Recent Developments in CRISPR/Cas9 Genome-Editing Technology Related to Plant Disease Resistance and Abiotic Stress Tolerance
Abstract
Simple Summary
Abstract
1. Introduction
2. Genome Editing Tools and Their Comparison
3. Arise of CRISPR/Cas9 Technology
4. CRISPR/Cas-Mediated Genome Editing for Disease Resistance
5. CRISPR/Cas-Mediated Genome Editing for Abiotic Stress Tolerance
5.1. Drought Tolerance
5.2. Salt Tolerance
5.3. Cold Tolerance
5.4. Heat Tolerance
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging Infectious Diseases of Plants: Pathogen Pollution, Climate Change and Agrotechnology Drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global Food Demand and the Sustainable Intensification of Agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Bebber, D.P.; Gurr, S.J. Crop-Destroying Fungal and Oomycete Pathogens Challenge Food Security. Fungal Genet. Biol. 2015, 74, 62–64. [Google Scholar] [CrossRef]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Field, C.B.; Barros, V.R. (Eds.) Climate Change 2014–Impacts, Adaptation and Vulnerability: Regional Aspects; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Scheben, A.; Wolter, F.; Batley, J.; Puchta, H.; Edwards, D. Towards CRISPR/Cas Crops–Bringing Together Genomics and Genome Editing. New Phytol. 2017, 216, 682–698. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, M.; Sakina, A.; Wani, S.H.; Shikari, A.B.; Tripathi, P.; Zaid, A.; Salgotra, R.K. Harnessing genome editing techniques to engineer disease resistance in plants. Front. Plant Sci. 2019, 10, 550. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Kumar, R.; Kumar, V.; Won, S.Y.; Shukla, P. Engineering disease resistant plants through CRISPR-Cas9 technology. GM Crops Food 2021, 12, 125–144. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Mulla, S.I.; Lee, K.J.; Chae, J.C.; Shukla, P. VOCs-Mediated Hormonal Signaling and Crosstalk with Plant Growth Promoting Microbes. Crit. Rev. Biotechnol. 2018, 38, 1277–1296. [Google Scholar] [CrossRef] [PubMed]
- Vannier, N.; Agler, M.; Hacquard, S. Microbiota-mediated disease resistance in plants. PLoS Pathog. 2019, 15, e1007740. [Google Scholar] [CrossRef]
- Yin, K.; Qiu, J.L. Genome Editing for Plant Disease Resistance: Applications and Perspectives. Philos. Trans. R. Soc. B 2019, 374, 20180322. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Wei, X.; Sheng, Z.; Hu, P.; Tang, S. CRISPR/Cas9 for development of disease resistance in plants: Recent progress, limitations and future prospects. Brief. Funct. Genom. 2020, 19, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, N.; Douches, D.; Dhingra, A.; Glenn, K.C.; Herzig, P.R.; Stowe, E.C.; Swarup, S. The Role of Conventional Plant Breeding in Ensuring Safe Levels of Naturally Occurring Toxins in Food Crops. Trends Food Sci. Technol. 2020, 100, 51–66. [Google Scholar] [CrossRef]
- Foley, J.A.; Ramankutty, N.; Brauman, K.A.; Cassidy, E.S.; Gerber, J.S.; Johnston, M.; Zaks, D.P. Solutions for a Cultivated Planet. Nature 2011, 478, 337–342. [Google Scholar] [CrossRef]
- Zhang, Y.; Massel, K.; Godwin, I.D.; Gao, C. Applications and Potential of Genome Editing in Crop Improvement. Genome Biol. 2018, 19, 210. [Google Scholar] [CrossRef] [PubMed]
- Prado, J.R.; Segers, G.; Voelker, T.; Carson, D.; Dobert, R.; Phillips, J.; Martino-Catt, S. Genetically Engineered Crops: From Idea to Product. Annu. Rev. Plant Biol. 2014, 65, 769–790. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Gal-On, A. Development of Broad Virus Resistance in Non-transgenic Cucumber Using CRISPR/Cas9 Technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 System for Plant Genome Editing and Beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Pallarès Masmitjà, M.; Knödlseder, N.; Güell, M. CRISPR-GRNA Design. In CRISPR Gene Editing: Methods and Protocols; Humana: New York, NY, USA, 2019; pp. 3–11. [Google Scholar]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Almendros, C. Short Motif Sequences Determine the Targets of the Prokaryotic CRISPR Defence System. Microbiology 2009, 155, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wen, Y.; Guo, X. CRISPR/Cas9 for Genome Editing: Progress, Implications and Challenges. Hum. Mol. Genet. 2014, 23, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Karginov, F.V.; Hannon, G.J. The CRISPR System: Small RNA-Guided Defense in Bacteria and Archaea. Mol. Cell 2010, 37, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhu, Q.; Chen, Y.; Liu, Y.G. CRISPR/Cas9 Platforms for Genome Editing in Plants: Developments and Applications. Mol. Plant 2016, 9, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Kamburova, V.S.; Nikitina, E.V.; Shermatov, S.E.; Buriev, Z.T.; Kumpatla, S.P.; Emani, C.; Abdurakhmonov, I.Y. Genome Editing in Plants: An Overview of Tools and Applications. Int. J. Agron. 2017, 2017, 7315351. [Google Scholar] [CrossRef]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid Restriction Enzymes: Zinc Finger Fusions to Fok I Cleavage Domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Voytas, D.F. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef]
- Jankele, R.; Svoboda, P. TAL Effectors: Tools for DNA Targeting. Brief. Funct. Genom. 2014, 13, 409–419. [Google Scholar] [CrossRef]
- Palpant, N.J.; Dudzinski, D. Zinc Finger Nucleases: Looking toward Translation. Gene Ther. 2013, 20, 121–127. [Google Scholar] [CrossRef]
- Khandagale, K.; Nadaf, A. Genome Editing for Targeted Improvement of Plants. Plant Biotechnol. Rep. 2016, 10, 327–343. [Google Scholar] [CrossRef]
- Shah, T.; Andleeb, T.; Lateef, S.; Noor, M.A. Genome Editing in Plants: Advancing Crop Transformation and Overview of Tools. Plant Physiol. Biochem. 2018, 131, 12–21. [Google Scholar] [CrossRef]
- Gratz, S.J.; Harrison, M.M.; Wildonger, J.; O’Connor-Giles, K.M. Precise Genome Editing of Drosophila with CRISPR RNA-Guided Cas9. Methods Mol Biol. 2015, 1311, 335–348. [Google Scholar]
- Ledford, H. CRISPR, the Disruptor. Nature 2015, 522, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous Modification of Three Homoeologs of Ta EDR 1 by Genome Editing Enhances Powdery Mildew Resistance in Wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, S.; Xu, J.; Sui, C.; Wei, J. Application of CRISPR/Cas9 in plant biology. Acta Pharm. Sin. B 2017, 7, 292–302. [Google Scholar] [CrossRef]
- Collias, D.; Beisel, C.L. CRISPR technologies and the search for the PAM-free nuclease. Nat. Commun. 2021, 12, 555. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Osakabe, K.; Osakabe, Y. Expanding the plant genome editing toolbox with recently developed CRISPR–Cas systems. Plant Physiol. 2022, 188, 1825–1837. [Google Scholar] [CrossRef] [PubMed]
