New Insights into NF-κB Signaling in Innate Immunity: Focus on Immunometabolic Crosstalks
Abstract
Simple Summary
Abstract
1. Introduction
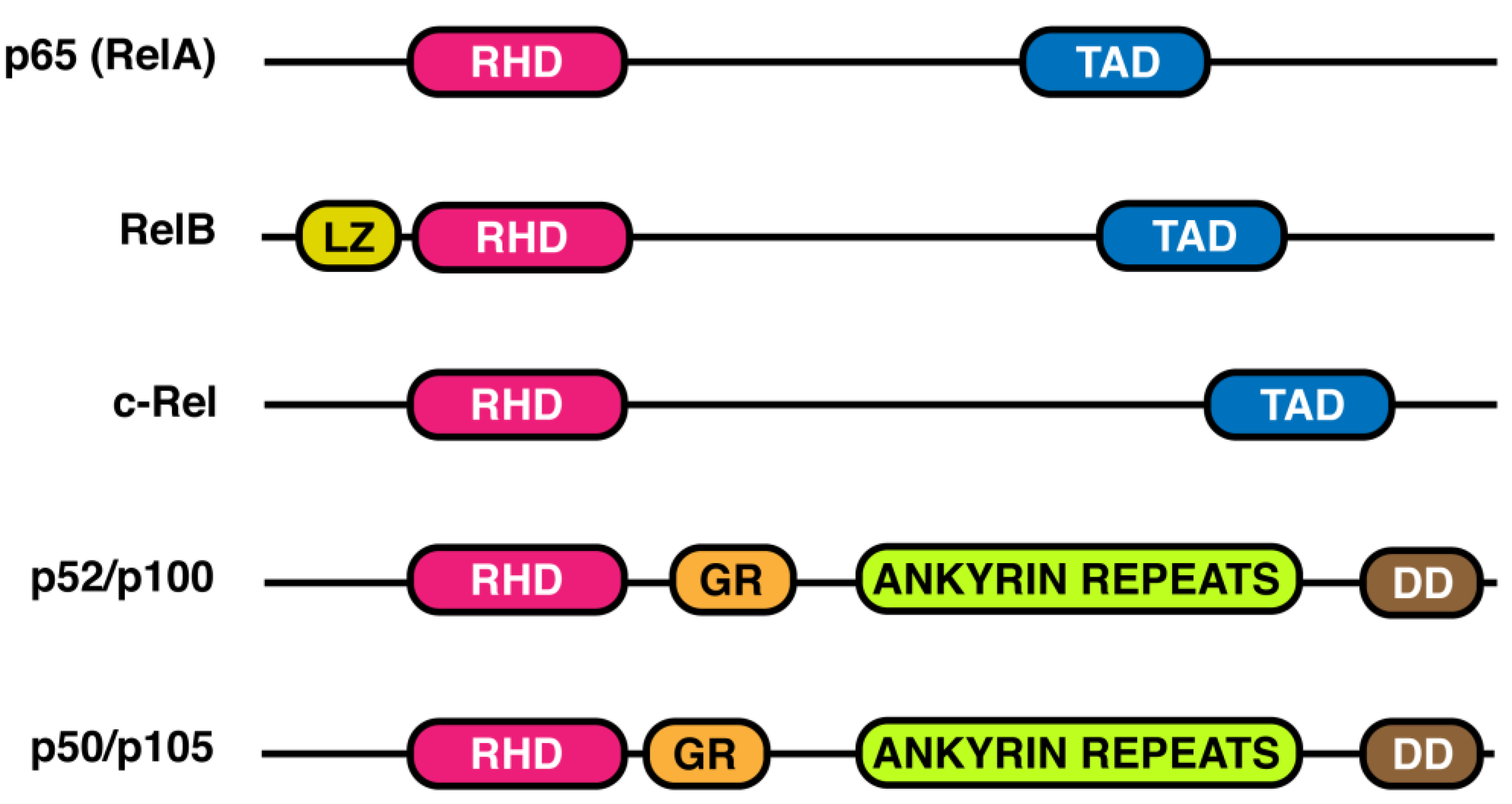
2. NF-κB Transcription Factor Family
3. IkB Proteins
4. Ubiquitin
5. NF-κB Activation Mechanisms
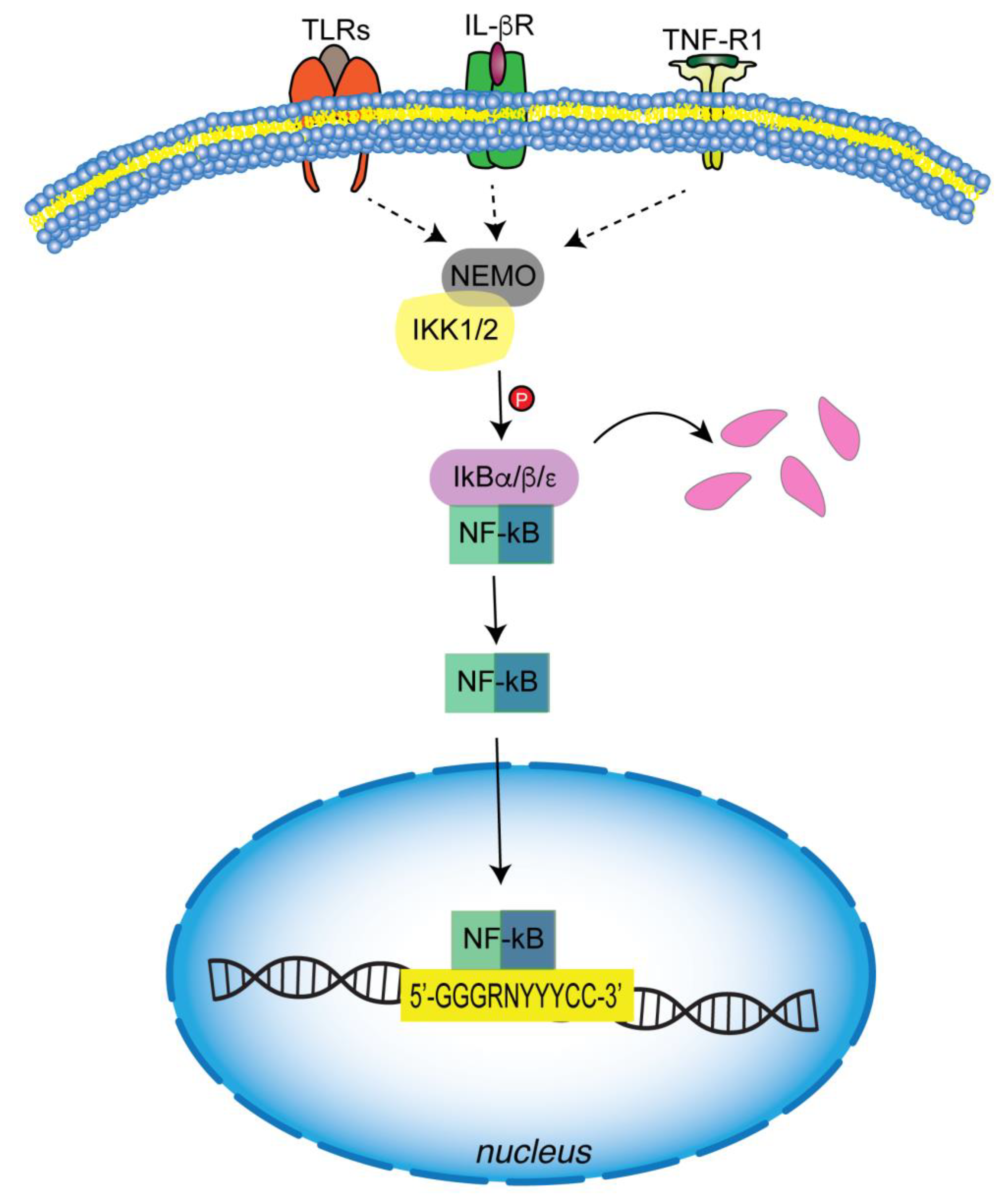
5.1. Canonical Pathway
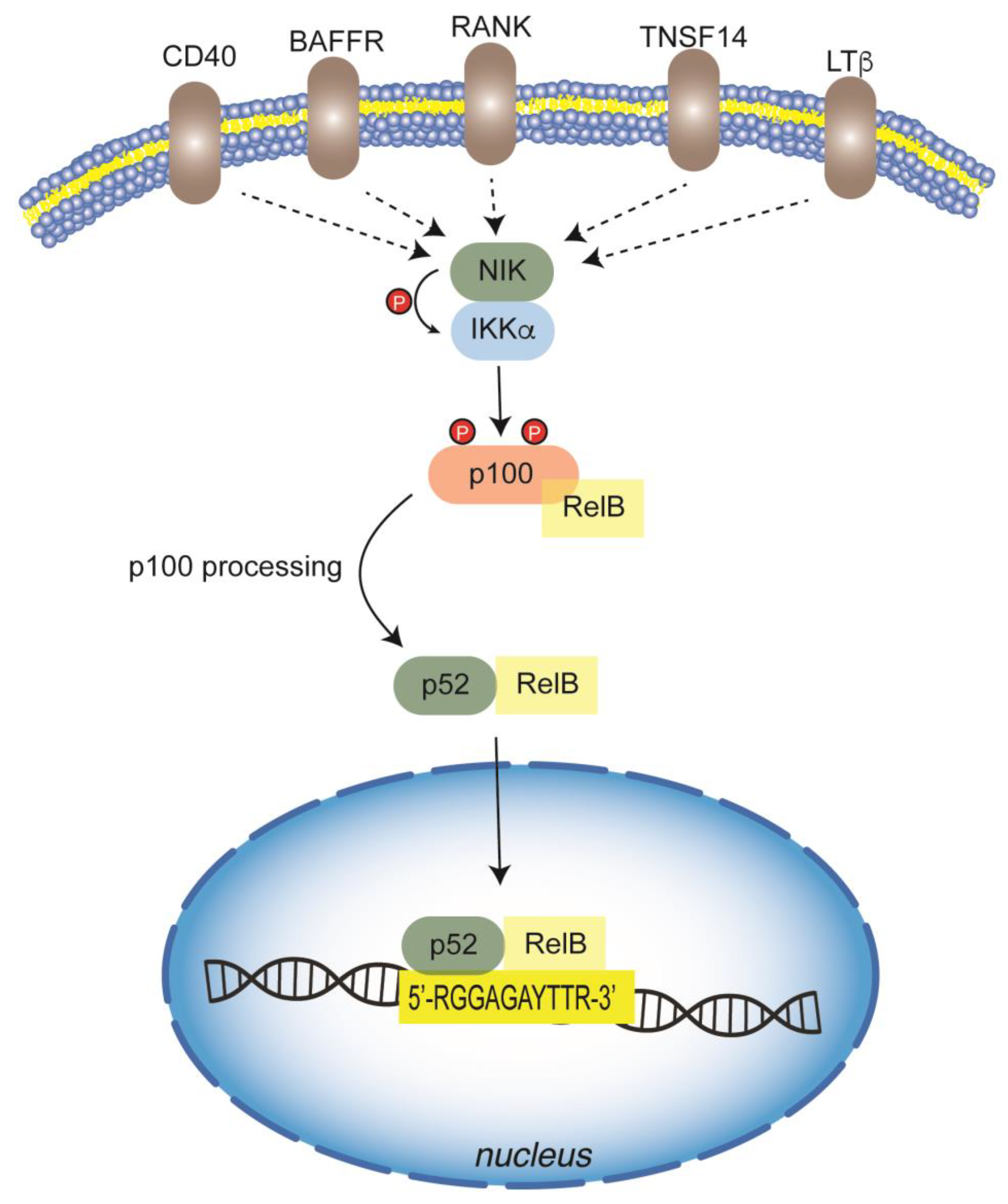
5.2. Non-Canonical Pathway
6. NF-κB in the Innate Immune Cells
7. NF-κB and Inflammasome Regulation through Metabolic Signals
8. Crosstalk between NF-κB and Metabolism in Innate Immune Cells
8.1. Metabolic-Linked NF-κB Post-Translational Modifications
8.2. NF-κB and Immunometabolic Interplay
9. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target 2017, 2, 17023. [Google Scholar] [CrossRef]
- Zinatizadeh, M.R.; Schock, B.; Chalbatani, G.M.; Zarandi, P.K.; Jalali, S.A.; Miri, S.R. The Nuclear Factor Kappa B (NF-κB) signaling in cancer development and immune diseases. Genes Dis. 2021, 8, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Palsson-McDermott, E.M.; O’Neill, L.A. The Warburg effect then and now: From cancer to inflammatory diseases. Bioessays 2013, 35, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Infantino, V.; Pierri, C.L.; Iacobazzi, V. Metabolic Routes in Inflammation: The Citrate Pathway and its Potential as Therapeutic Target. Curr. Med. Chem. 2019, 26, 7104–7116. [Google Scholar] [CrossRef] [PubMed]
- Infantino, V.; Dituri, F.; Convertini, P.; Santarsiero, A.; Palmieri, F.; Todisco, S.; Mancarella, S.; Giannelli, G.; Iacobazzi, V. Epigenetic upregulation and functional role of the mitochondrial aspartate/glutamate carrier isoform 1 in hepatocellular carcinoma. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 38–47. [Google Scholar] [CrossRef]
- Todisco, S.; Convertini, P.; Iacobazzi, V.; Infantino, V. TCA Cycle Rewiring as Emerging Metabolic Signature of Hepatocellular Carcinoma. Cancers 2019, 12, 68. [Google Scholar] [CrossRef]
- Corcoran, S.E.; O’Neill, L.A. HIF1alpha and metabolic reprogramming in inflammation. J. Clin. Investig. 2016, 126, 3699–3707. [Google Scholar] [CrossRef]
