Methods for Inferring Cell Cycle Parameters Using Thymidine Analogues
Abstract
Simple Summary
Abstract
1. Introduction
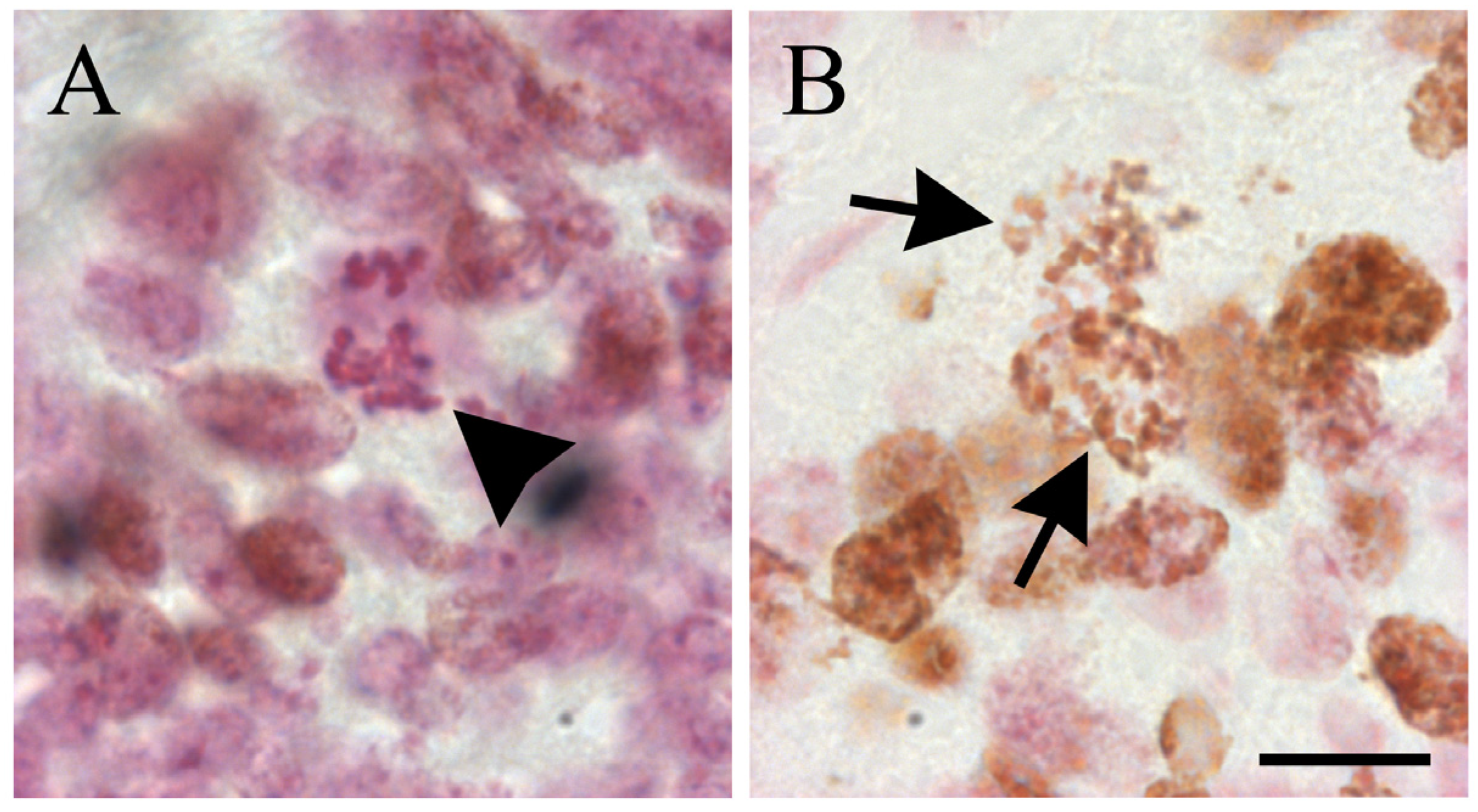
2. Thymidine Analogues and Cell Toxicity
3. Saturating S-Phase
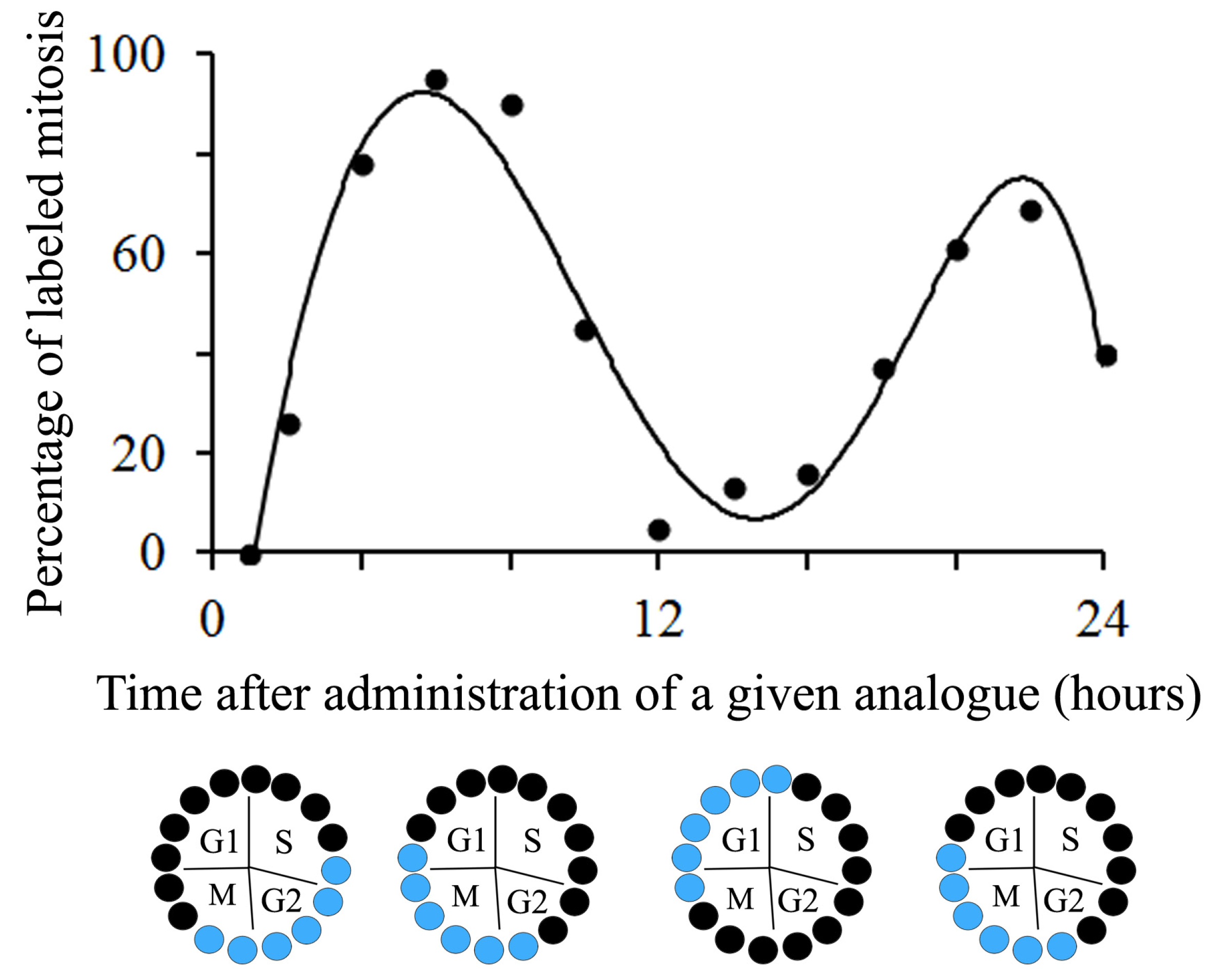
4. Estimating the CELL Cycle Time after a Single Administration of a Given Thymidine Analogue