- Nayeemul Bari, S.M.; Hatoum-Aslan, A. CRISPR-Cas10 assisted editing of virulent staphylococcal phages. Methods Enzymol. 2019, 616, 385–409. [Google Scholar]
- Hillary, V.E.; Ceasar, S.A. A Review on the Mechanism and Applications of CRISPR/Cas9/Cas12/Cas13/Cas14 Proteins Utilized for Genome Engineering. Mol. Biotechnol. 2023, 65, 311–325. [Google Scholar] [CrossRef]
- Pan, C.; Li, G.; Malzahn, A.A.; Cheng, Y.; Leyson, B.; Sretenovic, S.; Gurel, F.; Coleman, G.D.; Qi, Y. Boosting plant genome editing with a versatile CRISPR-Combo system. Nat. Plants 2022, 8, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Díez-Villaseñor, C.; Soria, E.; Juez, G. Biological Significance of a Family of Regularly Spaced Repeats in the Genomes of Archaea, Bacteria and Mitochondria. Mol. Microbiol. 2000, 36, 244–246. [Google Scholar] [CrossRef]
- Jansen, R.; Embden, J.D.V.; Gaastra, W.; Schouls, L.M. Identification of Genes That Are Associated with DNA Repeats in Prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening Sequences of Regularly Spaced Prokaryotic Repeats Derive from Foreign Genetic Elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered Regularly Interspaced Short Palindrome Repeats (CRISPRs) Have Spacers of Extrachromosomal Origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef]
- Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR Elements in Yersinia Pestis Acquire New Repeats by Preferential Uptake of Bacteriophage DNA, and Provide Additional Tools for Evolutionary Studies. Microbiology 2005, 151, 653–663. [Google Scholar] [CrossRef]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing Plant Genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Charpentier, E. CRISPR RNA Maturation by Trans-Encoded Small RNA and Host Factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef]
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef]
- Deb, S.; Choudhury, A.; Kharbyngar, B.; Satyawada, R.R. Applications of CRISPR/Cas9 technology for modification of the plant genome. Genetica 2022, 150, 1–12. [Google Scholar] [CrossRef]
- Kor, S.D.; Chowdhury, N.; Keot, A.K.; Yogendra, K.; Chikkaputtaiah, C.; Sudhakar Reddy, P. RNA Pol III promoters—Key players in precisely targeted plant genome editing. Front. Genet. 2023, 13, 989199. [Google Scholar] [CrossRef]
- Savary, S.; Ficke, A.; Aubertot, J.N.; Hollier, C. Crop Losses Due to Diseases and Their Implications for Global Food Production Losses and Food Security. Food Secur. 2012, 4, 519–537. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- de Almeida Engler, J.; Favery, B.; Engler, G.; Abad, P. Loss of Susceptibility as an Alternative for Nematode Resistance. Curr. Opin. Biotechnol. 2005, 16, 112–117. [Google Scholar] [CrossRef]
- Schie, C.C.; Takken, F.L. Susceptibility Genes 101: How to Be a Good Host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef]
- Pavan, S.; Jacobsen, E.; Visser, R.G.; Bai, Y. Loss of Susceptibility as a Novel Breeding Strategy for Durable and Broad-Spectrum Resistance. Mol. Breed. 2010, 25, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, N.A. Plant Disease Susceptibility Genes? Plant Cell 2002, 14, 1983–1986. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, I.H. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 1992, 63, 141–152. [Google Scholar] [CrossRef]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; Schulze-Lefert, P. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef]
- Devoto, A.; Piffanelli, P.; Nilsson, I.; Wallin, E.; Panstruga, R.; Heijne, G.; Schulze-Lefert, P. Topology, Subcellular Localization, and Sequence Diversity of the Mlo Family in Plants. J. Biol. Chem. 1999, 274, 34993–35004. [Google Scholar] [CrossRef]
- Devoto, A.; Hartmann, H.A.; Piffanelli, P.; Elliott, C.; Simmons, C.; Taramino, G.; Panstruga, R. Molecular Phylogeny and Evolution of the Plant-Specific Seven-Transmembrane MLO Family. J. Mol. Evol. 2003, 56, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Gao, C. Targeted Genome Modification of Crop Plants Using a CRISPR-Cas System. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous Editing of Three Homoeoalleles in Hexaploid Bread Wheat Confers Heritable Resistance to Powdery Mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid Generation of a Transgene-Free Powdery Mildew Resistant Tomato by Genome Deletion. Sci. Rep. 2017, 7, 482. [Google Scholar] [CrossRef] [PubMed]
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Panstruga, R. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 2006, 38, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Pavan, S.; Zheng, Z.; Zappel, N.F.; Reinstädler, A.; Lotti, C.; Panstruga, R. Naturally Occurring Broad-Spectrum Powdery Mildew Resistance in a Central American Tomato Accession Is Caused by Loss of Mlo Function. Mol. Plant-Microbe Interact. 2008, 21, 30–39. [Google Scholar] [CrossRef]
- Humphry, M.; Reinstaedler, A.; Ivanov, S.; Bisseling, T.O.N.; Panstruga, R. Durable Broad-spectrum Powdery Mildew Resistance in Pea Er1 Plants Is Conferred by Natural Loss-of-function Mutations in PsMLO1. Mol. Plant Pathol. 2011, 12, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Nonomura, T.; Appiano, M.; Pavan, S.; Matsuda, Y.; Toyoda, H.; Bai, Y. Loss of Function in Mlo Orthologs Reduces Susceptibility of Pepper and Tomato to Powdery Mildew Disease Caused by Leveillula Taurica. PLoS ONE 2013, 8, e70723. [Google Scholar] [CrossRef]
- Appiano, M.; Pavan, S.; Catalano, D.; Zheng, Z.; Bracuto, V.; Lotti, C.; Bai, Y. Identification of Candidate MLO Powdery Mildew Susceptibility Genes in Cultivated Solanaceae and Functional Characterization of Tobacco NtMLO1. Transgenic Res. 2015, 24, 847–858. [Google Scholar] [CrossRef] [PubMed]
- Feechan, A.; Jermakow, A.M.; Torregrosa, L.; Panstruga, R.; Dry, I.B. Identification of Grapevine MLO Gene Candidates Involved in Susceptibility to Powdery Mildew. Funct. Plant Biol. 2008, 35, 1255–1266. [Google Scholar] [CrossRef]
- Zaidi, S.S.E.A.; Mukhtar, M.S.; Mansoor, S. Genome Editing: Targeting Susceptibility Genes for Plant Disease Resistance. Trends Biotechnol. 2018, 36, 898–906. [Google Scholar] [CrossRef]
- Peterson, B.A.; Haak, D.C.; Nishimura, M.T.; Teixeira, P.J.; James, S.R.; Dangl, J.L.; Nimchuk, Z.L. Genome-Wide Assessment of Efficiency and Specificity in CRISPR/Cas9 Mediated Multiple Site Targeting in Arabidopsis. PLoS ONE 2016, 11, e0162169. [Google Scholar] [CrossRef]