- Convertini, P.; Todisco, S.; De Santis, F.; Pappalardo, I.; Iacobazzi, D.; Castiglione Morelli, M.A.; Fondufe-Mittendorf, Y.N.; Martelli, G.; Palmieri, F.; Infantino, V. Transcriptional Regulation Factors of the Human Mitochondrial Aspartate/Glutamate Carrier Gene, Isoform 2 (SLC25A13): USF1 as Basal Factor and FOXA2 as Activator in Liver Cells. Int. J. Mol. Sci. 2019, 20, 1888. [Google Scholar] [CrossRef]
- Infantino, V.; Iacobazzi, V.; Menga, A.; Avantaggiati, M.L.; Palmieri, F. A key role of the mitochondrial citrate carrier (SLC25A1) in TNFalpha- and IFNgamma-triggered inflammation. Biochim. Biophys. Acta 2014, 1839, 1217–1225. [Google Scholar] [CrossRef]
- Santarsiero, A.; Convertini, P.; Todisco, S.; Pierri, C.L.; De Grassi, A.; Williams, N.C.; Iacobazzi, D.; De Stefano, G.; O’Neill, L.A.J.; Infantino, V. ACLY Nuclear Translocation in Human Macrophages Drives Proinflammatory Gene Expression by NF-kappaB Acetylation. Cells 2021, 10, 2962. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, S.M.; Zhang, F.; Ding, C.; Montoya-Durango, D.E.; Hu, X.; Yang, C.; Wang, Z.; Yuan, F.; Fox, M.; Zhang, H.G.; et al. Tumor-derived exosomes drive immunosuppressive macrophages in a pre-metastatic niche through glycolytic dominant metabolic reprogramming. Cell Metab. 2021, 33, 2040–2058.e10. [Google Scholar] [CrossRef] [PubMed]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, G.; Van Duyne, G.; Ghosh, S.; Sigler, P.B. Structure of NF-kappa B p50 homodimer bound to a kappa B site. Nature 1995, 373, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.W.; Rey, F.A.; Sodeoka, M.; Verdine, G.L.; Harrison, S.C. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature 1995, 373, 311–317. [Google Scholar] [CrossRef]
- Chen, F.E.; Huang, D.B.; Chen, Y.Q.; Ghosh, G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature 1998, 391, 410–413. [Google Scholar] [CrossRef]
- Dobrzanski, P.; Ryseck, R.P.; Bravo, R. Both N- and C-terminal domains of RelB are required for full transactivation: Role of the N-terminal leucine zipper-like motif. Mol. Cell. Biol. 1993, 13, 1572–1582. [Google Scholar] [PubMed]