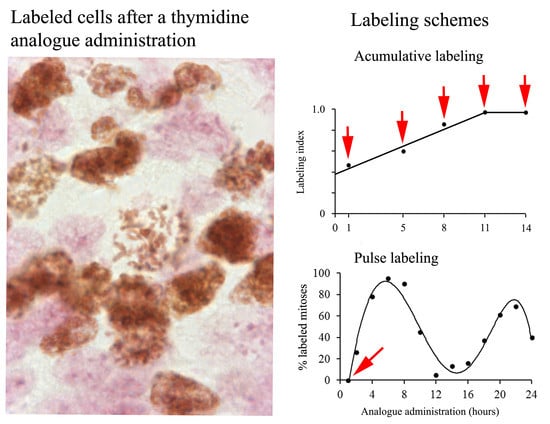
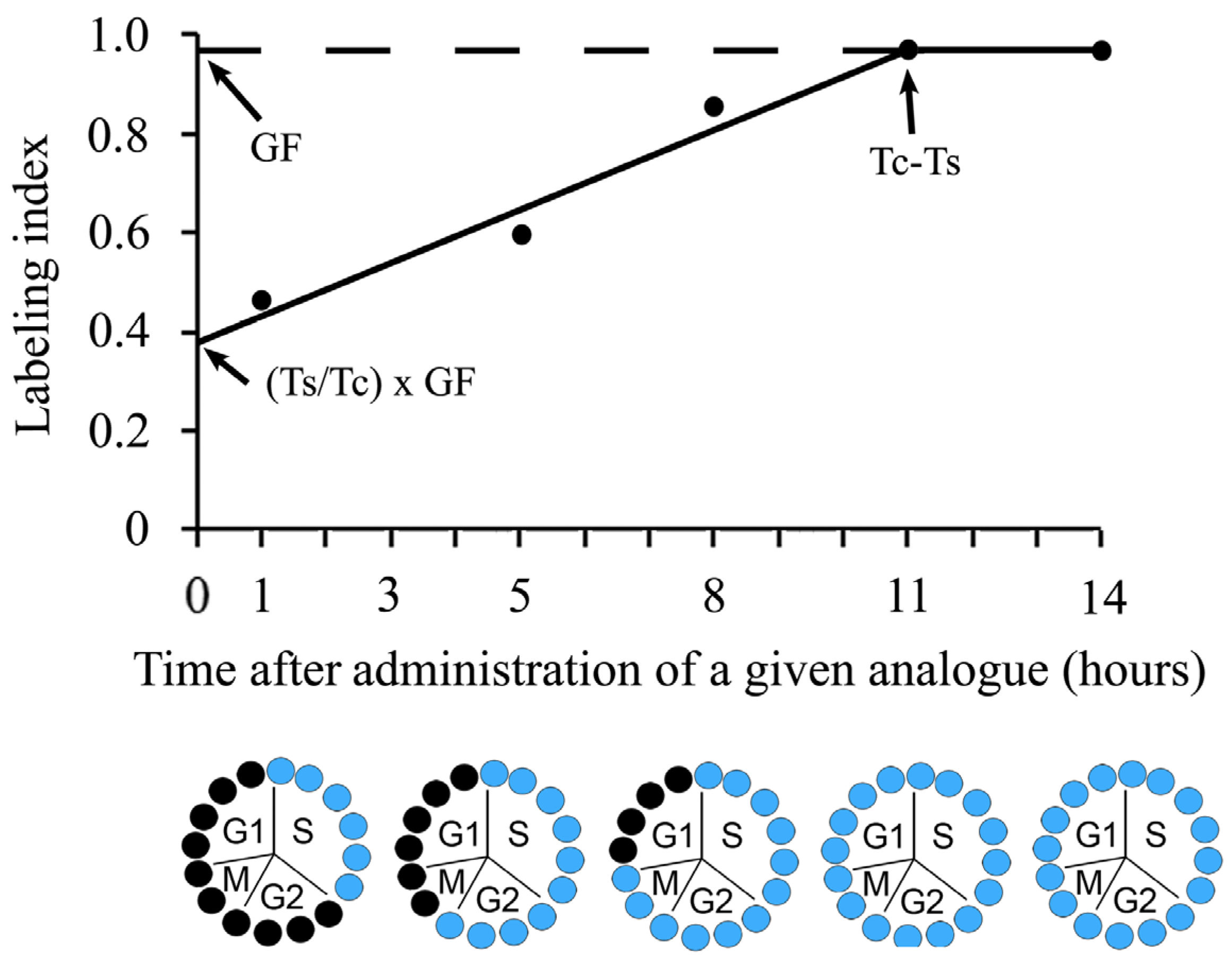
5. Inferring Duration and Phases of the Cell Cycle after Cumulative Labeling
6. Estimating the Cell Cycle Time and Duration of the Synthetic Phase after Double Labeling
- This approach has been proposed by Hwang et al. [60]. In this case, EdU administration is followed by the BrdU administration after a known time interval. The rationale of this approach is that during the EdU administration, some cells leave the S-phase. This fraction of cells is identified by the absence of the BrdU signal. The following equation is used for inferring Tc:
- 2.
