Understanding Snail Mucus Biosynthesis and Shell Biomineralisation through Genomic Data Mining of the Reconstructed Carbohydrate and Glycan Metabolic Pathways of the Giant African Snail (Achatina fulica)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Achatina fulica Genomic Raw Read Data
2.2. Genome Annotation
2.3. Filtration of the Annotated Proteins for Carbohydrate and Glycan-Related Enzymes
2.4. Reconstruction of the Carbohydrate and Glycan-Related Metabolic Pathways of A. fulica
2.5. Sequence and Structural Analysis of the Missing Enzyme Candidates
2.6. Finalising the Reconstructed Metabolic Pathways and Selection of Those Related to Mucous Biosynthesis and Shell Biomineralisation
2.7. Preliminary Confirmation of Specific Biochemical Pathways Based on Transcriptomic Analysis of A. fulica Tissues
3. Results
3.1. Reconstruction of the Carbohydrate and Glycan Metabolic Pathways from the A. fulica Genomic Data
3.2. Classification of Carbohydrate-Active Enzyme Families from the Reconstructed Carbohydrate and Glycan Metabolic Pathways of A. fulica
3.3. Additional Biochemical Pathways That Could Be Important to Mucus Biosynthesis and Shell Biomineralisation in the A. fulica Snail
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Raut, S.; Barker, G. Molluscs as Crop Pests; CABI Publishing, CAB International: Wallingford, UK, 2002; pp. 55–114. [Google Scholar]
- Thiengo, S.C.; Faraco, F.A.; Salgado, N.C.; Cowie, R.H.; Fernandez, M.A. Rapid spread of an invasive snail in south america: The giant african snail, Achatina fulica, in brasil. Biol. Invasions 2006, 9, 693–702. [Google Scholar] [CrossRef]
- Teng, Y.; Yin, Q.; Ding, M.; Zhao, F. Purification and characterization of a novel endo-β-1,4-glucanase, afeg22, from the giant snail, Achatina fulica frussac. Acta Biochim. Biophys. Sin. 2010, 42, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Gbadeyan, O.J.; Adali, S.; Bright, G.; Sithole, B. The investigation of reinforcement properties of nano-caco3 synthesized from Achatina fulica snail shell through mechanochemical methods on epoxy nanocomposites. Nanocomposites 2021, 7, 79–86. [Google Scholar] [CrossRef]
- Hart, A. Mini-review of waste shell-derived materials’ applications. Waste Manag. Res. 2020, 38, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Tamjidi, S.; Ameri, A. A review of the application of sea material shells as low cost and effective bio-adsorbent for removal of heavy metals from wastewater. Environ. Sci. Pollut. Res. 2020, 27, 31105–31119. [Google Scholar] [CrossRef]
- Puspitasari, P.; Fauzi, A.F.; Susanto, H.; Permanasari, A.A.; Gayatri, R.W.; Razak, J.A.; Abdillah Pratama, M.M. Phase identification and morphology of caco 3/cao from Achatina fulica snail shell as the base material for hydroxyapatite. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1034, 012128. [Google Scholar] [CrossRef]
- Gondek, M.; Knysz, P.; Lechowski, J.; Ziomek, M.; Drozd, Ł.; Szkucik, K. Content of vitamin c in edible tissues of snails obtained in poland. Med. Weter. 2020, 76, 580–584. [Google Scholar] [CrossRef]
- de la Secreción, P. Assessment of antimicrobial activity and healing potential of mucous secretion of Achatina fulica. Int. J. Morphol. 2012, 30, 365–373. [Google Scholar]
- Dn, H.M.; Kriswandini, I.L.; Ester Arijani, R. Antimicrobial proteins of snail mucus (Achatina fulica) against streptococcus mutans and aggregatibacter actinomycetemcomitans. Dent. J. (Maj. Kedokt. Gigi) 2014, 47, 31. [Google Scholar] [CrossRef]
- Ito, S.; Shimizu, M.; Nagatsuka, M.; Kitajima, S.; Honda, M.; Tsuchiya, T.; Kanzawa, N. High molecular weight lectin isolated from the mucus of the giant african snail Achatina fulica. Biosci. Biotechnol. Biochem. 2011, 75, 20–25. [Google Scholar] [CrossRef]