- Ghosh, G.; Wang, V.Y.; Huang, D.B.; Fusco, A. NF-kappaB regulation: Lessons from structures. Immunol. Rev. 2012, 246, 36–58. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. Shared principles in NF-kappaB signaling. Cell 2008, 132, 344–362. [Google Scholar] [CrossRef]
- Vallabhapurapu, S.; Karin, M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009, 27, 693–733. [Google Scholar] [CrossRef]
- Xiao, G.; Harhaj, E.W.; Sun, S.C. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol. Cell 2001, 7, 401–409. [Google Scholar] [CrossRef]
- Tsui, R.; Kearns, J.D.; Lynch, C.; Vu, D.; Ngo, K.A.; Basak, S.; Ghosh, G.; Hoffmann, A. IkappaBbeta enhances the generation of the low-affinity NFkappaB/RelA homodimer. Nat. Commun. 2015, 6, 7068. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.; Kerr, L.D.; Kakizuka, A.; Verma, I.M. I kappa B gamma, a 70 kd protein identical to the C-terminal half of p110 NF-kappa B: A new member of the I kappa B family. Cell 1992, 68, 1109–1120. [Google Scholar] [CrossRef]
- Gerondakis, S.; Morrice, N.; Richardson, I.B.; Wettenhall, R.; Fecondo, J.; Grumont, R.J. The activity of a 70 kilodalton I kappa B molecule identical to the carboxyl terminus of the p105 NF-kappa B precursor is modulated by protein kinase A. Cell Growth Differ. 1993, 4, 617–627. [Google Scholar]
- Malek, S.; Huang, D.B.; Huxford, T.; Ghosh, S.; Ghosh, G. X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex. J. Biol. Chem. 2003, 278, 23094–23100. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Natoli, G.; Ghosh, G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene 2006, 25, 6706–6716. [Google Scholar] [CrossRef] [PubMed]
- Courtois, G.; Pescatore, A.; Gautheron, J.; Fusco, F.; Ursini, M.V.; Senegas, A. NF-κB-Related Genetic Diseases; Springer International Publishing: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Hurley, J.H.; Lee, S.; Prag, G. Ubiquitin-binding domains. Biochem. J. 2006, 399, 361–372. [Google Scholar] [CrossRef]
