu, X.; Wang, W. Advances in Crop Proteomics: PTMs of Proteins under Abiotic Stress. Proteomics 2016, 16, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, S.; Corpas, F.J.; Rodríguez-Ruiz, M.; Valderrama, R.; Barroso, J.B.; Borsani, O.; Monza, J. Drought Stress Triggers the Accumulation of NO and SNOs in Cortical Cells of Lotus japonicus L. Roots and the Nitration of Proteins with Relevant Metabolic Function. Environ. Exp. Bot. 2019, 161, 228–241. [Google Scholar] [CrossRef]
- Ruiz-May, E.; Segura-Cabrera, A.; Elizalde-Contreras, J.M.; Shannon, L.M.; Loyola-Vargas, V.M. A Recent Advance in the Intracellular and Extracellular Redox Post-Translational Modification of Proteins in Plants. J. Mol. Recognit. 2019, 32, e2754. [Google Scholar] [CrossRef] [PubMed]
- Sehrawat, A.; Deswal, R. Proteomics Approach to Uncover Key Signalling Pathways in Brassica Juncea in Abiotic and Biotic Stress. In The Brassica juncea Genome; Kole, C., Mohapatra, T., Eds.; Compendium of Plant Genomes; Springer International Publishing: Cham, Switzerland, 2022; pp. 337–347. ISBN 978-3-030-91507-0. [Google Scholar]
- Chaki, M.; Valderrama, R.; Fernández-Ocaña, A.M.; Carreras, A.; Gómez-Rodríguez, M.V.; Pedrajas, J.R.; Begara-Morales, J.C.; Sánchez-Calvo, B.; Luque, F.; Leterrier, M.; et al. Mechanical Wounding Induces a Nitrosative Stress by Down-Regulation of GSNO Reductase and an Increase in S-Nitrosothiols in Sunflower (Helianthus annuus) Seedlings. J. Exp. Bot. 2011, 62, 1803–1813. [Google Scholar] [CrossRef]
- Gupta, K.J.; Kolbert, Z.; Durner, J.; Lindermayr, C.; Corpas, F.J.; Brouquisse, R.; Barroso, J.B.; Umbreen, S.; Palma, J.M.; Hancock, J.T.; et al. Regulating the Regulator: Nitric Oxide Control of Post-Translational Modifications. New Phytol. 2020, 227, 1319–1325. [Google Scholar] [CrossRef]
- Maszkowska, J.; Szymańska, K.P.; Kasztelan, A.; Krzywińska, E.; Sztatelman, O.; Dobrowolska, G. The Multifaceted Regulation of SnRK2 Kinases. Cells 2021, 10, 2180. [Google Scholar] [CrossRef]
- Witoń, D.; Sujkowska-Rybkowska, M.; Dąbrowska-Bronk, J.; Czarnocka, W.; Bernacki, M.; Szechyńska-Hebda, M.; Karpiński, S. MITOGEN-ACTIVATED PROTEIN KINASE 4 Impacts Leaf Development, Temperature, and Stomatal Movement in Hybrid Aspen. Plant Physiol. 2021, 186, 2190–2204. [Google Scholar] [CrossRef]
- Frederickson Matika, D.E.; Loake, G.J. Redox Regulation in Plant Immune Function. Antioxid. Redox. Signal 2014, 21, 1373–1388. [Google Scholar] [CrossRef]
- Gietler, M.; Fidler, J.; Labudda, M.; Nykiel, M. Abscisic Acid—Enemy or Savior in the Response of Cereals to Abiotic and Biotic Stresses? Int. J. Mol. Sci. 2020, 21, 4607. [Google Scholar] [CrossRef]
- Kundu, S.; Gantait, S. Abscisic Acid Signal Crosstalk during Abiotic Stress Response. Plant Gene 2017, 11, 61–69. [Google Scholar] [CrossRef]
- Ali, A.; Pardo, J.M.; Yun, D.-J. Desensitization of ABA-Signaling: The Swing from Activation to Degradation. Front. Plant Sci. 2020, 11, 379. [Google Scholar] [CrossRef]
- Wang, P.; Du, Y.; Hou, Y.-J.; Zhao, Y.; Hsu, C.-C.; Yuan, F.; Zhu, X.; Tao, W.A.; Song, C.-P.; Zhu, J.-K. Nitric Oxide Negatively Regulates Abscisic Acid Signaling in Guard Cells by S-Nitrosylation of OST1. Proc. Natl. Acad. Sci. USA 2015, 112, 613–618. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, J.-K.; Lang, Z. Nitric Oxide Suppresses the Inhibitory Effect of Abscisic Acid on Seed Germination by S-Nitrosylation of SnRK2 Proteins. Plant Signal. Behav. 2015, 10, e1031939. [Google Scholar] [CrossRef]
- Lanteri, M.L.; Pagnussat, G.C.; Lamattina, L. Calcium and Calcium-Dependent Protein Kinases Are Involved in Nitric Oxide- and Auxin-Induced Adventitious Root Formation in Cucumber. J. Exp. Bot. 2006, 57, 1341–1351. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Huang, D.; Zhang, X.; Zhou, J.; Zhu, S. Nitric Oxide Signaling in Plants. In Postharvest Biology and Nanotechnology; Paliyath, G., Subramanian, J., Lim, L.-T., Subramanian, K.S., Handa, A.K., Mattoo, A.K., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; pp. 103–127. ISBN 978-1-119-28947-0. [Google Scholar]