- Nantarat, N.; Tragoolpua, Y.; Gunama, P. Antibacterial activity of the mucus extract from the giant african snail (Lissachatina fulica) and golden apple snail (Pomacea canaliculata) against pathogenic bacteria causing skin diseases. Trop. Nat. Hist. 2019, 19, 103–112. [Google Scholar]
- Noothuan, N.; Apitanyasai, K.; Panha, S.; Tassanakajon, A. Snail mucus from the mantle and foot of two land snails, Lissachatina fulica and hemiplecta distincta, exhibits different protein profile and biological activity. BMC Res. Notes 2021, 14, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Cui, Y.; Hao, L.; Zhu, J.; Yi, J.; Kang, Q.; Huang, J.; Lu, J. Wound-healing activity of glycoproteins from white jade snail (Achatina fulica) on experimentally burned mice. Int. J. Biol. Macromol. 2021, 175, 313–321. [Google Scholar] [CrossRef]
- Tri Nuryana, C.; Haryana, S.M.; Wirohadidjojo, Y.W.; Arfian, N. Achatina fulica mucous improves cell viability and increases collagen deposition in uvb-irradiated human fibroblast culture. J. Stem Cells Regen. Med. 2020, 16, 26. [Google Scholar] [CrossRef]
- Zhong, J.; Wang, W.; Yang, X.; Yan, X.; Liu, R. A novel cysteine-rich antimicrobial peptide from the mucus of the snail of Achatina fulica. Peptides 2012, 39, 1–5. [Google Scholar] [CrossRef]
- E-Kobon, T.; Thongararm, P.; Roytrakul, S.; Meesuk, L.; Chumnanpuen, P. Prediction of anticancer peptides against mcf-7 breast cancer cells from the peptidomes of Achatina fulica mucus fractions. Comput. Struct. Biotechnol. J. 2016, 14, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Leśków, A.; Tarnowska, M.; Szczuka, I.; Diakowska, D. The effect of biologically active compounds in the mucus of slugs limax maximus and arion rufus on human skin cells. Sci. Rep. 2021, 11, 18660. [Google Scholar] [CrossRef]
- El Mubarak, M.A.S.; Lamari, F.N.; Kontoyannis, C. Simultaneous determination of allantoin and glycolic acid in snail mucus and cosmetic creams with high performance liquid chromatography and ultraviolet detection. J. Chromatogr. A 2013, 1322, 49–53. [Google Scholar] [CrossRef]
- Vieira, T.C.; Costa-Filho, A.; Salgado, N.C.; Allodi, S.; Valente, A.P.; Nasciutti, L.E.; Silva, L.C.F. Acharan sulfate, the new glycosaminoglycan from Achatina fulica bowdich 1822: Structural heterogeneity, metabolic labeling and localization in the body, mucus and the organic shell matrix. Eur. J. Biochem. 2004, 271, 845–854. [Google Scholar] [CrossRef]
- Lee, Y.S.; Yang, H.O.; Shin, K.H.; Choi, H.S.; Jung, S.H.; Kim, Y.M.; Oh, D.K.; Linhardt, R.J.; Kim, Y.S. Suppression of tumor growth by a new glycosaminoglycan isolated from the african giant snail Achatina fulica. Eur. J. Pharmacol. 2003, 465, 191–198. [Google Scholar] [CrossRef]
- Chase, R.; Pryer, K.; Baker, R.; Madison, D. Responses to conspecific chemical stimuli in the terrestrial snail Achatina fulica (pulmonata: Sigmurethra). Behav. Biol. 1978, 22, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Cook, A. The Biology of Terrestrial Molluscs; CABI: Wallingford, UK, 2001; pp. 447–487. [Google Scholar]
- Struthers, M.; Rosair, G.; Buckman, J.; Viney, C. The physical and chemical microstructure of the Achatina fulica epiphragm. J. Molluscan Stud. 2002, 68, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Suwannapan, W.; Ngankoh, S.; E-Kobon, T.; Chumnanpuen, P. Mucous cell distribution and mucus production during early growth periods of the giant african snail (Achatina fulica). Agric. Nat. Resour. 2019, 53, 423–428. [Google Scholar] [CrossRef]
- Murad, S.; Grove, D.; Lindberg, K.; Reynolds, G.; Sivarajah, A.; Pinnell, S. Regulation of collagen synthesis by ascorbic acid. Proc. Natl. Acad. Sci. USA 1981, 78, 2879–2882. [Google Scholar] [CrossRef]