- Rahighi, S.; Ikeda, F.; Kawasaki, M.; Akutsu, M.; Suzuki, N.; Kato, R.; Kensche, T.; Uejima, T.; Bloor, S.; Komander, D.; et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 2009, 136, 1098–1109. [Google Scholar] [CrossRef]
- Laplantine, E.; Fontan, E.; Chiaravalli, J.; Lopez, T.; Lakisic, G.; Veron, M.; Agou, F.; Israel, A. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 2009, 28, 2885–2895. [Google Scholar] [CrossRef]
- Shimizu, Y.; Taraborrelli, L.; Walczak, H. Linear ubiquitination in immunity. Immunol. Rev. 2015, 266, 190–207. [Google Scholar] [CrossRef]
- Sasaki, K.; Iwai, K. Roles of linear ubiquitinylation, a crucial regulator of NF-kappaB and cell death, in the immune system. Immunol. Rev. 2015, 266, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Kensche, T.; Tokunaga, F.; Ikeda, F.; Goto, E.; Iwai, K.; Dikic, I. Analysis of nuclear factor-kappaB (NF-kappaB) essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-kappaB. J. Biol. Chem. 2012, 287, 23626–23634. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Tokunaga, A.; Shimizu, S.; Whiting, A.L.; Aguilar-Alonso, F.; Takagi, K.; Walinda, E.; Sasaki, Y.; Shimokawa, T.; Mizushima, T.; et al. Cooperative Domain Formation by Homologous Motifs in HOIL-1L and SHARPIN Plays A Crucial Role in LUBAC Stabilization. Cell Rep. 2018, 23, 1192–1204. [Google Scholar] [CrossRef]
- Emmerich, C.H.; Ordureau, A.; Strickson, S.; Arthur, J.S.; Pedrioli, P.G.; Komander, D.; Cohen, P. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci. USA 2013, 110, 15247–15252. [Google Scholar] [CrossRef]
- Kelsall, I.R.; Zhang, J.; Knebel, A.; Arthur, J.S.C.; Cohen, P. The E3 ligase HOIL-1 catalyses ester bond formation between ubiquitin and components of the Myddosome in mammalian cells. Proc. Natl. Acad. Sci. USA 2019, 116, 13293–13298. [Google Scholar] [CrossRef]
- Aiba, Y.; Nakamura, M. The role of TL1A and DR3 in autoimmune and inflammatory diseases. Mediat. Inflamm. 2013, 2013, 258164. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Parent, L.; Maniatis, T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell 1996, 84, 853–862. [Google Scholar] [CrossRef]