- Hasanuzzaman, M.; Oku, H.; Nahar, K.; Bhuyan, M.H.M.B.; Mahmud, J.A.; Baluska, F.; Fujita, M. Nitric Oxide-Induced Salt Stress Tolerance in Plants: ROS Metabolism, Signaling, and Molecular Interactions. Plant Biotechnol. Rep. 2018, 12, 77–92. [Google Scholar] [CrossRef]
- Fujita, M.; Hasanuzzaman, M. Approaches to Enhancing Antioxidant Defense in Plants. Antioxidants 2022, 11, 925. [Google Scholar] [CrossRef]
- Sies, H. Hydrogen Peroxide as a Central Redox Signaling Molecule in Physiological Oxidative Stress: Oxidative Eustress. Redox. Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef]
- Muszyńska, E.; Labudda, M.; Różańska, E.; Hanus-Fajerska, E.; Znojek, E. Heavy Metal Tolerance in Contrasting Ecotypes of Alyssum Montanum. Ecotoxicol. Environ. Saf. 2018, 161, 305–317. [Google Scholar] [CrossRef]
- Muszyńska, E.; Labudda, M.; Różańska, E.; Hanus-Fajerska, E.; Koszelnik-Leszek, A. Structural, Physiological and Genetic Diversification of Silene Vulgaris Ecotypes from Heavy Metal-Contaminated Areas and Their Synchronous in Vitro Cultivation. Planta 2019, 249, 1761–1778. [Google Scholar] [CrossRef]
- Labudda, M. Ascorbate-Glutathione Pathway as an Important Player in Redox Regulation in Nematode-Infested Plants: What We Have Learned so Far. Physiol. Mol. Plant Pathol. 2018, 103, 47–53. [Google Scholar] [CrossRef]
- Labudda, M.; Różańska, E.; Cieśla, J.; Sobczak, M.; Dzik, J.M. Arginase Activity in Arabidopsis Thaliana Infected with Heterodera Schachtii. Plant Pathol. 2016, 65, 1529–1538. [Google Scholar] [CrossRef]
- Labudda, M.; Różańska, E.; Szewińska, J.; Sobczak, M.; Dzik, J.M. Protease Activity and Phytocystatin Expression in Arabidopsis Thaliana upon Heterodera Schachtii Infection. Plant Physiol. Biochem. 2016, 109, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Labudda, M.; Azam, F.M.S. Glutathione-Dependent Responses of Plants to Drought: A Review. Acta Soc. Bot. Pol. 2014, 83, 3–12. [Google Scholar] [CrossRef]
- Labudda, M.; Różańska, E.; Czarnocka, W.; Sobczak, M.; Dzik, J.M. Systemic Changes in Photosynthesis and Reactive Oxygen Species Homeostasis in Shoots of Arabidopsis thaliana Infected with the Beet Cyst Nematode Heterodera Schachtii. Mol. Plant Pathol. 2018, 19, 1690–1704. [Google Scholar] [CrossRef]
- Scheler, C.; Durner, J.; Astier, J. Nitric Oxide and Reactive Oxygen Species in Plant Biotic Interactions. Curr. Opin. Plant Biol. 2013, 16, 534–539. [Google Scholar] [CrossRef]
- Lindermayr, C.; Sell, S.; Müller, B.; Leister, D.; Durner, J. Redox Regulation of the NPR1-TGA1 System of Arabidopsis thaliana by Nitric Oxide. Plant Cell 2010, 22, 2894–2907. [Google Scholar] [CrossRef]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant Immunity Requires Conformational Charges of NPR1 via S-Nitrosylation and Thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef]
- Arnaiz, A.; Rosa-Diaz, I.; Romero-Puertas, M.C.; Sandalio, L.M.; Diaz, I. Nitric Oxide, an Essential Intermediate in the Plant–Herbivore Interaction. Front. Plant Sci. 2021, 11, 620086. [Google Scholar] [CrossRef]
- Besson-Bard, A.; Pugin, A.; Wendehenne, D. New Insights into Nitric Oxide Signaling in Plants. Annu. Rev. Plant Biol. 2008, 59, 21–39. [Google Scholar] [CrossRef]