- Adema, C.M.; Hillier, L.W.; Jones, C.S.; Loker, E.S.; Knight, M.; Minx, P.; Oliveira, G.; Raghavan, N.; Shedlock, A.; Amaral, L.R.D.; et al. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun. 2017, 8, 15451. [Google Scholar] [CrossRef]
- Sun, J.; Mu, H.; Ip, J.C.H.; Li, R.; Xu, T.; Accorsi, A.; Alvarado, A.S.; Ross, E.; Lan, Y.; Sun, Y.; et al. Signatures of divergence, invasiveness, and terrestrialization revealed by four apple snail genomes. Mol. Biol. Evol. 2019, 36, 1507–1520. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, Y.; Liu, Q.; Huang, Y.; Mao, G.; Yue, Z.; Abe, E.M.; Li, J.; Wu, Z.; Li, S.; et al. A chromosomal-level genome assembly for the giant african snail Achatina fulica. Gigascience 2019, 8, giz124. [Google Scholar] [CrossRef]
- Zeng, X.; Zhang, Y.; Meng, L.; Fan, G.; Bai, J.; Chen, J.; Song, Y.; Seim, I.; Wang, C.; Shao, Z.; et al. Genome sequencing of deep-sea hydrothermal vent snails reveals adaptions to extreme environments. Gigascience 2020, 9, giaa139. [Google Scholar] [CrossRef]
- Liu, C.; Ren, Y.; Li, Z.; Hu, Q.; Yin, L.; Wang, H.; Qiao, X.; Zhang, Y.; Xing, L.; Xi, Y.; et al. Giant african snail genomes provide insights into molluscan whole-genome duplication and aquatic–terrestrial transition. Mol. Ecol. Resour. 2021, 21, 478–494. [Google Scholar] [CrossRef]
- Chueca, L.J.; Schell, T.; Pfenninger, M. De novo genome assembly of the land snail candidula unifasciata (mollusca: Gastropoda). G3 Genes Genomes Genet. 2021, 11, jkab180. [Google Scholar] [CrossRef]
- Saenko, S.V.; Groenenberg, D.S.J.; Davison, A.; Schilthuizen, M. The draft genome sequence of the grove snail cepaea nemoralis. G3 Genes Genomes Genet. 2021, 11, jkaa071. [Google Scholar] [CrossRef] [PubMed]
- Tachapuripunya, V.; Roytrakul, S.; Chumnanpuen, P.; E-kobon, T. Unveiling putative functions of mucus proteins and their tryptic peptides in seven gastropod species using comparative proteomics and machine learning-based bioinformatics predictions. Molecules 2021, 26, 3475. [Google Scholar] [CrossRef] [PubMed]
- Chalongkulasak, S.; E-kobon, T.; Chumnanpuen, P. Prediction of antibacterial peptides against propionibacterium acnes from the peptidomes of Achatina fulica mucus fractions. Molecules 2022, 27, 2290. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.J.; Getachew, A.T.; Saravana, P.S.; Chun, B.S. Optimization and characterization of polysaccharides extraction from giant african snail (Achatina fulica) using pressurized hot water extraction (phwe). Bioact. Carbohydr. Diet. Fibre 2019, 18, 100179. [Google Scholar] [CrossRef]
- Andrews, S.S.A.; Fastqc. A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 18 January 2020).
- Zimin, A.V.; Marçais, G.; Puiu, D.; Roberts, M.; Salzberg, S.L.; Yorke, J.A. The masurca genome assembler. Bioinformatics 2013, 29, 2669–2677. [Google Scholar] [CrossRef]
- Adema, C.M.; Hillier, L.W.; Jones, C.S.; Loker, E.S.; Knight, M.; Minx, P.; Oliveira, G.; Raghavan, N.; Shedlock, A.; Amaral, L.R.D.; et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014, 24, 1384–1395. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. Quast: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. Busco: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. Augustus: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Kegg as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. Blastkoala and ghostkoala: Kegg tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Graham, E. Importing Ghostkoala/Kegg Annotations into Anvi’o. Available online: https://anvio.org/help/7/programs/anvi-run-kegg-kofams/ (accessed on 3 August 2020).