- Mercurio, F.; Zhu, H.; Murray, B.W.; Shevchenko, A.; Bennett, B.L.; Li, J.; Young, D.B.; Barbosa, M.; Mann, M.; Manning, A.; et al. IKK-1 and IKK-2: Cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 1997, 278, 860–866. [Google Scholar] [CrossRef]
- Woronicz, J.D.; Gao, X.; Cao, Z.; Rothe, M.; Goeddel, D.V. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science 1997, 278, 866–869. [Google Scholar] [CrossRef]
- Zandi, E.; Chen, Y.; Karin, M. Direct phosphorylation of IkappaB by IKKalpha and IKKbeta: Discrimination between free and NF-kappaB-bound substrate. Science 1998, 281, 1360–1363. [Google Scholar] [CrossRef]
- May, M.J.; D’Acquisto, F.; Madge, L.A.; Glockner, J.; Pober, J.S.; Ghosh, S. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science 2000, 289, 1550–1554. [Google Scholar] [CrossRef] [PubMed]
- May, M.J.; Marienfeld, R.B.; Ghosh, S. Characterization of the Ikappa B-kinase NEMO binding domain. J. Biol. Chem. 2002, 277, 45992–46000. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, H. Targeting of TAK1 in inflammatory disorders and cancer. Trends Pharm. Sci. 2012, 33, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Christian, F.; Smith, E.L.; Carmody, R.J. The Regulation of NF-kappaB Subunits by Phosphorylation. Cells 2016, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Pradere, J.P.; Hernandez, C.; Koppe, C.; Friedman, R.A.; Luedde, T.; Schwabe, R.F. Negative regulation of NF-kappaB p65 activity by serine 536 phosphorylation. Sci. Signal. 2016, 9, ra85. [Google Scholar] [CrossRef]
- Hu, Y.; Baud, V.; Delhase, M.; Zhang, P.; Deerinck, T.; Ellisman, M.; Johnson, R.; Karin, M. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science 1999, 284, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Baud, V.; Oga, T.; Kim, K.I.; Yoshida, K.; Karin, M. IKKalpha controls formation of the epidermis independently of NF-kappaB. Nature 2001, 410, 710–714. [Google Scholar] [CrossRef]