- Borrelli, V.M.; Brambilla, V.; Rogowsky, P.; Marocco, A.; Lanubile, A. The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front. Plant Sci. 2018, 9, 1245. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, L.; Huang, X. Genome Modification by CRISPR/Cas9. FEBS J. 2014, 281, 5186–5193. [Google Scholar] [CrossRef]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Chen, Q.J. A CRISPR/Cas9 Toolkit for Multiplex Genome Editing in Plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large Chromosomal Deletions and Heritable Small Genetic Changes Induced by CRISPR/Cas9 in Rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Yang, J. Rapid Improvement of Grain Weight via Highly Efficient CRISPR/Cas9-Mediated Multiplex Genome Editing in Rice. J. Genet. Genom. = Yi Chuan Xue Bao 2016, 43, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Pyott, D.E.; Sheehan, E.; Molnar, A. Engineering of CRISPR/Cas9-mediated Potyvirus Resistance in Transgene-free Arabidopsis Plants. Mol. Plant Pathol. 2016, 17, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Chen, J.; Dai, X.; Zhang, D.; Zhao, Y. An Effective Strategy for Reliably Isolating Heritable and Cas9-Free Arabidopsis Mutants Generated by CRISPR/Cas9-Mediated Genome Editing. Plant Physiol. 2016, 171, 1794–1800. [Google Scholar] [CrossRef]
- Bastet, A.; Robaglia, C.; Gallois, J.L. eIF4E Resistance: Natural Variation Should Guide Gene Editing. Trends Plant Sci. 2017, 22, 411–419. [Google Scholar] [CrossRef]
- Stover, E.D.; Driggers, R.; Richardson, M.L.; Hall, D.G.; Duan, Y.; Lee, R.F. Incidence and Severity of Asiatic Citrus Canker on Diverse Citrus and Citrus-Related Germplasm in a Florida Field Planting. HortScience 2014, 49, 4–9. [Google Scholar] [CrossRef]
- Zhou, J.; Peng, Z.; Long, J.; Sosso, D.; Liu, B.O.; Eom, J.S.; Yang, B. Gene Targeting by the TAL Effector PthXo2 Reveals Cryptic Resistance Gene for Bacterial Blight of Rice. Plant J. 2015, 82, 632–643. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Nagamangala Kanchiswamy, C. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Santillán Martínez, M.I.; Bracuto, V.; Koseoglou, E.; Appiano, M.; Jacobsen, E.; Visser, R.G.F.; Wolters, A.A.; Bai, Y. CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 2020, 20, 284. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 2015, 1, 15144. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, J.N.; Ntui, V.O.; Ron, M.; Muiruri, S.K.; Britt, A.; Tripathi, L. CRISPR/Cas9 Editing of Endogenous Banana Streak Virus in the B Genome of Musa spp. Overcomes a Major Challenge in Banana Breeding. Commun. Biol. 2019, 2, 46. [Google Scholar] [CrossRef] [PubMed]
- Kis, A.; Hamar, É.; Tholt, G.; Bán, R.; Havelda, Z. Creating Highly Efficient Resistance against Wheat Dwarf Virus in Barley by Employing CRISPR/Cas9 System. Plant Biotechnol. J. 2019, 17, 1004. [Google Scholar] [CrossRef]
- Fister, A.S.; Landherr, L.; Maximova, S.N.; Guiltinan, M.J. Transient Expression of CRISPR/Cas9 Machinery Targeting TcNPR3 Enhances Defense Response in Theobroma cacao. Front. Plant Sci. 2018, 9, 268. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, Y.; Orbović, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome Editing of the Disease Susceptibility Gene Cs LOB 1 in Citrus Confers Resistance to Citrus Canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Zou, X. Engineering Canker-resistant Plants through CRISPR/Cas9-targeted Editing of the Susceptibility Gene Cs LOB 1 Promoter in Citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, X. CRISPR/Cas9-mediated Efficient Targeted Mutagenesis in Grape in the First Generation. Plant Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef]
- Gumtow, R.; Wu, D.; Uchida, J.; Tian, M. A Phytophthora Palmivora Extracellular Cystatin-like Protease Inhibitor Targets Papain to Contribute to Virulence on Papaya. Mol. Plant-Microbe Interact. 2018, 31, 363–373. [Google Scholar] [CrossRef]
- Oliva, R.; Ji, C.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Yang, B. Broad-Spectrum Resistance to Bacterial Blight in Rice Using Genome Editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef]
- Kim, Y.A.; Moon, H.; Park, C.J. CRISPR/Cas9-Targeted Mutagenesis of Os8N3 in Rice to Confer Resistance to Xanthomonas oryzae pv. oryzae. Rice 2019, 12, 67. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, X.; Gong, Q.; Li, Z.; Li, Y.; Wang, S.; Chen, G. Engineering Broad-Spectrum Bacterial Blight Resistance by Simultaneously Disrupting Variable TALE-Binding Elements of Multiple Susceptibility Genes in Rice. Mol. Plant 2019, 12, 1434–1446. [Google Scholar] [CrossRef]
- Li, S.; Shen, L.; Hu, P.; Liu, Q.; Zhu, X.; Qian, Q.; Wang, Y. Developing Disease-resistant Thermosensitive Male Sterile Rice by Multiplex Gene Editing. J. Integr. Plant Biol. 2019, 61, 1201–1205. [Google Scholar] [CrossRef]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Zhao, K. Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef]
- Xie, K.; Yang, Y. RNA-Guided Genome Editing in Plants Using a CRISPR–Cas System. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef]
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Čermák, T.; Chadha-Mohanty, P. Novel Alleles of Rice EIF4G Generated by CRISPR/Cas9-targeted Mutagenesis Confer Resistance to Rice Tungro Spherical Virus. Plant Biotechnol. J. 2018, 16, 1918–1927. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Han, T.; Xie, K.; Zhao, J.; Song, J.; Liu, Y. Engineer Complete Resistance to Cotton Leaf Curl Multan Virus by the CRISPR/Cas9 System in Nicotiana benthamiana. Phytopathol. Res. 2019, 1, 9. [Google Scholar] [CrossRef]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-Mediated Viral Interference in Plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef]
- Baltes, N.J.; Hummel, A.W.; Konecna, E.; Cegan, R.; Bruns, A.N.; Bisaro, D.M.; Voytas, D.F. Conferring Resistance to Geminiviruses with the CRISPR–Cas Prokaryotic Immune System. Nat. Plants 2015, 1, 15145. [Google Scholar] [CrossRef] [PubMed]
- Tashkandi, M.; Ali, Z.; Aljedaani, F.; Shami, A.; Mahfouz, M.M. Engineering Resistance against Tomato Yellow Leaf Curl Virus via the CRISPR/Cas9 System in Tomato. Plant Signal. Behav. 2018, 13, 1525996. [Google Scholar] [CrossRef]
- Thomazella, D.P.T.; Seong, K.; Mackelprang, R.; Dahlbeck, D.; Geng, Y.; Gill, U.S.; Qi, T.; Pham, J.; Giuseppe, P.; Lee, C.Y.; et al. Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2021, 6, 118. [Google Scholar] [CrossRef]
- Ortigosa, A.; Gimenez-Ibanez, S.; Leonhardt, N.; Solano, R. Design of a Bacterial Speck Resistant Tomato by CRISPR/Cas9-mediated Editing of Sl JAZ 2. Plant Biotechnol. J. 2019, 17, 665–673. [Google Scholar] [CrossRef]
- Prihatna, C.; Barbetti, M.J.; Barker, S.J. A Novel Tomato Fusarium Wilt Tolerance Gene. Front. Microbiol. 2018, 9, 1226. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Zhao, R.; Yu, W.; Li, R.; Li, Y.; Shen, L. Knockout of SlMAPK3 Reduced Disease Resistance to Botrytis cinerea in Tomato Plants. J. Agric. Food Chem. 2018, 66, 8949–8956. [Google Scholar] [CrossRef]