- This procedure has been proposed by Martynova et al. [69]. In this case, pregnant dams were injected with IdU and then 1.5 h later to BrdU. Animals were sacrificed after 30 min. The rationale of this method is the proportion of cells tagged by the IdU administration as well as all cells labeled by the second administration served for estimating the TS. The length of this phase is based on the presupposition that the ratio between both fractions of cells is equal to the ratio between the length of the time interval between both labeling pulses and the TS. From the following equation, TS can be inferred.
- 3.
- This procedure has been proposed by Bialic et al. [70]. The principle of this method is to label S-phase cells with EdU, chase them with thymidine for varying time periods, and label the cells that are still in S-phase with BrdU. At regular intervals of time, from example at 0.5, 1.5, 3.5, 5.5 h, samples are collected. The rationale of this method is that as some cells tagged with EdU left the S-phase during the chase period, the proportion of cells co-labeled with both markers decreased with the prolongation of the chase length [6,70]. The time required for the earliest S-phase cells labeled with EdU and not tagged with BrdU corresponds to the duration of the TsS [70].
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Greenberg, A.; Simon, I.S. Phase Duration Is Determined by Local Rate and Global Organization of Replication. Biology 2022, 11, 718. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Andrés, D.; Holl, K.; Antonin, W. The second half of mitosis and its implications in cancer biology. Semin. Cancer Biol. 2023, 88, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.X.; Fakhreddin, R.I.; Shimerov, H.K.; Kedziora, K.M.; Kumar, R.J.; Perez, J.; Limas, J.C.; Grant, G.D.; Cook, J.G.; Gupta, G.P.; et al. Evidence that the human cell cycle is a series of uncoupled, memoryless phases. Mol. Syst. Biol. 2019, 15, e8604. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. Cell Cycle Progression and Synchronization: An Overview. Methods Mol. Biol. 2022, 2579, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.S.; Cook, J.G. Cell cycle exits and U-turns: Quiescence as multiple reversible forms of arrest. Fac. Rev. 2023, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Ligasová, A.; Frydrych, I.; Koberna, K. Basic Methods of Cell Cycle Analysis. Int. J. Mol. Sci. 2023, 24, 3674. [Google Scholar] [CrossRef]
- Vanzyl, E.J.; Rick, K.R.C.; Blackmore, A.B.; MacFarlane, E.M.; McKay, B.C. Flow cytometric analysis identifies changes in S and M phases as novel cell cycle alterations induced by the splicing inhibitor isoginkgetin. PLoS ONE 2018, 13, e0191178. [Google Scholar] [CrossRef]
- Taylor, J.H.; Woods, P.S.; Hughes, W.L. The organization and duplication of chromosomes as revealed by autoradiographic studies using tritium-labeled thymidinee. Proc. Natl. Acad. Sci. USA 1957, 43, 122–128. [Google Scholar] [CrossRef]
- Messier, B.; Leblond, C.P.; Smart, I. Presence of DNA synthesis and mitosis in the brain of young adult mice. Exp. Cell Res. 1958, 14, 224–226. [Google Scholar] [CrossRef]
- Martí, J.; Wills, K.V.; Ghetti, B.; Bayer, S.A. A combined immunohistochemical and autoradiographic method to detect midbrain dopaminergic neurons and determine their time of origin. Brain Res. Protoc. 2002, 9, 197–205. [Google Scholar] [CrossRef]
- Duque, A.; Rakic, P. Different effects of bromodeoxyuridine and [3H]thymidine incorporation into DNA on cell proliferation, position, and fate. J. Neurosci. 2011, 31, 15205–15217. [Google Scholar] [CrossRef]
- Gratzner, H.G. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science 1982, 218, 474–475. [Google Scholar] [CrossRef]