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. Dbcan: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.; Grüning, B.A.; Paszkiewicz, K.; Pritchard, L. Galaxy tools and workflows for sequence analysis with applications in molecular plant pathology. PeerJ 2013, 1, e167. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The interpro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.; Rempfer, C.; Bordoli, L.; et al. Swiss-model: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Capasso, L.; Aranda, M.; Cui, G.; Pousse, M.; Tambutté, S.; Zoccola, D. Investigating calcification-related candidates in a non-symbiotic scleractinian coral, tubastraea spp. Sci. Rep. 2022, 12, 13515. [Google Scholar] [CrossRef]
- Przybyło, M.; Langner, M. On the physiological and cellular homeostasis of ascorbate. Cell. Mol. Biol. Lett. 2020, 25, 1–17. [Google Scholar] [CrossRef]
- Sleight, V.A.; Antczak, P.; Falciani, F.; Clark, M.S. Computationally predicted gene regulatory networks in molluscan biomineralization identify extracellular matrix production and ion transportation pathways. Bioinformatics 2020, 36, 1326–1332. [Google Scholar] [CrossRef] [PubMed]
- Louis, V.; Besseau, L.; Lartaud, F. Step in time: Biomineralisation of bivalve’s shell. Front. Mar. Sci. 2022, 9, 3–5. [Google Scholar] [CrossRef]
- Liao, Q.; Qin, Y.; Zhou, Y.; Shi, G.; Li, X.; Li, J.; Mo, R.; Zhang, Y.; Yu, Z. Characterization and functional analysis of a chitinase gene: Evidence of ch-chit participates in the regulation of biomineralization in crassostrea hongkongensis. Aquac. Rep. 2021, 21, 100852. [Google Scholar] [CrossRef]
- Gordon-Thomson, C.; Kumari, A.; Tomkins, L.; Holford, P.; Djordjevic, J.T.; Wright, L.C.; Sorrell, T.C.; Moore, G. Chitotriosidase and gene therapy for fungal infections. Cell. Mol. Life Sci. 2009, 66, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Van den Burg, H.A.; Harrison, S.J.; Joosten, M.H.; Vervoort, J.; de Wit, P.J. Cladosporium fulvum avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant-Microbe Interact. 2006, 19, 1420–1430. [Google Scholar] [CrossRef] [PubMed]
- Din, N.; Damude, H.G.; Gilkes, N.R.; Miller, R.C., Jr.; Warren, R.; Kilburn, D. C1-cx revisited: Intramolecular synergism in a cellulase. Proc. Natl. Acad. Sci. USA 1994, 91, 11383–11387. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Shen, Y.-H.; Yang, W.-J.; Cao, Y.-F.; Xiang, Z.-H.; Zhang, Z. Expansion of the silkworm gmc oxidoreductase genes is associated with immunity. Insect Biochem. Mol. Biol. 2012, 42, 935–945. [Google Scholar] [CrossRef]
- Ebringerová, A.; Hromádková, Z.; Heinze, T. Polysaccharides i: Structure, Characterization and Use; Heinze, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–67. [Google Scholar]