- Wang, Z.; Hardcastle, T.J.; Pastor, A.C.; Yip, W.H.; Tang, S.; Baulcombe, D.C. A Novel DCL2-Dependent MiRNA Pathway in Tomato Affects Susceptibility to RNA Viruses. Genes Dev. 2018, 32, 1155–1160. [Google Scholar] [CrossRef]
- Polle, A. Dissecting the Superoxide Dismutase-Ascorbate-Glutathione-Pathway in Chloroplasts by Metabolic Modeling. Computer Simulations as a Step towards Flux Analysis. Plant Physiol. 2001, 126, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xiong, L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liang, Z. CRISPR Technology for Abiotic Stress Resistant Crop Breeding. Plant Growth Regul. 2021, 94, 115–129. [Google Scholar] [CrossRef]
- Zafar, S.A.; Zaidi, S.S.E.A.; Gaba, Y.; Singla-Pareek, S.L.; Dhankher, O.P.; Li, X.; Pareek, A. Engineering Abiotic Stress Tolerance via CRISPR/Cas-Mediated Genome Editing. J. Exp. Bot. 2020, 71, 470–479. [Google Scholar] [CrossRef]
- Ilyas, M.; Nisar, M.; Khan, N.; Hazrat, A.; Khan, A.H.; Hayat, K.; Fahad, S.; Khan, A.; Ullah, A. Drought Tolerance Strategies in Plants: A Mechanistic Approach. J. Plant Growth Regul. 2021, 40, 926–944. [Google Scholar] [CrossRef]
- Hanna, S.H.S.; Osborne-Lee, I.W.; Cesaretti, G.P.; Misso, R.; Khalil, M.T. Ecological Agro-Ecosystem Sustainable Development in Relationship to Other Sectors in the Economic System, and Human Ecological Footprint and Imprint. Agric. Agric. Sci. Procedia 2016, 8, 17–30. [Google Scholar] [CrossRef]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 Genome Editing in Wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 Confers Abscisic Acid Sensitivity and Tolerance to Drought Stress in Rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liu, C.; Zhao, R.; Wang, L.; Chen, L.; Yu, W.; Shen, L. CRISPR/Cas9-Mediated SlNPR1 Mutagenesis Reduces Tomato Plant Drought Tolerance. BMC Plant Biol. 2019, 19, 38. [Google Scholar] [CrossRef] [PubMed]
- Ganie, S.A.; Wani, S.H.; Henry, R.; Hensel, G. Improving Rice Salt Tolerance by Precision Breeding in a New Era. Curr. Opin. Plant Biol. 2021, 60, 101996. [Google Scholar] [CrossRef]
- Santosh Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 Mediated Genome Editing of Drought and Salt Tolerance (OsDST) Gene in Indica Mega Rice Cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Mishra, R.; Joshi, R.K.; Zhao, K. Genome editing in rice: Recent advances, challenges, and future implications. Front. Plant Sci. 2018, 9, 1361. [Google Scholar] [CrossRef]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Habben, J.E. ARGOS 8 Variants Generated by CRISPR-Cas9 Improve Maize Grain Yield under Field Drought Stress Conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef]
- Wang, C.T.; Ru, J.N.; Liu, Y.W.; Yang, J.F.; Li, M.; Xu, Z.S.; Fu, J.D. The Maize WRKY Transcription Factor ZmWRKY40 Confers Drought Resistance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2580. [Google Scholar] [CrossRef]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced Drought Tolerance by CRISPR/Cas9-Mediated SlMAPK3 Mutagenesis in Tomato Plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Testerink, C. Tuning Plant Signaling and Growth to Survive Salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.R.; Fu, C.Y.; Fan, Z.L.; Chen, Y.; Chen, W.F.; Zhang, J.; Li, C. Genomic and Transcriptomic Analysis Reveal Molecular Basis of Salinity Tolerance in a Novel Strong Salt-Tolerant Rice Landrace Changmaogu. Rice 2019, 12, 99. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.B.; Li, J.; Qin, R.Y.; Xu, R.F.; Li, H.; Yang, Y.C.; Yang, J.B. Identification of a Regulatory Element Responsible for Salt Induction of Rice OsRAV2 through Ex Situ and in Situ Promoter Analysis. Plant Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Ma, Y.Z. Identification and Characterization of GmMYB118 Responses to Drought and Salt Stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Yu, D. The Sucrose Non-Fermenting-1-Related Protein Kinases SAPK1 and SAPK2 Function Collaboratively as Positive Regulators of Salt Stress Tolerance in Rice. BMC Plant Biol. 2018, 18, 203. [Google Scholar] [CrossRef] [PubMed]
- Bouzroud, S.; Gasparini, K.; Hu, G.; Barbosa, M.A.M.; Rosa, B.L.; Fahr, M.; Zouine, M. Down Regulation and Loss of Auxin Response Factor 4 Function Using CRISPR/Cas9 Alters Plant Growth, Stomatal Function and Improves Tomato Tolerance to Salinity and Osmotic Stress. Genes 2020, 11, 272. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Chen, S.; Cheng, Y.; Zhang, N.; Ma, Y.; Wang, W.; Wang, S. Cell Wall/Vacuolar Inhibitor of Fructosidase 1 Regulates ABA Response and Salt Tolerance in Arabidopsis. Plant Signal. Behav. 2020, 15, 1744293. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Wang, L.; Chen, L.; Zhao, R.; Sheng, J.; Shen, L. Reduction of Tomato-Plant Chilling Tolerance by CRISPR–Cas9-Mediated SlCBF1 Mutagenesis. J. Agric. Food Chem. 2018, 66, 9042–9051. [Google Scholar] [CrossRef]
- Shen, C.; Que, Z.; Xia, Y.; Tang, N.; Li, D.; He, R.; Cao, M. Knock out of the Annexin Gene OsAnn3 via CRISPR/Cas9-Mediated Genome Editing Decreased Cold Tolerance in Rice. J. Plant Biol. 2017, 60, 539–547. [Google Scholar] [CrossRef]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 with the CRISPR–Cas9 System. Front. Plant Sci. 2020, 10, 1663. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.; Singh, M.; Rai, A.C. Effect of Heat-Shock Induced Oxidative Stress Is Suppressed in BcZAT12 Expressing Drought Tolerant Tomato. Phytochemistry 2013, 95, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Kang, S.; He, L.; Zhao, J.; Zhang, S.; Hu, J.; Zhu, L. The Newly Identified Heat-Stress Sensitive Albino 1 Gene Affects Chloroplast Development in Rice. Plant Sci. 2018, 267, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Qin, K.; Song, X.; Zhang, Q.; Zhou, Y.; Xia, X.; Yu, J. BZR1 Transcription Factor Regulates Heat Stress Tolerance through FERONIA Receptor-like Kinase-Mediated Reactive Oxygen Species Signaling in Tomato. Plant Cell Physiol. 2018, 59, 2239–2254. [Google Scholar] [CrossRef] [PubMed]
- Klap, C.; Yeshayahou, E.; Bolger, A.M.; Arazi, T.; Gupta, S.K.; Shabtai, S.; Barg, R. Tomato Facultative Parthenocarpy Results from Sl AGAMOUS-LIKE 6 Loss of Function. Plant Biotechnol. J. 2017, 15, 634–647. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Si, X.; Tian, Y.; Chen, K.; Liu, J.; Gao, C. Generation of Thermosensitive Male-Sterile Maize by Targeted Knockout of the ZmTMS5 Gene. J. Genet. Genom. = Yi Chuan Xue Bao 2017, 44, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhong, Z.; Wang, X.; Han, X.; Yu, D.; Wang, C.; Zhang, Y. Knockout of the OsNAC006 Transcription Factor Causes Drought and Heat Sensitivity in Rice. Int. J. Mol. Sci. 2020, 21, 2288. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yan, G.; Duan, Z.; Wang, Z.; Kang, C.; Guo, L.; Dai, C. Roles of the Brassica napus DELLA Protein BnaA6.RGA, in Modulating Drought Tolerance by Interacting with the ABA Signaling Component BnaA10.ABF2. Front. Plant Sci. 2020, 11, 577. [Google Scholar] [CrossRef]