- Solius, G.M.; Maltsev, D.I.; Belousov, V.V.; Podgorny, O.V. Recent advances in nucleotide analogue-based techniques for tracking dividing stem cells: An overview. J. Biol. Chem. 2021, 297, 101345. [Google Scholar] [CrossRef]
- Molina, V.; Rodríguez-Vázquez, L.; Owen, D.; Valero, O.; Martí, J. Cell cycle analysis in the rat external granular layer evaluated by several bromodeoxyuridine immunoperoxidase staining protocols. Histochem. Cell Biol. 2017, 148, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Dolbeare, F. Bromodeoxyuridine: A diagnostic tool in biology and medicine, Part I: Historical perspectives, histochemical methods and cell kinetics. Histochem. J. 1995, 27, 339–369. [Google Scholar] [CrossRef] [PubMed]
- Ffrench, M.; Morel, F.; Souchier, C.; Benchaib, M.; Catallo, R.; Bryon, P.A. Choice of fixation and denaturation for the triple labelling of intra-cytoplasmic antigen, bromodeoxyuridine and DNA. Application to bone marrow plasma cells. Histochemistry 1994, 101, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Wang, H.; Naumann, R.K. Developmental Patterning and Neurogenetic Gradients of Nurr1 Positive Neurons in the Rat Claustrum and Lateral Cortex. Front. Neuroanat. 2021, 15, 786329. [Google Scholar] [CrossRef] [PubMed]
- Apraiz, A.; Mitxelena, J.; Zubiaga, A. Studying Cell Cycle-regulated Gene Expression by Two Complementary Cell Synchronization Protocols. J. Vis. Exp. 2017, 124, 55745. [Google Scholar] [CrossRef]
- Ligasová, A.; Koberna, K. Strengths and Weaknesses of Cell Synchronization Protocols Based on Inhibition of DNA Synthesis. Int. J. Mol. Sci. 2021, 22, 10759. [Google Scholar] [CrossRef]
- Newton, H.B. Hydroxyurea chemotherapy in the treatment of meningiomas. Neurosurg. Focus 2007, 23, E11. [Google Scholar] [CrossRef]
- Rodríguez-Vázquez, L.; Martí, J. Administration of 5-bromo-2′-deoxyuridine interferes with neuroblast proliferation and promotes apoptotic cell death in the rat cerebellar neuroepithelium. J. Comp. Neurol. 2021, 529, 1081–1096. [Google Scholar] [CrossRef]
- Wang, L.; Cao, X.; Yang, Y.; Kose, C.; Kawara, H.; Lindsey-Boltz, L.A.; Selby, C.P.; Sancar, A. Nucleotide excision repair removes thymidine analog 5-ethynyl-2′-deoxyuridine from the mammalian genome. Proc. Natl. Acad. Sci. USA 2022, 119, e2210176119. [Google Scholar] [CrossRef]
- En, A.; Watanabe, K.; Ayusawa, D.; Fujii, M. The key role of a basic domain of histone H2B N-terminal tail in the action of 5-bromodeoxyuridine to induce cellular senescence. FEBS J. 2023, 290, 692–711. [Google Scholar] [CrossRef]
- Brandon, M.L.; Mi, L.; Chaung, W.; Teebor, G.; Boorstein, R.J. 5-chloro-2′-deoxyuridine cytotoxicity results from base excision repair of uracil subsequent to thymidylate synthase inhibition. Mutat. Res. 2000, 459, 161–169. [Google Scholar] [CrossRef]
- Taverna, P.; Hwang, H.S.; Schupp, J.E.; Radivoyevitch, T.; Session, N.N.; Reddy, G.; Zarling, D.A.; Kinsella, T.J. Inhibition of base excision repair potentiates iododeoxyuridine-induced cytotoxicity and radiosensitization. Cancer Res. 2003, 63, 838–846. [Google Scholar]
- Breunig, J.J.; Arellano, J.I.; Macklis, J.D.; Rakic, P. Everything that glitters isn’t gold: A critical review of postnatal neural precursor analyses. Cell Stem Cell 2007, 1, 612–627. [Google Scholar] [CrossRef] [PubMed]
- Taupin, P. BrdU immunohistochemistry for studying adult neurogenesis: Paradigms, pitfalls, limitations, and validation. Brain Res. Rev. 2007, 53, 198–214. [Google Scholar] [CrossRef] [PubMed]