- Linhardt, R.J. Polysaccharides i: Structure, Characterization and Use. Advances in Polymer Science, 186 Edited by Thomas Heinze (Friedrich-Schiller-Universität Jena, Germany); Springer: Berlin/Heidelberg, Germany; ACS Publications: New York, NY, USA, 2006; ISBN 3-540-26112-5. [Google Scholar]
- Barsett, H.; Ebringerová, A.; Harding, S.; Heinze, T.; Hromádková, Z.; Muzzarelli, C.; Muzzraelli, R.; Paulsen, B.; Elseoud, O. Polysaccharides i: Structure, Characterisation and Use; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Dimarogona, M.; Topakas, E. New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2016; pp. 171–190. [Google Scholar]
- Axelsson, E.; Ratnakumar, A.; Arendt, M.L.; Maqbool, K.; Webster, M.T.; Perloski, M.; Liberg, O.; Arnemo, J.M.; Hedhammar, Å.; Lindblad-Toh, K. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 2013, 495, 360–364. [Google Scholar] [CrossRef]
- Liu, S.; Li, L.; Wang, W.; Li, B.; Zhang, G. Characterization, fluctuation and tissue differences in nutrient content in the pacific oyster (Crassostrea gigas) in qingdao, northern china. Aquac. Res. 2020, 51, 1353–1364. [Google Scholar] [CrossRef]
- Nieland, M.L.; Goudsmit, E.M. Ultrastructure of galactogen in the albumen gland of helix pomatia. J. Ultrastruct. Res. 1969, 29, 119–140. [Google Scholar] [CrossRef]
- Dreon, M.S.; Heras, H.; Pollero, R.J. Characterization of the major egg glycolipoproteins from the perivitellin fluid of the apple snail Pomacea canaliculata. Mol. Reprod. Dev. Inc. Gamete Res. 2004, 68, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Holst, O.; Mayer, H.; Okotore, R.O.; König, W.A. Structural studies on the galactan from the albumin gland of Achatina fulica. Z. Für Nat. C 1984, 39, 1063–1065. [Google Scholar] [CrossRef]
- du Souich, P.; García, A.G.; Vergés, J.; Montell, E. Immunomodulatory and anti-inflammatory effects of chondroitin sulphate. J. Cell. Mol. Med. 2009, 13, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Weigel, P.H.; DeAngelis, P.L. Hyaluronan synthases: A decade-plus of novel glycosyltransferases. J. Biol. Chem. 2007, 282, 36777–36781. [Google Scholar] [CrossRef] [PubMed]
- Turley, S.; West, C.; Horton, B. The role of ascorbic acid in the regulation of cholesterol metabolism and in the pathogenesis of atherosclerosis. Atherosclerosis 1976, 24, 1–18. [Google Scholar] [CrossRef]
- Yang, H. Conserved or lost: Molecular evolution of the key gene gulo in vertebrate vitamin c biosynthesis. Biochem. Genet. 2013, 51, 413–425. [Google Scholar] [CrossRef]
- Rivas, C.; Zuniga, F.; Salas-Burgos, A.; Mardones, L.; Ormazabal, V.; Vera, J. Vitamin c transporters. J. Physiol. Biochem. 2008, 64, 357–375. [Google Scholar] [CrossRef]
- Machałowski, T.; Jesionowski, T. Hemolymph of molluscan origin: From biochemistry to modern biomaterials science. Appl. Phys. A 2021, 127, 1–22. [Google Scholar] [CrossRef]