- Liao, S.; Qin, X.; Luo, L.; Han, Y.; Wang, X.; Usman, B.; Li, R. CRISPR/Cas9-Induced Mutagenesis of Semi-Rolled Leaf1, 2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.). Agronomy 2019, 9, 728. [Google Scholar] [CrossRef]
- Shim, J.S.; Oh, N.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Kim, J.K. Overexpression of OsNAC14 Improves Drought Tolerance in Rice. Front. Plant Sci. 2018, 9, 310. [Google Scholar]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Zhu, J.K. The CRISPR/C As9 System Produces Specific and Homozygous Targeted Gene Editing in Rice in One Generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Roca Paixão, J.F.; Gillet, F.X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; Grossi-de-Sa, M.F. Improved Drought Stress Tolerance in Arabidopsis by CRISPR/DCas9 Fusion with a Histone AcetylTransferase. Sci. Rep. 2019, 9, 8080. [Google Scholar] [CrossRef]
- Park, J.J.; Dempewolf, E.; Zhang, W.; Wang, Z.Y. RNA-Guided Transcriptional Activation via CRISPR/DCas9 Mimics Overexpression Phenotypes in Arabidopsis. PLoS ONE 2017, 12, e0179410. [Google Scholar] [CrossRef]
- Osakabe, Y.; Watanabe, T.; Sugano, S.S.; Ueta, R.; Ishihara, R.; Shinozaki, K.; Osakabe, K. Optimization of CRISPR/Cas9 Genome Editing to Modify Abiotic Stress Responses in Plants. Sci. Rep. 2016, 6, 26685. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational Evidence for the Critical Role of CBF Transcription Factors in Cold Acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.; Wang, X.; Han, X.; An, Y.; Lin, S.; Xia, X. Root-specific NF-Y Family Transcription Factor, PdNF-YB21, Positively Regulates Root Growth and Drought Resistance by Abscisic Acid-mediated Indoylacetic Acid Transport in Populus. New Phytol. 2020, 227, 407–426. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, Contribute to Cold, Salt and Drought Stress Tolerance via Modulating Anthocyanin Accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, G.; Han, Y.; Usman, B.; Liu, F.; Qin, B.; Li, R. Knockout of OsPRP1, a Gene Encoding Proline-Rich Protein, Confers Enhanced Cold Sensitivity in Rice (Oryza sativa L.) at the Seedling Stage. 3 Biotech 2019, 9, 254. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, B.J.; Park, S.; Jameel, M.I.; Kraft, J.C.; Thomashow, M.F.; Schemske, D.W.; Oakley, C.G. Genetic and Physiological Mechanisms of Freezing Tolerance in Locally Adapted Populations of a Winter Annual. Am. J. Bot. 2020, 107, 250–261. [Google Scholar] [CrossRef]
- Park, S.; Gilmour, S.J.; Grumet, R.; Thomashow, M.F. CBF-Dependent and CBF-Independent Regulatory Pathways Contribute to the Differences in Freezing Tolerance and Cold-Regulated Gene Expression of Two Arabidopsis Ecotypes Locally Adapted to Sites in Sweden and Italy. PLoS ONE 2018, 13, e0207723. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Huang, J.; Sun, T.; Wang, X.; Zhu, C.; Ai, Y.; Gu, H. The Precise Regulation of Different COR Genes by Individual CBF Transcription Factors in Arabidopsis thaliana. J. Integr. Plant Biol. 2017, 59, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The Cbfs Triple Mutants Reveal the Essential Functions of CBF s in Cold Acclimation and Allow the Definition of CBF Regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, D.; Shan, T.; Xu, S.; Qin, R.; Li, H.; Li, J. The Trihelix Transcription Factor OsGTγ-2 Is Involved Adaption to Salt Stress in Rice. Plant Mol. Biol. 2020, 103, 545–560. [Google Scholar] [CrossRef]
- Mo, W.; Tang, W.; Du, Y.; Jing, Y.; Bu, Q.; Lin, R. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 Interaction Controls Seedling Growth under Salt Stress. Plant Physiol. 2020, 184, 506–517. [Google Scholar] [CrossRef]
- Alfatih, A.; Wu, J.; Jan, S.U.; Zhang, Z.S.; Xia, J.Q.; Xiang, C.B. Loss of Rice PARAQUAT TOLERANCE 3 Confers Enhanced Resistance to Abiotic Stresses and Increases Grain Yield in Field. Plant Cell Environ. 2020, 43, 2743–2754. [Google Scholar] [CrossRef]
- Yin, W.; Xiao, Y.; Niu, M.; Meng, W.; Li, L.; Zhang, X.; Tong, H. ARGONAUTE2 Enhances Grain Length and Salt Tolerance by Activating BIG GRAIN3 to Modulate Cytokinin Distribution in Rice. Plant Cell 2020, 32, 2292–2306. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Hu, J.; Dong, L.; Zeng, D.; Guo, L.; Zhang, G.; Qian, Q. The Tolerance of Salinity in Rice Requires the Presence of a Functional Copy of FLN2. Biomolecules 2019, 10, 17. [Google Scholar] [CrossRef]
- Wang, W.C.; Lin, T.C.; Kieber, J.; Tsai, Y.C. Response Regulators 9 and 10 Negatively Regulate Salinity Tolerance in Rice. Plant Cell Physiol. 2019, 60, 2549–2563. [Google Scholar] [CrossRef]
- Qin, H.; Wang, J.; Chen, X.; Wang, F.; Peng, P.; Zhou, Y.; Huang, R. Rice Os DOF 15 Contributes to Ethylene-inhibited Primary Root Elongation under Salt Stress. New Phytol. 2019, 223, 798–813. [Google Scholar] [CrossRef]
- Lan, T.; Zheng, Y.; Su, Z.; Yu, S.; Song, H.; Zheng, X.; Wu, W. OsSPL10, a SBP-Box Gene, Plays a Dual Role in Salt Tolerance and Trichome Formation in Rice (Oryza sativa L.). G3 Genes Genomes Genet. 2019, 9, 4107–4114. [Google Scholar] [CrossRef]
- Liu, J.; Cui, L.; Xie, Z.; Zhang, Z.; Liu, E.; Peng, X. Two NCA1 Isoforms Interact with Catalase in a Mutually Exclusive Manner to Redundantly Regulate Its Activity in Rice. BMC Plant Biol. 2019, 19, 105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Luo, L. Enhanced Rice Salinity Tolerance via CRISPR/Cas9-Targeted Mutagenesis of the OsRR22 Gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhong, Z.; Zhang, H.; Wang, X.; Liu, B.; Yang, L.; Han, X.; Yu, D.; Zheng, X.; Wang, C.; et al. Targeted Mutagenesis of NAC Transcription Factor Gene, OsNAC041, Leading to Salt Sensitivity in Rice. Rice Sci. 2019, 26, 98–108. [Google Scholar]
- Zhang, C.; Srivastava, A.K.; Sadanandom, A. Targeted Mutagenesis of the SUMO Protease, Overly Tolerant to Salt1 in Rice through CRISPR/Cas9-Mediated Genome Editing Reveals a Major Role of This SUMO Protease in Salt Tolerance. BioRxiv 2019, 555706. [Google Scholar] [CrossRef]