- Lehner, B.; Sandner, B.; Marschallinger, J.; Lehner, C.; Furtner, T.; Couillard-Despres, S.; Rivera, F.J.; Brockhoff, G.; Bauer, H.C.; Weidner, N.; et al. The dark side of BrdU in neural stem cell biology: Detrimental effects on cell cycle, differentiation and survival. Cell Tissue Res. 2011, 345, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Martí-Clúa, J. Incorporation of 5-Bromo-2′-deoxyuridine into DNA and Proliferative Behavior of Cerebellar Neuroblasts: All That Glitters Is Not Gold. Cells 2021, 10, 1453. [Google Scholar] [CrossRef]
- Schneider, L.; d’Adda di Fagagna, F. Neural stem cells exposed to BrdU lose their global DNA methylation and undergo astrocytic differentiation. Nucleic Acids Res. 2012, 40, 5332–5342. [Google Scholar] [CrossRef]
- Ross, H.H.; Levkoff, L.H.; Marshall, G. P 2nd.; Caldeira, M.; Steindler, D.A.; Reynolds, B.A.; Laywell, E.D. Bromodeoxyuridine induces senescence in neural stem and progenitor cells. Stem Cells 2008, 26, 3218–3227. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.M.; Domon, O.E.; McGarrity, L.J.; Kodell, R.L.; Casciano, D.A. Effect of bromodeoxyuridine on the proliferation and growth of ethyl methanesulfonate-exposed P3 cells: Relationship to the induction of sister-chromatid exchanges. Cell Biol. Toxicol. 1992, 8, 75–87. [Google Scholar] [CrossRef]
- Hervás, J.P.; Martí-Clúa, J.; Muñoz-García, A.; Santa-Cruz, M.C. Proliferative activity in the cerebellar external granular layer evaluated by bromodeoxyuridine labeling. Biotech. Histochem. 2002, 77, 27–35. [Google Scholar] [CrossRef]
- Martí, J.; Rodríguez-Vázquez, L. An immunocytochemical approach to the analysis of the cell division cycle in the rat cerebellar neuroepithelium. Cell Cycle 2020, 19, 2451–2459. [Google Scholar] [CrossRef]
- Leuner, B.; Glasper, E.R.; Gould, E. Thymidine analog methods for studies of adult neurogenesis are not equally sensitive. J. Comp. Neurol. 2009, 517, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Nowakowski, R.S.; Caviness, V.S., Jr. BUdR as an S-phase marker for quantitative studies of cytokinetic behaviour in the murine cerebral ventricular zone. J. Neurocytol. 1992, 21, 185–197. [Google Scholar] [CrossRef]
- Lewis, P.D.; Balázs, R.; Patel, A.J.; Johnson, A.L. The effect of undernutrition in early life on cell generation in the rat brain. Brain Res. 1975, 83, 235–247. [Google Scholar] [CrossRef]
- Lewis, P.D.; Patel, A.J.; Johnson, A.L.; Balázs, R. Effect of thyroid deficiency on cell acquistion in the postnatal rat brain: A quantitative histological study. Brain Res. 1976, 104, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Mares, V.; Lodin, Z.; Srajer, J. The cellular kinetics of the developing mouse cerebellum. I. The generation cycle, growth fraction and rate of proliferation of the external granular layer. Brain Res. 1970, 23, 323–342. [Google Scholar] [CrossRef]
- Fujita, S.; Shimada, M.; Nakamura, T. H3-thymidine autoradiographic studies on the cell proliferation and differentiation in the external and the internal granular layers of the mouse cerebellum. J. Comp. Neurol. 1966, 128, 191–208. [Google Scholar] [CrossRef]
- Böswald, M.; Harasim, S.; Maurer-Schultze, B. Tracer dose and availability time of thymidine and bromodeoxyuridine: Application of bromodeoxyuridine in cell kinetic studies. Cell Prolif. 1990, 23, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Cawood, A.H.; Savage, J.R. A comparison of the use of bromodeoxyuridine and [3H]thymidine in studies of the cell cycle. Cell Tissue Kinet. 1983, 16, 51–57. [Google Scholar] [PubMed]
- Hamre, K.M.; Goldowitz, D. Analysis of gene action in the meander tail mutant mouse: Examination of cerebellar phenotype and mitotic activity of granule cell neuroblasts. J. Comp. Neurol. 1996, 368, 304–315. [Google Scholar] [CrossRef]
- Schorl, C.; Sedivy, J.M. Analysis of cell cycle phases and progression in cultured mammalian cells. Methods 2007, 41, 143–150. [Google Scholar] [CrossRef]