- Beg, A.A.; Sha, W.C.; Bronson, R.T.; Baltimore, D. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 1995, 9, 2736–2746. [Google Scholar] [CrossRef]
- Li, Q.; Van Antwerp, D.; Mercurio, F.; Lee, K.F.; Verma, I.M. Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science 1999, 284, 321–325. [Google Scholar] [CrossRef]
- Li, Z.W.; Chu, W.; Hu, Y.; Delhase, M.; Deerinck, T.; Ellisman, M.; Johnson, R.; Karin, M. The IKKbeta subunit of IkappaB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J. Exp. Med. 1999, 189, 1839–1845. [Google Scholar] [CrossRef]
- Rudolph, D.; Yeh, W.C.; Wakeham, A.; Rudolph, B.; Nallainathan, D.; Potter, J.; Elia, A.J.; Mak, T.W. Severe liver degeneration and lack of NF-kappaB activation in NEMO/IKKgamma-deficient mice. Genes Dev. 2000, 14, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Makris, C.; Godfrey, V.L.; Krahn-Senftleben, G.; Takahashi, T.; Roberts, J.L.; Schwarz, T.; Feng, L.; Johnson, R.S.; Karin, M. Female mice heterozygous for IKK gamma/NEMO deficiencies develop a dermatopathy similar to the human X-linked disorder incontinentia pigmenti. Mol. Cell 2000, 5, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.C.; Ley, S.C. New insights into NF-kappaB regulation and function. Trends Immunol. 2008, 29, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.; Kim, H.; Kearns, J.D.; Tergaonkar, V.; O’Dea, E.; Werner, S.L.; Benedict, C.A.; Ware, C.F.; Ghosh, G.; Verma, I.M.; et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell 2007, 128, 369–381. [Google Scholar] [CrossRef]
- Fusco, A.J.; Huang, D.B.; Miller, D.; Wang, V.Y.; Vu, D.; Ghosh, G. NF-kappaB p52:RelB heterodimer recognizes two classes of kappaB sites with two distinct modes. EMBO Rep. 2009, 10, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Foster, S.L.; Hargreaves, D.C.; Medzhitov, R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 2007, 447, 972–978. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Paul, W.E. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 2010, 327, 1098–1102. [Google Scholar] [CrossRef]