- Adeboye, S.; Ogundajo, A.; Ajayi, O.; Oluba, O.M. Archachatina marginata haemolymph potentiates hypoglycemic effect by mimicking insulin in streptozotocin-induced diabetic rat. J. Pharm. Res. Int. 2017, 18, 1–8. [Google Scholar] [CrossRef]
- Rousseau, M.; Plouguerne, E.; Wan, G.; Wan, R.; Lopez, E.; Fouchereau-Peron, M. Biomineralisation markers during a phase of active growth in pinctada margaritifera. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2003, 135, 271–278. [Google Scholar] [CrossRef]
- Song, X.; Liu, Z.; Wang, L.; Song, L. Recent advances of shell matrix proteins and cellular orchestration in marine molluscan shell biomineralization. Front. Mar. Sci. 2019, 6, 41. [Google Scholar] [CrossRef]
- Rosenthal, J.; Roberson, L.; Vazquez, N. A possible role for vitamin c in coral calcification. Am. Geophys. Union 2016, 2016, AH11A-02. [Google Scholar]
- Flores, R.L.; Livingston, B.T. The skeletal proteome of the sea star patiria miniata and evolution of biomineralization in echinoderms. BMC Evol. Biol. 2017, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.F.; Chen, A.-P.; Parker, M.D.; Boron, W.F. The slc4 family of bicarbonate (hco3-) transporters. Mol. Asp. Med. 2013, 34, 159–182. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.V. A Novel Function of Invertebrate Collagen in the Biomineralization Process during the Shell Repair of Eastern Oyster, Crassostrea Virginica. Master’s Thesis, Clemson University, Clemson, SC, USA, 13 December 2004. [Google Scholar]
- Marin, F.; Le Roy, N.; Marie, B. The formation and mineralization of mollusk shell. Front. Biosci. -Sch. 2012, 4, 1099–1125. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.Y.; Stifler, C.A.; Chopdekar, R.V.; Schmidt, C.A.; Parida, G.; Schoeppler, V.; Fordyce, B.I.; Brau, J.H.; Mass, T.; Tambutté, S.; et al. From particle attachment to space-filling coral skeletons. Proc. Natl. Acad. Sci. USA 2020, 117, 30159–30170. [Google Scholar] [CrossRef]
- Aguilera, F.; McDougall, C.; Degnan, B.M. Evolution of the tyrosinase gene family in bivalve molluscs: Independent expansion of the mantle gene repertoire. Acta Biomater. 2014, 10, 3855–3865. [Google Scholar] [CrossRef]
- Hardies, K.; De Kovel, C.G.; Weckhuysen, S.; Asselbergh, B.; Geuens, T.; Deconinck, T.; Azmi, A.; May, P.; Brilstra, E.; Becker, F.; et al. Recessive mutations in slc13a5 result in a loss of citrate transport and cause neonatal epilepsy, developmental delay and teeth hypoplasia. Brain 2015, 138, 3238–3250. [Google Scholar] [CrossRef]






| Pathways | Number of Missing Enzymes | Number of Enzymes That Passed the Filtering Criteria | Number of Enzymes Added After the First Gap Filling | Number of Enzymes Added After the Second Gap Filling | Completed Pathways/Remained Missing Enzymes |
|---|---|---|---|---|---|
| Carbohydrate metabolic pathways | |||||
| Pentose phosphate | 12 | 8 | 4 | - | ✓ |
| Pentose and glucuronate interconversions | 9 | 8 | - | 1 | ✓ |