- Mur, L.A.J.; Kenton, P.; Lloyd, A.J.; Ougham, H.; Prats, E. The Hypersensitive Response; The Centenary Is upon Us but How Much Do We Know? J. Exp. Bot. 2008, 59, 501–520. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Verma, S. Hypersensitive Responses in Plants- A Review. Agric. Rev. 2019, 40, 113–120. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H. Reactive Oxygen Species and Nitric Oxide as Mediators in Plant Hypersensitive Response and Stomatal Closure. Plant Signal. Behav. 2021, 16, 1985860. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, J. Rapid Accumulation of NO Regulates ABA Catabolism and Seed Dormancy during Imbibition in Arabidopsis. Plant Signal Behav. 2009, 4, 905–907. [Google Scholar] [CrossRef]
- Castillo, M.-C.; Lozano-Juste, J.; González-Guzmán, M.; Rodriguez, L.; Rodriguez, P.L.; León, J. Inactivation of PYR/PYL/RCAR ABA Receptors by Tyrosine Nitration May Enable Rapid Inhibition of ABA Signaling by Nitric Oxide in Plants. Sci. Signal 2015, 8, ra89. [Google Scholar] [CrossRef]
- Albertos, P.; Romero-Puertas, M.C.; Tatematsu, K.; Mateos, I.; Sánchez-Vicente, I.; Nambara, E.; Lorenzo, O. S-Nitrosylation Triggers ABI5 Degradation to Promote Seed Germination and Seedling Growth. Nat. Commun. 2015, 6, 8669. [Google Scholar] [CrossRef]
- Correa-Aragunde, N.; Graziano, M.; Lamattina, L. Nitric Oxide Plays a Central Role in Determining Lateral Root Development in Tomato. Planta 2004, 218, 900–905. [Google Scholar] [CrossRef]
- Yu, M.; Lamattina, L.; Spoel, S.H.; Loake, G.J. Nitric Oxide Function in Plant Biology: A Redox Cue in Deconvolution. New Phytol. 2014, 202, 1142–1156. [Google Scholar] [CrossRef]
- Piacentini, D.; Della Rovere, F.; Sofo, A.; Fattorini, L.; Falasca, G.; Altamura, M.M. Nitric Oxide Cooperates with Auxin to Mitigate the Alterations in the Root System Caused by Cadmium and Arsenic. Front. Plant Sci. 2020, 11, 1182. [Google Scholar] [CrossRef]
- Xu, J.; Wang, W.; Yin, H.; Liu, X.; Sun, H.; Mi, Q. Exogenous Nitric Oxide Improves Antioxidative Capacity and Reduces Auxin Degradation in Roots of Medicago Truncatula Seedlings under Cadmium Stress. Plant Soil 2009, 326, 321. [Google Scholar] [CrossRef]
- Zottini, M.; Costa, A.; De Michele, R.; Ruzzene, M.; Carimi, F.; Lo Schiavo, F. Salicylic Acid Activates Nitric Oxide Synthesis in Arabidopsis. J. Exp. Bot. 2007, 58, 1397–1405. [Google Scholar] [CrossRef]
- Kohli, S.K.; Khanna, K.; Bhardwaj, R.; Abd_Allah, E.F.; Ahmad, P.; Corpas, F.J. Assessment of Subcellular ROS and NO Metabolism in Higher Plants: Multifunctional Signaling Molecules. Antioxidants 2019, 8, 641. [Google Scholar] [CrossRef]
- Belt, K.; Huang, S.; Thatcher, L.F.; Casarotto, H.; Singh, K.B.; Van Aken, O.; Millar, A.H. Salicylic Acid-Dependent Plant Stress Signaling via Mitochondrial Succinate Dehydrogenase. Plant Physiol. 2017, 173, 2029–2040. [Google Scholar] [CrossRef] [PubMed]
- Prakash, V.; Singh, V.P.; Tripathi, D.K.; Sharma, S.; Corpas, F.J. Nitric Oxide (NO) and Salicylic Acid (SA): A Framework for Their Relationship in Plant Development under Abiotic Stress. Plant Biol. 2021, 23, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, I.; Durner, J.; Lindermayr, C. Crosstalk between Nitric Oxide and Glutathione Is Required for NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1)-Dependent Defense Signaling in Arabidopsis Thaliana. New Phytol. 2015, 208, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.; Colcombet, J. Sustained Incompatibility between MAPK Signaling and Pathogen Effectors. Int. J. Mol. Sci. 2020, 21, 7954. [Google Scholar] [CrossRef] [PubMed]
- Tajti, J.; Németh, E.; Glatz, G.; Janda, T.; Pál, M. Pattern of Changes in Salicylic Acid-Induced Protein Kinase (SIPK) Gene Expression and Salicylic Acid Accumulation in Wheat under Cadmium Exposure. Plant Biol. 2019, 21, 1176–1180. [Google Scholar] [CrossRef]
- Caarls, L.; Pieterse, C.M.J.; Van Wees, S.C.M. How Salicylic Acid Takes Transcriptional Control over Jasmonic Acid Signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef]