- Weinstein, G.D.; McCullough, J.L.; Ross, P. Cell proliferation in normal epidermis. J. Investig. Dermatol. 1984, 82, 623–628. [Google Scholar] [CrossRef]
- Pereira, P.D.; Serra-Caetano, A.; Cabrita, M.; Bekman, E.; Braga, J.; Rino, J.; Santus, R.; Filipe, P.L.; Sousa, A.E.; Ferreira, J.A. Quantification of cell cycle kinetics by EdU (5-ethynyl-2′-deoxyuridine)-coupled-fluorescence-intensity analysis. Oncotarget 2017, 8, 40514–40532. [Google Scholar] [CrossRef]
- Duthoo, E.; Vral, A.; Baeyens, A. An updated view into the cell cycle kinetics of human T lymphocytes and the impact of irradiation. Sci. Rep. 2022, 12, 7687. [Google Scholar] [CrossRef]
- Lauder, J.M. The effects of early hypo- and hyperthyroidism on the development of rat cerebellar cortex. III. Kinetics of cell proliferation in the external granular layer. Brain Res. 1977, 126, 31–51. [Google Scholar] [CrossRef]
- Schmahl, W. Kinetics of telencephalic neural cell proliferation during the fetal regeneration period following a single X-irradiation at the late organogenesis stage. II. Cycle times and the size of the functional compartment of neural epithelial cells of distinct lesion districts. Radiat. Environ. Biophys. 1983, 22, 95–115. [Google Scholar] [CrossRef]
- Knapp, P.E. The cell cycle of glial cells grown in vitro: An immunocytochemical method of analysis. J. Histochem. Cytochem. 1992, 40, 1405–1411. [Google Scholar] [CrossRef]
- Reznikov, K.; van der Kooy, D. Variability and partial synchrony of the cell cycle in the germinal zone of the early embryonic cerebral cortex. J. Comp. Neurol. 1995, 360, 536–554. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.S.; Miller, M.W. Cell cycle kinetics and immunohistochemical characterization of dissociated fetal neocortical cultures: Evidence that differentiated neurons have mitotic capacity. Dev. Brain Res. 2000, 122, 67–80. [Google Scholar] [CrossRef] [PubMed]
- McClain, J.A.; Hayes, D.M.; Morris, S.A.; Nixon, K. Adolescent binge alcohol exposure alters hippocampal progenitor cell proliferation in rats: Effects on cell cycle kinetics. J. Comp. Neurol. 2011, 519, 2697–2710. [Google Scholar] [CrossRef]
- Nowakowski, R.S.; Lewin, S.B.; Miller, M.W. Bromodeoxyuridine immunohistochemical determination of the lengths of the cell cycle and the DNA-synthetic phase for an anatomically defined population. J. Neurocytol. 1989, 18, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Nowakowski, R.S.; Caviness, V.S., Jr. Cell cycle parameters and patterns of nuclear movement in the neocortical proliferative zone of the fetal mouse. J. Neurosci. 1993, 13, 820–833. [Google Scholar] [CrossRef]
- Takahashi, T.; Nowakowski, R.S.; Caviness, V.S., Jr. The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J. Neurosci. 1995, 15, 6046–6057. [Google Scholar] [CrossRef]
- de Boer, B.A.; van den Berg, G.; Soufan, A.T.; de Boer, P.A.; Hagoort, J.; van den Hoff, M.J.; Moorman, A.F.; Ruijter, J.M. Measurement and 3D-visualization of cell-cycle length using double labelling with two thymidine analogues applied in early heart development. PLoS ONE 2012, 7, e47719. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.; Zalucki, O.; Piper, M. BrdU/EdU dual labeling to determine the cell-cycle dynamics of defined cellular subpopulations. J. Mol. Histol. 2018, 49, 229–234. [Google Scholar] [CrossRef]
- Itaman, S.; Enikolopov, G.; Podgorny, O.V. Detection of De Novo Dividing Stem Cells In Situ through Double Nucleotide Analogue Labeling. Cells 2022, 11, 4001. [Google Scholar] [CrossRef]
- Hwang, Y.; Futran, M.; Hidalgo, D.; Pop, R.; Iyer, D.R.; Scully, R.; Rhind, N.; Socolovsky, M. Global increase in replication fork speed during a p57KIP2-regulated erythroid cell fate switch. Sci. Adv. 2017, 3, e1700298. [Google Scholar] [CrossRef]