- Wang, N.; Liang, H.; Zen, K. Molecular mechanisms that influence the macrophage m1–m2 polarization balance. Front. Immunol. 2014, 5, 614. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Mosser, D.M. The many faces of macrophage activation. J. Leukoc. Biol. 2003, 73, 209–212. [Google Scholar] [CrossRef]
- Lu, Y.C.; Yeh, W.C.; Ohashi, P.S. LPS/TLR4 signal transduction pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhou, H.; Zhao, J.; Xiao, N.; Roychowdhury, S.; Schmitt, D.; Hu, B.; Ransohoff, R.M.; Harding, C.V.; Hise, A.G.; et al. MyD88-dependent interplay between myeloid and endothelial cells in the initiation and progression of obesity-associated inflammatory diseases. J. Exp. Med. 2014, 211, 887–907. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Tschopp, J. The inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Umemura, A.; Sanchez-Lopez, E.; Liang, S.; Shalapour, S.; Wong, J.; He, F.; Boassa, D.; Perkins, G.; Ali, S.R.; et al. NF-kappaB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell 2016, 164, 896–910. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, R.; Yu, Q.; Bi, Y.; Liu, G. Metabolic regulation of inflammasomes in inflammation. Immunology 2019, 157, 95–109. [Google Scholar] [CrossRef]
- Moon, J.S.; Hisata, S.; Park, M.A.; DeNicola, G.M.; Ryter, S.W.; Nakahira, K.; Choi, A.M.K. mTORC1-Induced HK1-Dependent Glycolysis Regulates NLRP3 Inflammasome Activation. Cell Rep. 2015, 12, 102–115. [Google Scholar] [CrossRef]
- Sanman, L.E.; Qian, Y.; Eisele, N.A.; Ng, T.M.; van der Linden, W.A.; Monack, D.M.; Weerapana, E.; Bogyo, M. Disruption of glycolytic flux is a signal for inflammasome signaling and pyroptotic cell death. eLife 2016, 5, e13663. [Google Scholar] [CrossRef]
- Finucane, O.M.; Sugrue, J.; Rubio-Araiz, A.; Guillot-Sestier, M.V.; Lynch, M.A. The NLRP3 inflammasome modulates glycolysis by increasing PFKFB3 in an IL-1beta-dependent manner in macrophages. Sci. Rep. 2019, 9, 4034. [Google Scholar] [CrossRef] [PubMed]