- Zeng, D.D.; Yang, C.C.; Qin, R.; Alamin, M.; Yue, E.K.; Jin, X.L.; Shi, C.H. A Guanine Insert in OsBBS1 Leads to Early Leaf Senescence and Salt Stress Sensitivity in Rice (Oryza sativa L.). Plant Cell Rep. 2018, 37, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Qiu, J.; Agarwal, G.; Wang, J.; Ren, X.; Xia, H.; Wang, X. Genome-Wide Discovery of Microsatellite Markers from Diploid Progenitor Species, Arachis duranensis and A. ipaensis, and Their Application in Cultivated Peanut (A. hypogaea). Front. Plant Sci. 2017, 8, 1209. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.Q.; Shi, J.; Liang, X.; Jiang, C. A Retrotransposon in an HKT1 Family Sodium Transporter Causes Variation of Leaf Na+ Exclusion and Salt Tolerance in Maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Qi, M.; Ding, X.; Zheng, Y.; Zhou, T.; Chen, Y.; Wang, J. The SAUR41 Subfamily of SMALL AUXIN UP RNA Genes Is Abscisic Acid Inducible to Modulate Cell Expansion and Salt Tolerance in Arabidopsis thaliana Seedlings. Ann. Bot. 2020, 125, 805–819. [Google Scholar] [CrossRef]
- Huang, Y.; Cao, H.; Yang, L.; Chen, C.; Shabala, L.; Xiong, M.; Shabala, S. Tissue-Specific Respiratory Burst Oxidase Homolog-Dependent H2O2 Signaling to the Plasma Membrane H+-ATPase Confers Potassium Uptake and Salinity Tolerance in Cucurbitaceae. J. Exp. Bot. 2019, 70, 5879–5893. [Google Scholar] [CrossRef]
- Vlcko, T.; Ohnoutkova, L. Allelic Variants of CRISPR/Cas9 Induced Mutation in an Inositol Trisphosphate 5/6 Kinase Gene Manifest Different Phenotypes in Barley. Plants 2020, 9, 195. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xun, H.; Wang, W.; Ding, X.; Tian, H.; Hussain, S.; Wang, S. Mutation of GmAITR Genes by CRISPR/Cas9 Genome Editing Results in Enhanced Salinity Stress Tolerance in Soybean. Front. Plant Sci. 2021, 12, 779598. [Google Scholar] [CrossRef]
- Dubiel, M.; Beeckman, T.; Smagghe, G.; Damme, E.J. Arabidopsis Lectin EULS3 Is Involved in ABA Signaling in Roots. Front. Plant Sci. 2020, 11, 437. [Google Scholar] [CrossRef] [PubMed]
- Nandy, S.; Pathak, B.; Zhao, S.; Srivastava, V. Heat-shock-inducible CRISPR/Cas9 System Generates Heritable Mutations in Rice. Plant Direct 2019, 3, 00145. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xuan, H.; Yang, C.; Guo, N.; Wang, H.; Zhao, J.; Xing, H. GmHsp90A2 Is Involved in Soybean Heat Stress as a Positive Regulator. Plant Sci. 2019, 285, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Li, J.; Ding, S.; Cheng, F.; Li, X.; Jiang, Y.; Shi, K. The Protein Kinase CPK28 Phosphorylates Ascorbate Peroxidase and Enhances Thermotolerance in Tomato. Plant Physiol. 2021, 186, 1302–1317. [Google Scholar] [CrossRef] [PubMed]
- Bertier, L.D.; Ron, M.; Huo, H.; Bradford, K.J.; Britt, A.B.; Michelmore, R.W. High-Resolution Analysis of the Efficiency, Heritability, and Editing Outcomes of CRISPR/Cas9-Induced Modifications of NCED4 in Lettuce (Lactuca sativa). G3 Genes Genomes Genet. 2018, 8, 1513–1521. [Google Scholar] [CrossRef]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J.K. Mutations in a Subfamily of Abscisic Acid Receptor Genes Promote Rice Growth and Productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef]
- Zhang, D.; Li, Z.; Li, J.F. Targeted Gene Manipulation in Plants Using the CRISPR/Cas Technology. J. Genet. Genom. 2016, 43, 251–262. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Gao, C. Efficient and Transgene-Free Genome Editing in Wheat through Transient Expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef]
- Ceasar, S.A.; Rajan, V.; Prykhozhij, S.V.; Berman, J.N.; Ignacimuthu, S. Insert, Remove or Replace: A Highly Advanced Genome Editing System Using CRISPR/Cas9. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2016, 1863, 2333–2344. [Google Scholar] [CrossRef]
- Pan, C.; Ye, L.; Qin, L.; Liu, X.; He, Y.; Wang, J.; Lu, G. CRISPR/Cas9-Mediated Efficient and Heritable Targeted Mutagenesis in Tomato Plants in the First and Later Generations. Sci. Rep. 2016, 6, 24765. [Google Scholar] [CrossRef]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.; Østergaard, L.; Patron, N.; Harwood, W. Induction of Targeted, Heritable Mutations in Barley and Brassica Oleracea Using RNA-Guided Cas9 Nuclease. Genome Biol. 2015, 16, 258. [Google Scholar] [CrossRef]
- Paul, J.W.; Qi, Y. CRISPR/Cas9 for Plant Genome Editing: Accomplishments, Problems and Prospects. Plant Cell Rep. 2016, 35, 1417–1427. [Google Scholar] [CrossRef]
- Rani, R.; Yadav, P.; Barbadikar, K.M.; Baliyan, N.; Malhotra, E.V.; Singh, B.K.; Singh, D. CRISPR/Cas9: A Promising Way to Exploit Genetic Variation in Plants. Biotechnol. Lett. 2016, 38, 1991–2006. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, J.S. DNA-Free Genome Editing in Plants with Preassembled CRISPR-Cas9 Ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Svitashev, S.; Schwartz, C.; Lenderts, B.; Young, J.K.; Mark Cigan, A. Genome Editing in Maize Directed by CRISPR–Cas9 Ribonucleoprotein Complexes. Nat. Commun. 2016, 7, 13274. [Google Scholar] [CrossRef] [PubMed]
- Capdeville, N.; Schindele, P.; Puchta, H. Getting better all the time—Recent progress in the development of CRISPR/Cas-based tools for plant genome engineering. Curr. Opin. Biotechnol. 2023, 79, 102854. [Google Scholar] [CrossRef] [PubMed]
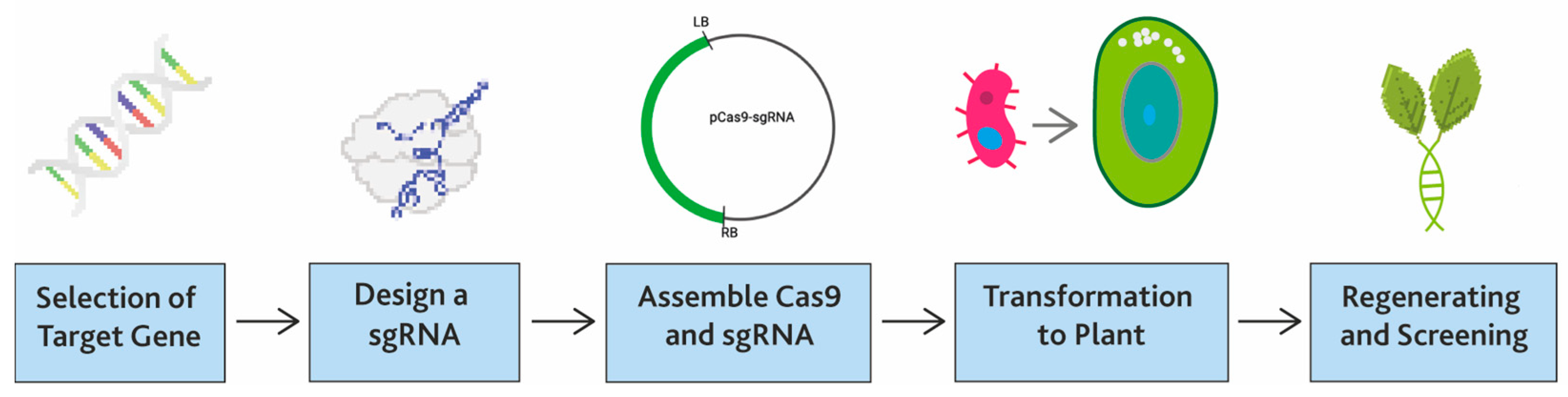
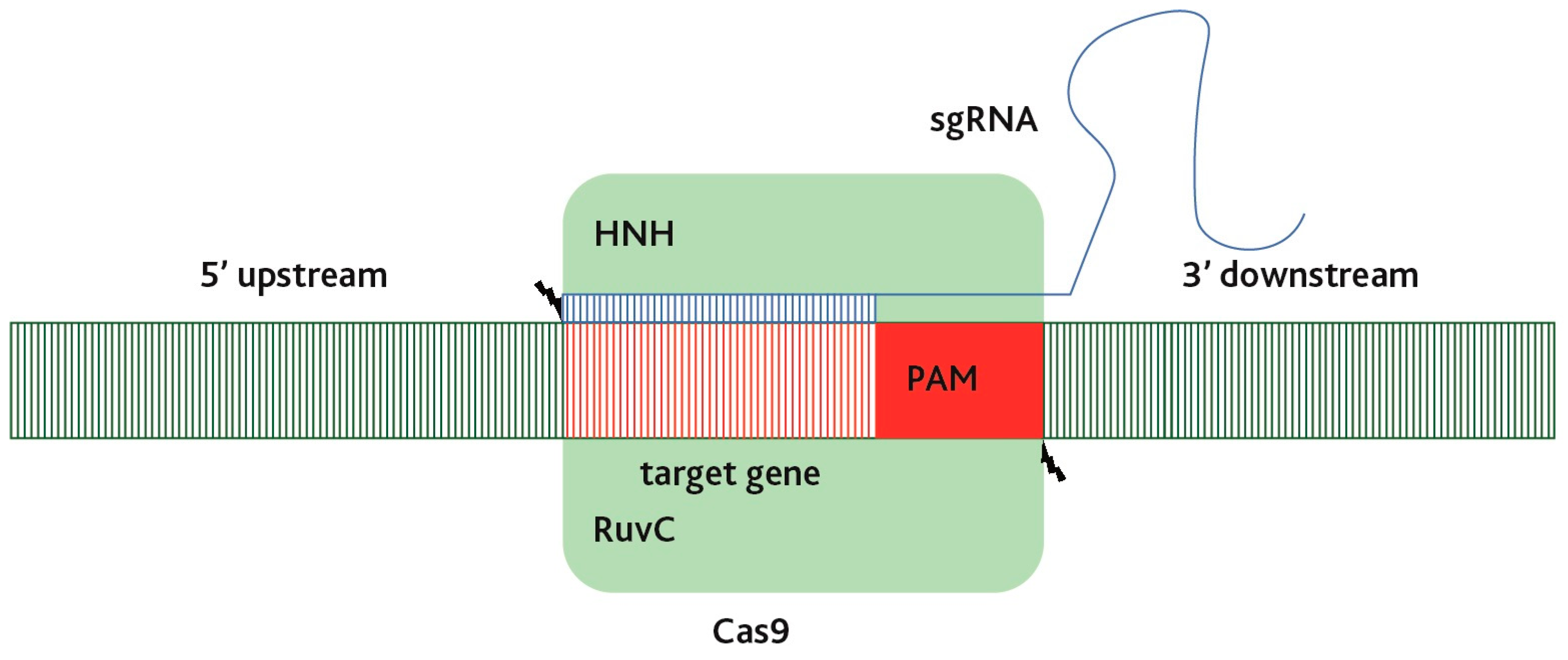
| Genome Editing Tools | Target Site (bp) | Off Targeting | Enzyme | DNA Binding Mediator | Binding Specifity | DNA Cleavage | Usage | Origin |
|---|---|---|---|---|---|---|---|---|
| CRISPR/Cas9 | 20 | Variable | Cas9 | crRNA/ sgRNA | 1:1 nucleotide pairing | RNA-dependent | Easy | Bacteria/Archaea |
| ZFNs | 18–36 | High | FokI | Zinc-finger protein | 3 nucleotides | Protein-dependent | Highly difficult | Eukaryotes |
| TALENs | 30–40 | Low | FokI | Transcription activator-like effector | 1 nucleotide | Protein-dependent | Difficult | Bacteria |
| Host Plant | Pathogen | Disease | Targeted Gene | Delivery Method | Transgene-Free | Result | References |
|---|---|---|---|---|---|---|---|
| Apple | Erwinia amylovora | Fire blight | DIPM-1, DIPM-2 and DIPM-4 | Agrobacterium-mediated transformation | Yes | Enhanced disease resistance | [83] |
| Arabidopsis | Oidium neolycopersici | Powdery mildew | PMR4 | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [84] |
| Beet Severe Curly Top Virus (BSCTV) | DNA viral disease | IR, CP, Rep | Agrobacterium-mediated transformation | No | Geminivirus-resistant plants | [85] | |
| Turnip Mosaic Virus (TuMV) | RNA viral disease | Elf(iso)4E | Agrobacterium-mediated transformation | Yes | Potyvirus-resistant plants | [78] | |
| Banana | Banana Streak Virus (BSV) | DNA viral disease | eBSV | Agrobacterium-mediated transformation | Not defined | Inactivation of eBSV caused asymptomatic plants | [86] |