- Nomoto, M.; Skelly, M.J.; Itaya, T.; Mori, T.; Suzuki, T.; Matsushita, T.; Tokizawa, M.; Kuwata, K.; Mori, H.; Yamamoto, Y.Y.; et al. Suppression of MYC Transcription Activators by the Immune Cofactor NPR1 Fine-Tunes Plant Immune Responses. Cell Rep. 2021, 37, 110125. [Google Scholar] [CrossRef]
- Mur, L.A.J.; Mandon, J.; Persijn, S.; Cristescu, S.M.; Moshkov, I.E.; Novikova, G.V.; Hall, M.A.; Harren, F.J.M.; Hebelstrup, K.H.; Gupta, K.J. Nitric Oxide in Plants: An Assessment of the Current State of Knowledge. AoB Plants 2013, 5, pls052. [Google Scholar] [CrossRef]
- Palmieri, M.C.; Sell, S.; Huang, X.; Scherf, M.; Werner, T.; Durner, J.; Lindermayr, C. Nitric Oxide-Responsive Genes and Promoters in Arabidopsis Thaliana: A Bioinformatics Approach. J. Exp. Bot 2008, 59, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Romero-Puertas, M.C.; Campostrini, N.; Mattè, A.; Righetti, P.G.; Perazzolli, M.; Zolla, L.; Roepstorff, P.; Delledonne, M. Proteomic Analysis of S-Nitrosylated Proteins in Arabidopsis Thaliana Undergoing Hypersensitive Response. Proteomics 2008, 8, 1459–1469. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Zhou, Y.; Liu, M. Nitric Oxide Participates in the Regulation of the Ascorbate-Glutathione Cycle by Exogenous Jasmonic Acid in the Leaves of Wheat Seedlings under Drought Stress. Protoplasma 2015, 252, 1397–1405. [Google Scholar] [CrossRef]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent Developments in Enzymatic Antioxidant Defence Mechanism in Plants with Special Reference to Abiotic Stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef] [PubMed]
- Couto, N.; Wood, J.; Barber, J. The Role of Glutathione Reductase and Related Enzymes on Cellular Redox Homoeostasis Network. Free Radic. Biol. Med. 2016, 95, 27–42. [Google Scholar] [CrossRef]
- da-Silva, C.J.; Rodrigues, A.C.; Modolo, L.V. H2O2, NO, and H2S. In Reactive Oxygen, Nitrogen and Sulfur Species in Plants; John Wiley & Sons, Ltd.: Chichester, UK, 2019; pp. 841–856. ISBN 978-1-119-46867-7. [Google Scholar]
- Qiao, W.; Li, C.; Fan, L.-M. Cross-Talk between Nitric Oxide and Hydrogen Peroxide in Plant Responses to Abiotic Stresses. Environ. Exp. Bot. 2014, 100, 84–93. [Google Scholar] [CrossRef]
- Wünsche, H.; Baldwin, I.T.; Wu, J. S-Nitrosoglutathione Reductase (GSNOR) Mediates the Biosynthesis of Jasmonic Acid and Ethylene Induced by Feeding of the Insect Herbivore Manduca Sexta and Is Important for Jasmonate-Elicited Responses in Nicotiana Attenuata. J. Exp. Bot. 2011, 62, 4605–4616. [Google Scholar] [CrossRef]
- Fidler, J.; Graska, J.; Gietler, M.; Nykiel, M.; Prabucka, B.; Rybarczyk-Płońska, A.; Muszyńska, E.; Morkunas, I.; Labudda, M. PYR/PYL/RCAR Receptors Play a Vital Role in the Abscisic-Acid-Dependent Responses of Plants to External or Internal Stimuli. Cells 2022, 11, 1352. [Google Scholar] [CrossRef]
- Pii, Y.; Crimi, M.; Cremonese, G.; Spena, A.; Pandolfini, T. Auxin and Nitric Oxide Control Indeterminate Nodule Formation. BMC Plant Biol. 2007, 7, 21. [Google Scholar] [CrossRef]
- Terrile, M.C.; París, R.; Calderón-Villalobos, L.I.A.; Iglesias, M.J.; Lamattina, L.; Estelle, M.; Casalongué, C.A. Nitric Oxide Influences Auxin Signaling through S-Nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 Auxin Receptor. Plant J. 2012, 70, 492–500. [Google Scholar] [CrossRef]
- Morkunas, I.; Doğu, M.Z.; Woźniak, A.; Bednarski, W.; Kęsy, J.; Bocianowski, J.; Atar, Ş.H.; Ürün, İ.D.; Labudda, M.; Zydlik, Z.; et al. Profile of Semiquinone Radicals, Phytohormones and Sugars in Pistacia vera L. Cv. Kirmizi Development. Agronomy 2021, 11, 2115. [Google Scholar] [CrossRef]
- Arif, Y.; Sami, F.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salicylic Acid in Relation to Other Phytohormones in Plant: A Study towards Physiology and Signal Transduction under Challenging Environment. Environ. Exp. Bot. 2020, 175, 104040. [Google Scholar] [CrossRef]