| Fructose and mannose metabolism | 7 | 4 | 3 | - | ✓ |
| Galactose metabolism | 3 | 2 | 1 | - | ✓ |
| Ascorbate metabolism | 5 | 4 | 1 | - | ✓ |
| Starch and sucrose metabolism | 5 | 3 | 2 | - | ✓ |
| Amino sugar and nucleotide sugar metabolism | 3 | 3 | - | - | ✓ |
| Inositol phosphate metabolism | 8 | 5 | 3 | - | ✓ |
| Glyoxylate and dicarboxylate metabolism | 11 | 8 | 3 | - | ✓ |
| Propanoate metabolism | 6 | 3 | 3 | - | ✓ |
| Butanoate metabolism | 6 | 3 | 3 | - | ✓ |
| Pyruvate metabolism | 5 | 4 | - | 1 | ✓ |
| Glycolysis/gluconeogenesis | 4 | 1 | 3 | - | ✓ |
| Glycan metabolic pathways | |||||
| Chondroitin sulfate/dermatan sulfate biosynthesis | 4 | - | 2 | - | 2 |
| Galactose type of O-glycan biosynthesis | 1 | - | 1 | - | ✓ |
| Glycosylphosphatidylinositol (GPI) biosynthesis | 4 | - | 1 | - | 3 |
| Mannose type O-glycan biosynthesis CORE 1 | 5 | 4 | 1 | - | ✓ |
| Mannose type O-glycan biosynthesis CORE 2 | 3 | 1 | 2 | - | ✓ |
| Mucin type of O-glycan biosynthesis | 2 | 1 | 1 | - | ✓ |
| N-glycan biosynthesis | 1 | - | 1 | - | ✓ |
| Glycohormone type of N-glycan biosynthesis | 3 | - | 2 | - | ✓ |
| Hyaluronan biosynthesis | 1 | - | - | - | 1 |
| Keratan sulfate biosynthesis | 2 | - | - | 2 | ✓ |
| Total number of enzymes | 110 | 48 | 38 | 10 | 6 |
| Protein Family Names | Number of Protein Subfamilies | Protein Subfamily Names |
|---|---|---|
| Auxiliary activities (AAs) | 4 | AA1 (1), AA2 (1), AA3 (7), AA15(8) |
| Carbohydrate-binding modules (CBMs) | 6 | CBM2 (3), CBM13 (16), CBM14 (2), CBM43 (1), CBM48 (2), CBM57 (1) |
| Carbohydrate esterases (CEs) | 4 | CE8 (1), CE9 (1), CE10 (5), CE12 (1) |
| Glycoside hydrolases (GHs) | 31 | GH0 (1), GH1 (6),GH2 (8), GH7 (1), GH9 (7), GH13 (8), GH15 (1), GH18 (12), GH19 (1), GH20 (5) GH23 (1), GH25 (1), GH27 (2), GH29 (3), GH31 (10), GH35 (3), GH36 (1), GH37 (2), GH38 (8), GH39 (1), GH47 (4), GH48 (1), GH56 (1), GH57 (1), GH59 (1), GH63 (2), GH79 (1), GH84 (1), GH85 (2), GH89 (1), GH99(1) |
| Glycosyltransferases (GTs) | 38 | GT1 (10), GT2 (19), GT3 (1), GT4 (7), GT7 (14), GT8 (2), GT10 (3), GT11 (11), GT13 (2), GT14 (7), GT15 (1), GT16 (1), GT18 (1), GT20 (1), GT22 (4), GT23 (4), GT25 (1), GT27 (19), GT29 (1), GT30 (1), GT31 (22), GT33 (1), GT35 (3), GT39 (1), GT41 (4), GT47 (2), GT49 (1), GT54 (2), GT57 (2), GT59 (1), GT61 (1), GT64 (3), GT65 (1), GT66 (3), GT68 (1), GT92 (4), GT95 (2), GT98 (1) |
| Polysaccharide lyases (PLs) | 2 | PL1 (1), PL14 (1) |
| Protein Groups | Protein Names (Copy Number of Proteins) |
|---|---|
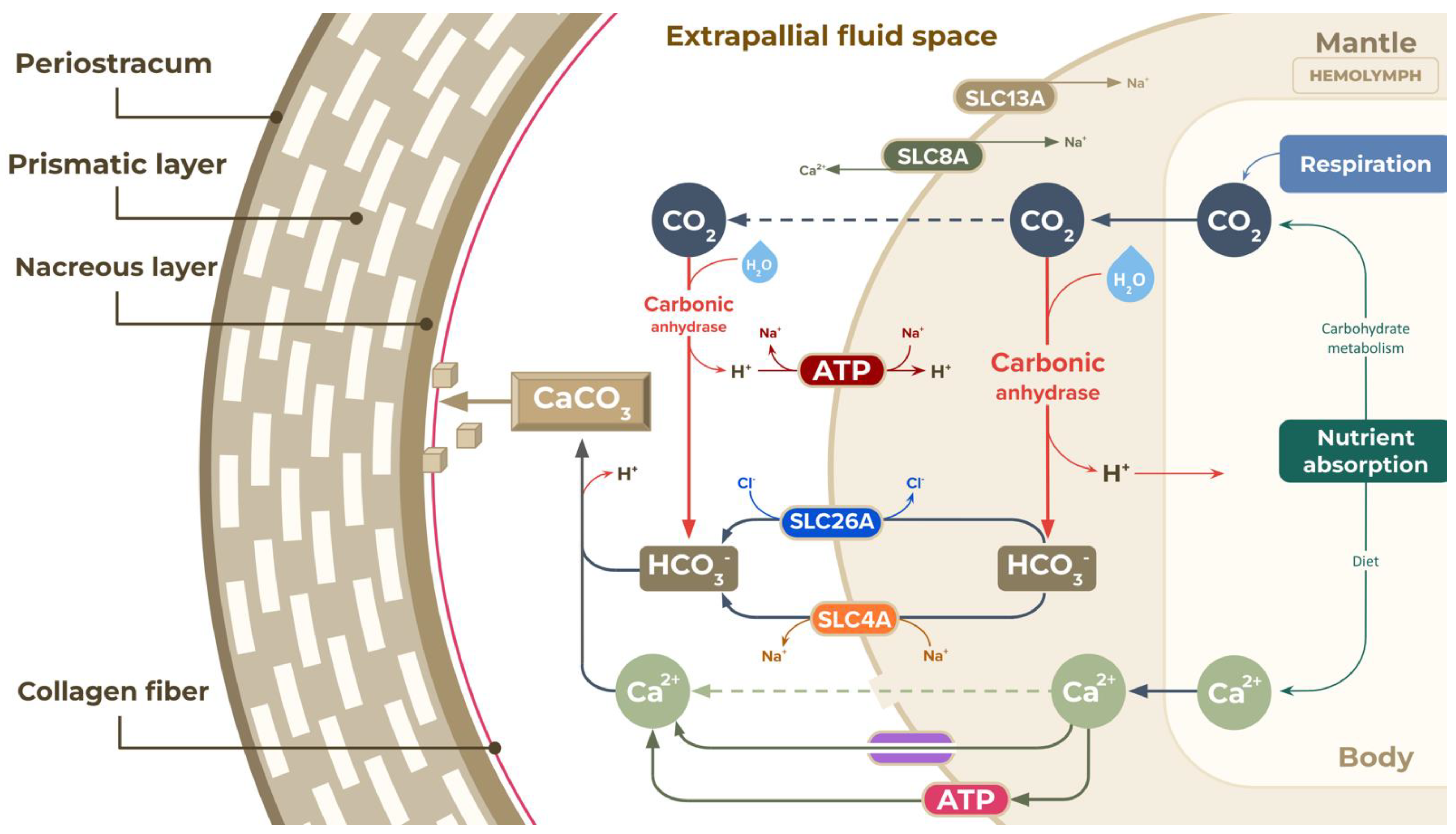
| Bicarbonate transporter a | Solute carrier family 4 (anion exchanger), member 2 (5), Solute carrier family 4 (sodium bicarbonate cotransporter), member 7 (2), Solute carrier family 4 (sodium bicarbonate cotransporter), member 8 (5), Solute carrier family 4 (sodium bicarbonate cotransporter), member 10 (3), Solute carrier family 4 (sodium bicarbonate cotransporter), member 11 (2) |