| Barley | Wheat Dwarf Virus (WDV) | DNA viral disease | MP, CP, Rep/Rep, IR | Agrobacterium-mediated transformation | No | No disease symptoms and virus presence | [87] |
| Cacao | Phytophthora tropicalis | Black pod rot | TcNPR3 | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [88] |
| Citrus | Xanthomonas citri subsp. citri | Citrus canker | CsLOB1 | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [89] |
| X. citri subsp. citri | Citrus canker | CsLOB1/promoter | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [90] | |
| Cucumber | Cucumber Vein Yellowing Virus (CVYV), Zucchini Yellow Mosaic Virus (ZYMV), and Papaya Ring Spot Mosaic Virus-W (PRSV-W) | RNA viral disease | elf4E | Agrobacterium-mediated transformation | Yes | Resistance to viruses | [18] |
| Grapevine | Erysiphe necator | Powdery mildew | Mlo-7 | Polyethylene glycol-mediated (PEG) protoplast transformation | Yes | Enhanced disease resistance | [83] |
| Botrytis cinerea | Gray mold | VvWRKY52 | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [91] | |
| Papaya | P. palmivora | Root, stem, and fruit rot | alEPIC8 | Agrobacterium-mediated transformation | Not defined | Enhanced disease resistance | [92] |
| Rice | X. oryzae pv. Oryzae | Bacterial Blight | SWEET11, SWEET13 and SWEET14/promoter | Agrobacterium-mediated transformation | Not defined | Enhanced broad-spectrum disease resistance | [93] |
| X. oryzae pv. Oryzae | Bacterial Blight | Os8N3/promoter | Agrobacterium-mediated transformation | Yes | Enhanced disease resistance | [94] | |
| X. oryzae pv. Oryzae | Bacterial Blight | OsSWEET11 and OsSWEET14/promoter | Agrobacterium-mediated transformation | No | Enhanced broad-spectrum disease resistance | [95] | |
| X. oryzae pv. Oryzae | Bacterial Blight and Rice Blast | Xa13 | Not defined | Yes | Enhanced disease resistance | [96] | |
| X. oryzae pv. Oryzae | Bacterial Blight | OsSWEET13 | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [82] | |
| Magnaporthe oryzae | Rice blast | OsERF922 | Agrobacterium-mediated transformation | Yes | Enhanced disease resistance | [97] | |
| M. oryzae | Rice blast | TMS5, Pi21, and Xa13 | Not defined | Yes | Enhanced disease resistance | [96] | |
| M. grisea | Rice blast | OsMPK5 | Protoplast transformation | No | Resistance not confirmed | [98] | |
| Rice tungro bacilliform virus | Rice tungro disease | eIF4G | Agrobacterium-mediated transformation | Yes | Enhanced disease resistance | [99] | |
| Tobacco | Cotton Leaf Curl Multan Virus (CLCuMuV) | DNA viral disease | IR and C1 | Agrobacterium-mediated transformation | No | Complete resistance to virus infection | [100] |
| Tomato Yellow Leaf Curly Virus (TYLCV), Beet Curly Top Virus (BCTV), and Merremia Mosaic Virus (MeMV) | DNA viral disease | IR, CP, RCRII | Agrobacterium-mediated transformation | No | No disease symptoms and reduced virus accumulation | [101] | |
| Bean Yellow Dwarf Virus (BeYDV) | DNA viral disease | LIR, Rep | Agrobacterium-mediated transformation | No | Reduced symptoms and virus load | [102] | |
| Beet Severe Curly Top Virus (BSCTV) | DNA viral disease | IR, CP, Rep | Agrobacterium-mediated transformation | No | Geminivirus-resistant plants | [85] | |
| Tomato Yellow Leaf Curl Virus (TYLCV) | DNA viral disease | CP, Rep | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [103] | |
| Tomato | Pseudomonas syringae | Bacterial speck | SIDMR6–1 | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [104] |
| P. capsici | Phytophthora blight | SIDMR6–1 | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [104] | |
| Xanthomonas spp. | Bacterial spot | SIDMR6–1 | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [104] | |
| P. syringae pv. tomato (Pto) DC3000 | Bacterial speck | SlJAZ2 | Agrobacterium-mediated transformation | Not defined | Enhanced disease resistance and defence trade-off solved | [105] | |
| O. neolycopersici | Powdery mildew | SlMlo1 | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [64] | |
| O. neolycopersici | Powdery mildew | SIPMR4 | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [84] | |
| Fusarium oxysporum f. sp. lycopersici | Fusarium wilt | Solyc08g075770 | Agrobacterium-mediated transformation | No | Enhanced disease susceptibility | [106] | |
| B. cinerea | Gray mold | SlMAPK3 | Agrobacterium-mediated transformation | No | Enhanced disease susceptibility | [107] | |
| PVX, TMV | RNA viral disease | DCL2 | Agrobacterium-mediated transformation | No | Resistance to PVX and TMV | [108] | |
| Tomato Yellow Leaf Curl Virus (TYLCV) | DNA viral disease | CP, Rep | Agrobacterium-mediated transformation | No | Enhanced disease resistance | [103] | |
| Wheat | Blumeria graminis f. sp. tritici | Powdery mildew | TaMlo | Protoplast and Biolistic transformation | Yes | Enhanced disease resistance | [63] |
| B. graminis f. sp. tritici | Powdery mildew | TaEDR1 | Biolistic transformation | No | Enhanced disease resistance | [35] |
| Abiotic Stresses | Plant Species | Targeted Gene | Delivery Method | Regulating Direction of Response to Stress Function | References |
|---|---|---|---|---|---|
| Drought | Oryza sativa | OsNAC006 | Agrobacterium -mediated transformation | Transcription factor | [139] |
| Brassica napus | BnaA6.RGA | Agrobacterium-mediated transformation | Transcription factor | [140] | |
| O. sativa | SRL1, SRL2 | Agrobacterium-mediated transformation | Rolling of leaf | [141] | |
| O. sativa subsp. indica | OsDST | Agrobacterium-mediated transformation | Drought and salt tolerance (DST) gene | [119] | |
| O. sativa | OsNAC14 | Agrobacterium-mediated transformation | Transcription factor | [142] | |
| O. sativa | OsSAPK2 | Agrobacterium-mediated transformation | ABA signaling | [116] | |
| O. sativa | OsMYB1, OsYSA, OsROC5, OsDERF1, OsPDS, OsPMS3, OsEPSPS, OsMSH1, OsMYB5, OsSPP | Agrobacterium-mediated transformation | Amino acid synthesis | [143] | |
| Solanum lycopersicum L. | SlNPR1 | Agrobacterium-mediated transformation | Drought tolerance | [117] | |
| S. lycopersicum L. | MAPK3 | Agrobacterium-mediated transformation | Growth and development | [123] | |
| Triticum aestivum | TaDREB2, TaERF3 | PEG-mediated transformation | Dehydration-responsive element-binding protein | [115] | |
| Zea mays | ARGOS8 | Particle bombardment | Ethylene-responsive gene family regulator | [121] | |
| Arabidopsis thaliana | AREB1 | Agrobacterium-mediated transformation | ABA signaling | [144] | |
| A. thaliana | AtAVP1, AtPAP1 | Agrobacterium-mediated transformation | Transcription factor | [145] | |
| A. thaliana | AtOST2 | Agrobacterium-mediated transformation | Stomatal movement | [146] | |
| A. thaliana | AtMIR169a, AtMIR827a, TFL1 | Agrobacterium-mediated transformation | Negative factor of drought tolerance | [147] | |