- Formela-Luboińska, M.; Chadzinikolau, T.; Drzewiecka, K.; Jeleń, H.; Bocianowski, J.; Kęsy, J.; Labudda, M.; Jeandet, P.; Morkunas, I. The Role of Sugars in the Regulation of the Level of Endogenous Signaling Molecules during Defense Response of Yellow Lupine to Fusarium Oxysporum. Int. J. Mol. Sci. 2020, 21, 4133. [Google Scholar] [CrossRef]
- Mohamed, H.I.; El-Shazly, H.H.; Badr, A. Role of Salicylic Acid in Biotic and Abiotic Stress Tolerance in Plants. In Plant Phenolics in Sustainable Agriculture: Volume 1; Lone, R., Shuab, R., Kamili, A.N., Eds.; Springer: Singapore, 2020; pp. 533–554. ISBN 9789811548901. [Google Scholar]
- Koo, Y.M.; Heo, A.Y.; Choi, H.W. Salicylic Acid as a Safe Plant Protector and Growth Regulator. Plant Pathol. J. 2020, 36, 1–10. [Google Scholar] [CrossRef]
- Domingos, P.; Prado, A.M.; Wong, A.; Gehring, C.; Feijo, J.A. Nitric Oxide: A Multitasked Signaling Gas in Plants. Mol. Plant 2015, 8, 506–520. [Google Scholar] [CrossRef]
- Moeder, W.; Phan, V.; Yoshioka, K. Ca2+ to the Rescue–Ca2+channels and Signaling in Plant Immunity. Plant Sci. 2019, 279, 19–26. [Google Scholar] [CrossRef]
- Al-Whaibi, M.H.; Siddiqui, M.H.; Basalah, M.O. Salicylic Acid and Calcium-Induced Protection of Wheat against Salinity. Protoplasma 2012, 249, 769–778. [Google Scholar] [CrossRef]
- Chen, L.; Wang, W.-S.; Wang, T.; Meng, X.-F.; Chen, T.; Huang, X.-X.; Li, Y.; Hou, B.-K. Methyl Salicylate Glucosylation Regulates Plant Defense Signaling and Systemic Acquired Resistance. Plant Physiol. 2019, 180, 2167–2181. [Google Scholar] [CrossRef]
- Gondor, O.K.; Pál, M.; Janda, T.; Szalai, G. The Role of Methyl Salicylate in Plant Growth under Stress Conditions. J. Plant Physiol. 2022, 277, 153809. [Google Scholar] [CrossRef]
- Durner, J.; Klessig, D.F. Nitric Oxide as a Signal in Plants. Curr. Opin. Plant Biol. 1999, 2, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.R.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic Acid-Induced Abiotic Stress Tolerance and Underlying Mechanisms in Plants. Front. Plant Sci. 2015, 6, 462. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Zhang, M.; Wei, S.; Zhang, J.; Wang, C.; Liao, W. Roles of Nitric Oxide in Heavy Metal Stress in Plants: Cross-Talk with Phytohormones and Protein S-Nitrosylation. Environ. Pollut. 2020, 259, 113943. [Google Scholar] [CrossRef] [PubMed]
- Bürger, M.; Chory, J. Stressed Out About Hormones: How Plants Orchestrate Immunity. Cell Host Microbe. 2019, 26, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of Jasmonic Acid in Plant Regulation and Response to Abiotic Stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef]
- Ghorbel, M.; Brini, F.; Sharma, A.; Landi, M. Role of Jasmonic Acid in Plants: The Molecular Point of View. Plant Cell Rep. 2021, 40, 1471–1494. [Google Scholar] [CrossRef]
- Seo, H.S.; Song, J.T.; Cheong, J.-J.; Lee, Y.-H.; Lee, Y.-W.; Hwang, I.; Lee, J.S.; Choi, Y.D. Jasmonic Acid Carboxyl Methyltransferase: A Key Enzyme for Jasmonate-Regulated Plant Responses. Proc. Natl. Acad. Sci. USA 2001, 98, 4788–4793. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The Roles of Methyl Jasmonate to Stress in Plants. Funct. Plant Biol. 2019, 46, 197. [Google Scholar] [CrossRef]
- Smith, J.L.; De Moraes, C.M.; Mescher, M.C. Jasmonate- and Salicylate-Mediated Plant Defense Responses to Insect Herbivores, Pathogens and Parasitic Plants. Pest Manag. Sci. 2009, 65, 497–503. [Google Scholar] [CrossRef]
- Gimenez-Ibanez, S.; Solano, R. Nuclear Jasmonate and Salicylate Signaling and Crosstalk in Defense against Pathogens. Front. Plant Sci. 2013, 4, 72. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van Loon, L.C. NPR1: The Spider in the Web of Induced Resistance Signaling Pathways. Curr. Opin. Plant Biol. 2004, 7, 456–464. [Google Scholar] [CrossRef]