| Pores ion channels a | Ammonium transporter (32), Glutathione S-transferase (23) |
| Na+/sulfate/carboxylate cotransporter a | Solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2/3/5 (10) |
| Facilitative GLUT transporter b | Solute carrier family 2 (facilitated glucose transporter), member 1 (3), Solute carrier family 2 (facilitated glucose transporter), member 8 (1), Solute carrier family 2 (facilitated glucose transporter), member 11 (1) |
| Na+/Ca2+ exchanger a | Solute carrier family 8 (sodium/calcium exchanger) (15) |
| Multifunctional anion exchanger a | Solute carrier family 26 (sulfate anion transporter), member 1 (1), Solute carrier family 26 (sulfate anion transporter), member 2 (1), Solute carrier family 26 (sulfate anion transporter), member 5 (2), Solute carrier family 26 (sulfate anion transporter), member 6 (4), Solute carrier family 26 (sulfate anion transporter), member 7 (2), Solute carrier family 26 (sulfate anion transporter), member 10 (1), Solute carrier family 26 (sulfate anion transporter), member 11 (2) |
| Hydrolyases c | Carbonic anhydrase (4.2.1.1) (30) |
| Ca2+ transporter d | P-type Ca2+ transporter type 2A (5), P-type Ca2+ transporter type 2B (3), P-type Ca2+ transporter type 2C (2) |
| Oxidoreductases c | Tyrosinase (16) |
| Na+-dependent ascorbic acid transporter a | Solute carrier family 23 (nucleobase transporter), member 1 (10) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nualnisachol, P.; Chumnanpuen, P.; E-kobon, T. Understanding Snail Mucus Biosynthesis and Shell Biomineralisation through Genomic Data Mining of the Reconstructed Carbohydrate and Glycan Metabolic Pathways of the Giant African Snail (Achatina fulica). Biology 2023, 12, 836. https://doi.org/10.3390/biology12060836
Nualnisachol P, Chumnanpuen P, E-kobon T. Understanding Snail Mucus Biosynthesis and Shell Biomineralisation through Genomic Data Mining of the Reconstructed Carbohydrate and Glycan Metabolic Pathways of the Giant African Snail (Achatina fulica). Biology. 2023; 12(6):836. https://doi.org/10.3390/biology12060836
Chicago/Turabian StyleNualnisachol, Pornpavee, Pramote Chumnanpuen, and Teerasak E-kobon. 2023. "Understanding Snail Mucus Biosynthesis and Shell Biomineralisation through Genomic Data Mining of the Reconstructed Carbohydrate and Glycan Metabolic Pathways of the Giant African Snail (Achatina fulica)" Biology 12, no. 6: 836. https://doi.org/10.3390/biology12060836
APA StyleNualnisachol, P., Chumnanpuen, P., & E-kobon, T. (2023). Understanding Snail Mucus Biosynthesis and Shell Biomineralisation through Genomic Data Mining of the Reconstructed Carbohydrate and Glycan Metabolic Pathways of the Giant African Snail (Achatina fulica). Biology, 12(6), 836. https://doi.org/10.3390/biology12060836