| Glycine max | GmMYB118 | Agrobacterium-mediated transformation | Transcription factor | [127] | |
| Populus clone NE-19 | PdNF-YB21 | Agrobacterium-mediated leaf disc method | Transcription factor ABA-mediated indoylacetic acid transport | [148] | |
| Cold | O. sativa | OsMYB30, OsPIN5b, GS3 | Agrobacterium-mediated transformation | Cold tolerance | [133] |
| A. thaliana | UGT79B2, UGT79B3 | Agrobacterium-mediated transformation | UDP-glycosyltransferases | [149] | |
| O. sativa subsp. indica | OsPRP1 | Agrobacterium-mediated transformation | Plant growth and stress response | [150] | |
| A. thaliana | AtCBF2 | Agrobacterium-mediated transformation | Encodes AP2/ERF (APETALA2/Ethylene-Responsive Factor)-type transcription factors) | [151] | |
| A. thaliana | AtCBF1, AtCBF2, AtCBF3 | Agrobacterium-mediated transformation | Encodes AP2/ERF (APETALA2/Ethylene-Responsive Factor)-type transcription factors) | [147,152,153] | |
| S. lycopersicum L. | SlCBF1 | Agrobacterium-mediated transformation | Transcription factor | [131] | |
| O. sativa | OsAnn3 | Agrobacterium-mediated transformation | Plant development and protection from environmental stresses | [132] | |
| A. thaliana | CBFs | Agrobacterium-mediated transformation | Transcription factor | [154] | |
| Salinity | O. sativa | OsGTg-2 | Agrobacterium-mediated transformation | Transcription factor | [155] |
| O. sativa | PIL14 | Agrobacterium-mediated transformation | Phytochrome-Interacting Factor | [156] | |
| O. sativa | OstPQT3 | Agrobacterium-mediated transformation | E3 ubiquitin ligase (enhances resistance to abiotic stresses) | [157] | |
| O. sativa | OsAGO2 | Agrobacterium-mediated transformation | Transcriptional transactivator (growth and development, stress and defense responses, alternative splicing, and DNA repair) | [158] | |
| O. sativa | OsDST | Agrobacterium-mediated transformation | Zinc-finger transcription factor | [119] | |
| O. sativa | FLN2 | Agrobacterium-mediated transformation | Sucrose metabolism fructokinase-like protein2 | [159] | |
| O. sativa | OsRR9 and OsRR10 | Agrobacterium-mediated transformation | Cytokinin signaling | [160] | |
| O. sativa | OsDOF15 | Agrobacterium-mediated transformation | Transcription factor (regulates cell proliferation in the root) | [161] | |
| O. sativa | OsSPL10 | Agrobacterium-mediated transformation | Transcription factor | [162] | |
| O. sativa | NCA1a, NCA1b | Agrobacterium-mediated transformation | Regulation of catalase activity | [163] | |
| O. sativa | RR22 | Agrobacterium-mediated transformation | Transcription factor (cytokinin signal transduction and metabolism) | [164] | |
| O. sativa | OsNAC041 | Agrobacterium-mediated transformation | Transcription factor | [165] | |
| O. sativa | OsOTS1 | Agrobacterium-mediated transformation | Salt stress response regulation | [166] | |
| O. sativa | OsSAPK1, OsSAPK2 | Agrobacterium-mediated transformation | ABA pathway regulator | [129] | |
| O. sativa | OsBBS1 | Agrobacterium-mediated transformation | Receptor-like cytoplasmic kinase | [167] | |
| O. sativa | OsMIR408, OsMIR528 | Agrobacterium -mediated transformation | Salt stress response regulation | [168] | |
| O. sativa | OsRAV2 | Agrobacterium-mediated transformation | Transcription factor | [126] | |
| Z. mays | HKT1 | Agrobacterium-mediated transformation | High-affinity potassium transporter | [169] | |
| A. thaliana | AtSAUR41 | Agrobacterium-mediated transformation | Auxin response gene | [170] | |
| Cucurbita moschata | RBOHD | Agrobacterium-mediated transformation | NADPH oxidase is a key member for H2O2 production | [171] | |
| A. thaliana | AtC/VIF1 | Agrobacterium-mediated transformation | Cell wall/vacuolar inhibitor of fructosidases | [130] | |
| Hordeum vulgare | HvITPK5/6 | Agrobacterium-mediated transformation | Sequential phosphorylation of inositol phosphate to inositol hexakisphosphate | [172] | |
| G. max L. | GmAITR | Agrobacterium-mediated transformation | Transcription factors that are involved in the regulation of ABA signaling | [173] | |
| S. lycopersicum L. | SlARF4 | Agrobacterium-mediated transformation | ARFs play a key role in regulating the expression of auxin response genes | [129] | |
| A. thaliana | ArathEULS3 | Agrobacterium-mediated transformation | Stress-responsive protein, stomatal closure | [174] | |
| Heat Stress | S. lycopersicum L. | BZR1 | Agrobacterium-mediated transformation | Brassinosteroid regulation | [136] |
| O sativa | OsPDS | Gene gun | Phytoene Desaturase gene encodes one of the important enzymes in the carotenoid biosynthesis pathway | [175] | |
| G. max L. | GmHsp90A2 | Agrobacterium-mediated transformation | Molecular chaperone and heat shock protein | [176] | |
| O. sativa | OsHSA1 | Agrobacterium-mediated transformation | Chloroplast development at early stages and functions can protect chloroplasts under heat stress at later stages | [135] | |
| S. lycopersicum L. | Slcpk28 | Agrobacterium-mediated transformation | Decreases the activity of antioxidant enzymes | [177] | |
| O. sativa | OsNAC006 | PEG-mediated | Mediates the process of photosynthesis and limits the activity of antioxidant enzymes | [139] | |
| Z. mays | TMS5 | Particle bombardment | Thermosensitive genic male sterile 5 | [138] | |
| Lactuca sativa | LsNCED4 | Agrobacterium-mediated transformation | Key regulatory enzyme in the biosynthesis of abscisic acid (ABA) | [178] | |
| O. sativa | OsPYL1/4/6 | Agrobacterium-mediated transformation | ABA receptor | [179] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erdoğan, İ.; Cevher-Keskin, B.; Bilir, Ö.; Hong, Y.; Tör, M. Recent Developments in CRISPR/Cas9 Genome-Editing Technology Related to Plant Disease Resistance and Abiotic Stress Tolerance. Biology 2023, 12, 1037. https://doi.org/10.3390/biology12071037
Erdoğan İ, Cevher-Keskin B, Bilir Ö, Hong Y, Tör M. Recent Developments in CRISPR/Cas9 Genome-Editing Technology Related to Plant Disease Resistance and Abiotic Stress Tolerance. Biology. 2023; 12(7):1037. https://doi.org/10.3390/biology12071037
Chicago/Turabian StyleErdoğan, İbrahim, Birsen Cevher-Keskin, Özlem Bilir, Yiguo Hong, and Mahmut Tör. 2023. "Recent Developments in CRISPR/Cas9 Genome-Editing Technology Related to Plant Disease Resistance and Abiotic Stress Tolerance" Biology 12, no. 7: 1037. https://doi.org/10.3390/biology12071037
APA StyleErdoğan, İ., Cevher-Keskin, B., Bilir, Ö., Hong, Y., & Tör, M. (2023). Recent Developments in CRISPR/Cas9 Genome-Editing Technology Related to Plant Disease Resistance and Abiotic Stress Tolerance. Biology, 12(7), 1037. https://doi.org/10.3390/biology12071037