- Telford, M.J.; Bourlat, S.J.; Economou, A.; Papillon, D.; Rota-Stabelli, O. The Evolution of the Ecdysozoa. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 1529–1537. [Google Scholar] [CrossRef]
- Howard, R.J.; Giacomelli, M.; Lozano-Fernandez, J.; Edgecombe, G.D.; Fleming, J.F.; Kristensen, R.M.; Ma, X.; Olesen, J.; Sørensen, M.V.; Thomsen, P.F.; et al. The Ediacaran Origin of Ecdysozoa: Integrating Fossil and Phylogenomic Data. J. Geol. Soc. 2022, 179, jgs2021–107. [Google Scholar] [CrossRef]
- Zhou, J.; Jia, F.; Shao, S.; Zhang, H.; Li, G.; Xia, X.; Zhou, Y.; Yu, J.; Shi, K. Involvement of Nitric Oxide in the Jasmonate-Dependent Basal Defense against Root-Knot Nematode in Tomato Plants. Front. Plant Sci. 2015, 6, 193. [Google Scholar] [CrossRef]
- Melillo, M.; Leonetti, P.; Leone, A.; Veronico, P.; Bleve-Zacheo, T. ROS and NO Production in Compatible and Incompatible Tomato-Meloidogyne Incognita Interactions. Eur. J. Plant Path. 2011, 130, 489–502. [Google Scholar] [CrossRef]
- He, H.; Oo, T.L.; Huang, W.; He, L.-F.; Gu, M. Nitric Oxide Acts as an Antioxidant and Inhibits Programmed Cell Death Induced by Aluminum in the Root Tips of Peanut (Arachis hypogaea L.). Sci. Rep. 2019, 9, 9516. [Google Scholar] [CrossRef]
- Leonetti, P.; Melillo, M.T.; Bleve-Zacheo, T. Nitric Oxide and Hydrogen Peroxide: Two Players in the Defence Response of Tomato Plants to Root-Knot Nematodes. Commun. Agric. Appl. Biol. Sci. 2011, 76, 371–381. [Google Scholar]
- Yu, L.-Z.; Wu, X.-Q.; Ye, J.-R.; Zhang, S.-N.; Wang, C. NOS-like-Mediated Nitric Oxide Is Involved in Pinus Thunbergii Response to the Invasion of Bursaphelenchus Xylophilus. Plant Cell Rep. 2012, 31, 1813–1821. [Google Scholar] [CrossRef]
- Labudda, M.; Różańska, E.; Gietler, M.; Fidler, J.; Muszyńska, E.; Prabucka, B.; Morkunas, I. Cyst Nematode Infection Elicits Alteration in the Level of Reactive Nitrogen Species, Protein S-Nitrosylation and Nitration, and Nitrosoglutathione Reductase in Arabidopsis Thaliana Roots. Antioxidants 2020, 9, 795. [Google Scholar] [CrossRef]
- Mai, V.C.; Drzewiecka, K.; Jeleń, H.; Narożna, D.; Rucińska-Sobkowiak, R.; Kęsy, J.; Floryszak-Wieczorek, J.; Gabryś, B.; Morkunas, I. Differential Induction of Pisum Sativum Defense Signaling Molecules in Response to Pea Aphid Infestation. Plant Sci. 2014, 221–222, 1–12. [Google Scholar] [CrossRef]
- Woźniak, A.; Formela, M.; Bilman, P.; Grześkiewicz, K.; Bednarski, W.; Marczak, Ł.; Narożna, D.; Dancewicz, K.; Mai, V.; Borowiak-Sobkowiak, B.; et al. The Dynamics of the Defense Strategy of Pea Induced by Exogenous Nitric Oxide in Response to Aphid Infestation. Int. J. Mol. Sci. 2017, 18, 329. [Google Scholar] [CrossRef]
- Xu, Y.; Qu, C.; Sun, X.; Jia, Z.; Xue, M.; Zhao, H.; Zhou, X. Nitric Oxide Boosts Bemisia Tabaci Performance Through the Suppression of Jasmonic Acid Signaling Pathway in Tobacco Plants. Front. Physiol. 2020, 11, 847. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Grewal, S.K.; Singh, R.; Bhardwaj, R.D. Induced Defense Dynamics in Plant Parts Is Requisite for Resistance to Helicoverpa Armigera (Hubner) Infestation in Chickpea. Phytoparasitica 2017, 45, 559–576. [Google Scholar] [CrossRef]
- Moloi, M.J.; van der Westhuizen, A.J. Involvement of Nitric Oxide in the Russian Wheat Aphid Resistance Response of Wheat. Cereal Res. Commun. 2014, 42, 119–125. [Google Scholar] [CrossRef]
- Alvarenga, R.; Auad, A.M.; Moraes, J.C.; Silva, S.E. Do Silicon and Nitric Oxide Induce Resistance to Mahanarva Spectabilis (Hemiptera: Cercopidae) in Forage Grasses? Pest Manag. Sci. 2019, 75, 3282–3292. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, J.; Jiang, L.; Wu, H.; Xiao, Y.; Liu, Y.; Li, G.; Du, Y.; Liu, C.; Wan, J. Nitric Oxide Production Is Associated with Response to Brown Planthopper Infestation in Rice. J. Plant Physiol. 2011, 168, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, M.E.; Arnaiz, A.; Rosa-Diaz, I.; González-Melendi, P.; Romero-Hernandez, G.; Ojeda-Martinez, D.A.; Garcia, A.; Contreras, E.; Martinez, M.; Diaz, I. Plant Defenses Against Tetranychus Urticae: Mind the Gaps. Plants 2020, 9, 464. [Google Scholar] [CrossRef]