- Shibui, S.; Hoshino, T.; Vanderlaan, M.; Gray, J.W. Double labeling with iodo- and bromodeoxyuridine for cell kinetics studies. J. Histochem. Cytochem. 1989, 37, 1007–1011. [Google Scholar] [CrossRef]
- Burns, K.A.; Kuan, C.Y. Low doses of bromo- and iododeoxyuridine produce near-saturation labeling of adult proliferative populations in the dentate gyrus. Eur. J. Neurosci. 2005, 21, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Maheshwari, Y.; Yasin, Z.; Mandava, N.; Mayers, G.; Preisler, H.D. A new method for studying cell cycle characteristics in ANLL using double-labeling with BrdU and 3HTdr. Leuk. Res. 1987, 11, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Schultze, B. Double Labeling Autoradiography. Cell Kinetic Studies with 3H- and 14C-Thymidine 1. J. Histochem. Cytochem. 1981, 29 (Suppl. 1A), 109–116. [Google Scholar] [CrossRef] [PubMed]
- Aten, J.A.; Bakker, P.J.; Stap, J.; Boschman, G.A.; Veenhof, C.H. DNA double labelling with IdUrd and CldUrd for spatial and temporal analysis of cell proliferation and DNA replication. Histochem. J. 1992, 24, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Merrick, C.J.; Jackson, D.; Diffley, J.F. Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 2004, 279, 20067–20075. [Google Scholar] [CrossRef]
- Seiler, J.A.; Conti, C.; Syed, A.; Aladjem, M.I.; Pommier, Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: Single-cell and -DNA fiber analyses. Mol. Cell Biol. 2007, 27, 5806–5818. [Google Scholar] [CrossRef]
- Liboska, R.; Ligasová, A.; Strunin, D.; Rosenberg, I.; Koberna, K. Most anti-BrdU antibodies react with 2′-deoxy-5-ethynyluridine—The method for the effective suppression of this cross-reactivity. PLoS ONE 2012, 7, e51679. [Google Scholar] [CrossRef]
- Martynoga, B.; Morrison, H.; Price, D.J.; Mason, J.O. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 2005, 283, 113–127. [Google Scholar] [CrossRef]
- Bialic, M.; Al Ahmad Nachar, B.; Koźlak, M.; Coulon, V.; Schwob, E. Measuring S-Phase Duration from Asynchronous Cells Using Dual EdU-BrdU Pulse-Chase Labeling Flow Cytometry. Genes 2022, 13, 408. [Google Scholar] [CrossRef]

| Survival Time after Bromodeoxyuridine (50 mg/Kg) Administration (Hours) | ||||||
|---|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 1.5 | 2 | 2.5 | 3 |
| A | S | |||||
| A | S | |||||
| A | S | |||||
| A | S | |||||
| A | S | |||||
| A | S | |||||
| Survival Time after Bromodeoxyuridine (50 mg/kg) Administration (h) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 2 | 4 | 6 | 8 | 10 | 12 | 14 | 16 | 18 | 20 | 22 | 24 |
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| A | S | |||||||||||||
| Survival Time after Bromodeoxyuridine (35 mg/kg) Administration of (hours) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 2 | 4 | 5 | 6 | 7 | 8 | 10 | 11 | 12 | 13 | 14 |
| A | S | ||||||||||||
| A | A | S | |||||||||||
| A | A | A | S | ||||||||||
| A | A | A | A | S | |||||||||
| A | A | A | A | A | S | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martí-Clúa, J. Methods for Inferring Cell Cycle Parameters Using Thymidine Analogues. Biology 2023, 12, 885. https://doi.org/10.3390/biology12060885
Martí-Clúa J. Methods for Inferring Cell Cycle Parameters Using Thymidine Analogues. Biology. 2023; 12(6):885. https://doi.org/10.3390/biology12060885
Chicago/Turabian StyleMartí-Clúa, Joaquín. 2023. "Methods for Inferring Cell Cycle Parameters Using Thymidine Analogues" Biology 12, no. 6: 885. https://doi.org/10.3390/biology12060885
APA StyleMartí-Clúa, J. (2023). Methods for Inferring Cell Cycle Parameters Using Thymidine Analogues. Biology, 12(6), 885. https://doi.org/10.3390/biology12060885