- Santamaría, M.E.; Arnaiz, A.; Velasco-Arroyo, B.; Grbic, V.; Diaz, I.; Martinez, M. Arabidopsis Response to the Spider Mite Tetranychus Urticae Depends on the Regulation of Reactive Oxygen Species Homeostasis. Sci. Rep. 2018, 8, 9432. [Google Scholar] [CrossRef]
- Golan, K.; Jurado, I.G.; Kot, I.; Górska-Drabik, E.; Kmieć, K.; Łagowska, B.; Skwaryło-Bednarz, B.; Kopacki, M.; Jamiołkowska, A. Defense Responses in the Interactions between Medicinal Plants from Lamiaceae Family and the Two-Spotted Spider Mite Tetranychus Urticae Koch (Acari: Tetranychidae). Agronomy 2021, 11, 438. [Google Scholar] [CrossRef]
- Labudda, M.; Tokarz, K.; Tokarz, B.; Muszyńska, E.; Gietler, M.; Górecka, M.; Różańska, E.; Rybarczyk-Płońska, A.; Fidler, J.; Prabucka, B.; et al. Reactive Oxygen Species Metabolism and Photosynthetic Performance in Leaves of Hordeum Vulgare Plants Co-Infested with Heterodera Filipjevi and Aceria Tosichella. Plant Cell Rep. 2020, 39, 1719–1741. [Google Scholar] [CrossRef]
- Dworak, A.; Nykiel, M.; Walczak, B.; Miazek, A.; Szworst-Łupina, D.; Zagdańska, B.; Kiełkiewicz, M. Maize Proteomic Responses to Separate or Overlapping Soil Drought and Two-Spotted Spider Mite Stresses. Planta 2016, 244, 939–960. [Google Scholar] [CrossRef]
- Arbona, V.; Ximénez-Embún, M.G.; Echavarri-Muñoz, A.; Martin-Sánchez, M.; Gómez-Cadenas, A.; Ortego, F.; González-Guzmán, M. Early Molecular Responses of Tomato to Combined Moderate Water Stress and Tomato Red Spider Mite Tetranychus Evansi Attack. Plants 2020, 9, 1131. [Google Scholar] [CrossRef]
- Homayoonzadeh, M.; Moeini, P.; Talebi, K.; Allahyari, H.; Torabi, E.; Michaud, J.P. Physiological Responses of Plants and Mites to Salicylic Acid Improve the Efficacy of Spirodiclofen for Controlling Tetranychus Urticae (Acari: Tetranychidae) on Greenhouse Tomatoes. Exp. Appl. Acarol. 2020, 82, 319–333. [Google Scholar] [CrossRef]
- Santamaria, M.E.; Garcia, A.; Arnaiz, A.; Rosa-Diaz, I.; Romero-Hernandez, G.; Diaz, I.; Martinez, M. Comparative Transcriptomics Reveals Hidden Issues in the Plant Response to Arthropod Herbivores. J. Integr. Plant Biol. 2021, 63, 312–326. [Google Scholar] [CrossRef]
- Paspati, A.; Urbaneja, A.; González-Cabrera, J. Transcriptomic Profile of the Predatory Mite Amblyseius Swirskii (Acari: Phytoseiidae) on Different Host Plants. Exp. Appl. Acarol. 2022, 86, 479–498. [Google Scholar] [CrossRef]
- Maserti, B.E.; Del Carratore, R.; Croce, C.M.D.; Podda, A.; Migheli, Q.; Froelicher, Y.; Luro, F.; Morillon, R.; Ollitrault, P.; Talon, M.; et al. Comparative Analysis of Proteome Changes Induced by the Two Spotted Spider Mite Tetranychus Urticae and Methyl Jasmonate in Citrus Leaves. J. Plant Physiol. 2011, 168, 392–402. [Google Scholar] [CrossRef]

| NO Crosstalk | Influence | Effect | References |
|---|---|---|---|
| ROS | NO-H2O2 modulation of transcription factors | putative R-SNO and cysteine residue oxidation | [172] |
| NO-H2O2—MAPK phosphorylation | PCD activation | [172] | |
| R-SNO of NPR1 protein | SAR activation | [172,173,174] | |
| GSNO production by GSNOR | presence of NO reservoir under pathogen attack | [172,175,176] | |
| HR gene expression regulation | HR and PCD | [177,178,179] | |
| ABA | induction of (+)-ABA 8′-hydroxylase expression | ABA signaling inhibition (breaking seed dormancy) | [180] |
| Tyr nitration of PYR/PYL/RCAR | PYR/PYL/RCAR degradation by UPS | [181] | |
| R-SNO of Cys residue of SnRK2 | inhibition of SnRK2 | [157,158] | |
| R-SNO of Cys residue of ABI5 | degradation of ABI5 by UPS | [182] | |
| IAA | phosphorylation of CDPK | lateral/primary root growth | [183,184] |
| IAA-overproduction by Sinorhizobium meliloti (in presence of NO) | nodulation in Medicago species | [185] | |
| production of ROS | oxidized IAA | [186] | |
| SA | induction of defense genes expression | regulation of SA level during biotic stress | [34] |
| induction of NOS-like activity | NO synthesis | [187] | |
| molecular regulation of NPR gene expression | induction of SAR via NO-SA crosstalk | [188,189] | |
| accumulation of NPR1 | activation of PR genes | [190,191] | |
| modulation of SIPK | development of resistance to pathogen | [192,193] | |
| JA | NPR1 and TGA modifications | suppression of JA-dependent genes | [194] |
| interaction of NPR1 and basic-helix-loop-helix transcription factors MYC2-mediator complex subunit 25 | suppression of JA-dependent genes | [195] | |
| induction gene expression of LOX2 and OPR | JA synthesis | [196,197] | |
| R-SNO of AOC | decreased JA synthesis | [196,198] | |
| activation of ascorbate-glutathione cycle | plant growth improvement under drought | [49,199] |
| Parasite | Plant | Response | References |
|---|---|---|---|
| Nematodes | |||
| Meloidogyne incognita | Solanum lycopersicon | increased expression of NO- and JA-induced genes | [231] |
| Meloidogyne incognita | Solanum lycopersicon | NO-H2O2 crosstalk, PCD activation | [232] |
| Meloidogyne incognita | Solanum lycopersicon | NO-ROS crosstalk, increased NOS-like activity | [234] |
| Bursaphelenchus xylophilus | Pinus thunbergii | increased NOS-like activity | [235] |
| Heterodera schachtii | Arabidopsis thaliana | alteration in the level of RNS, protein R-SNO and nitration, and GSNOR | [236] |
| Insects | |||
| Acyrthosiphon pisum | Pisum sativum | interconnection of NO production with JA, ET, SA, H2O2 synthesis | [237] |
| Acyrthosiphon pisum | Pisum sativum | restriction of aphids’ reproduction | [238] |
| Bemisia tabaci | Nicotiana tabacum | suppression of JA-defense responses and favoring B. tabaci reproduction | [239] |
| Helicoverpa armigera | Cicer arietinum | changes in antioxidants enzymes, NO, H2O2, phenols and trypsin inhibitor | [240] |
| Diuraphis noxia | Triticum aestivum | changes in NR and NiR activities, NO-dependent induction of β-1,3-glucanase and peroxidase | [241] |
| Manduca sexta | Nicotiana attenuata | GSNOR interconnection with NO- and JA-dependent responses | [204] |
| Mahanarva spectabilis | Brachiaria ruziziensis, Pennisetum purpureum and Digitaria sp. | increased content of phenols, lack of inhibition of pest development cycle | [242] |
| Nilaparvata lugens | Oryza sativa | increased NO content in resistant cultivar, increased expression of genes related to drought response | [243] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graska, J.; Fidler, J.; Gietler, M.; Prabucka, B.; Nykiel, M.; Labudda, M. Nitric Oxide in Plant Functioning: Metabolism, Signaling, and Responses to Infestation with Ecdysozoa Parasites. Biology 2023, 12, 927. https://doi.org/10.3390/biology12070927
Graska J, Fidler J, Gietler M, Prabucka B, Nykiel M, Labudda M. Nitric Oxide in Plant Functioning: Metabolism, Signaling, and Responses to Infestation with Ecdysozoa Parasites. Biology. 2023; 12(7):927. https://doi.org/10.3390/biology12070927
Chicago/Turabian StyleGraska, Jakub, Justyna Fidler, Marta Gietler, Beata Prabucka, Małgorzata Nykiel, and Mateusz Labudda. 2023. "Nitric Oxide in Plant Functioning: Metabolism, Signaling, and Responses to Infestation with Ecdysozoa Parasites" Biology 12, no. 7: 927. https://doi.org/10.3390/biology12070927
APA StyleGraska, J., Fidler, J., Gietler, M., Prabucka, B., Nykiel, M., & Labudda, M. (2023). Nitric Oxide in Plant Functioning: Metabolism, Signaling, and Responses to Infestation with Ecdysozoa Parasites. Biology, 12(7), 927. https://doi.org/10.3390/biology12070927