- Camilli, G.; Bohm, M.; Piffer, A.C.; Lavenir, R.; Williams, D.L.; Neven, B.; Grateau, G.; Georgin-Lavialle, S.; Quintin, J. Beta-Glucan-induced reprogramming of human macrophages inhibits NLRP3 inflammasome activation in cryopyrinopathies. J. Clin. Investig. 2020, 130, 4561–4573. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, J.Y.; Liu, J.Q.; Yang, J.; Liu, Y.; Wang, C.; Ma, X.N.; Liu, B.L.; Xin, G.Z.; Liu, L.F. Succinate/NLRP3 Inflammasome Induces Synovial Fibroblast Activation: Therapeutical Effects of Clematichinenoside AR on Arthritis. Front. Immunol. 2016, 7, 532. [Google Scholar] [CrossRef] [PubMed]
- Youm, Y.H.; Nguyen, K.Y.; Grant, R.W.; Goldberg, E.L.; Bodogai, M.; Kim, D.; D’Agostino, D.; Planavsky, N.; Lupfer, C.; Kanneganti, T.D.; et al. The ketone metabolite beta-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat. Med. 2015, 21, 263–269. [Google Scholar] [CrossRef]
- Hooftman, A.; Angiari, S.; Hester, S.; Corcoran, S.E.; Runtsch, M.C.; Ling, C.; Ruzek, M.C.; Slivka, P.F.; McGettrick, A.F.; Banahan, K.; et al. The Immunomodulatory Metabolite Itaconate Modifies NLRP3 and Inhibits Inflammasome Activation. Cell. Metab. 2020, 32, 468–478.e7. [Google Scholar] [CrossRef]
- Anand, P.K. Lipids, inflammasomes, metabolism, and disease. Immunol. Rev. 2020, 297, 108–122. [Google Scholar] [CrossRef]
- Shen, L.; Yang, Y.; Ou, T.; Key, C.C.; Tong, S.H.; Sequeira, R.C.; Nelson, J.M.; Nie, Y.; Wang, Z.; Boudyguina, E.; et al. Dietary PUFAs attenuate NLRP3 inflammasome activation via enhancing macrophage autophagy. J. Lipid Res. 2017, 58, 1808–1821. [Google Scholar] [CrossRef]
- Karasawa, T.; Kawashima, A.; Usui-Kawanishi, F.; Watanabe, S.; Kimura, H.; Kamata, R.; Shirasuna, K.; Koyama, Y.; Sato-Tomita, A.; Matsuzaka, T.; et al. Saturated Fatty Acids Undergo Intracellular Crystallization and Activate the NLRP3 Inflammasome in Macrophages. Arter. Thromb. Vasc. Biol. 2018, 38, 744–756. [Google Scholar] [CrossRef]
- Guo, C.; Chi, Z.; Jiang, D.; Xu, T.; Yu, W.; Wang, Z.; Chen, S.; Zhang, L.; Liu, Q.; Guo, X.; et al. Cholesterol Homeostatic Regulator SCAP-SREBP2 Integrates NLRP3 Inflammasome Activation and Cholesterol Biosynthetic Signaling in Macrophages. Immunity 2018, 49, 842–856.e7. [Google Scholar] [CrossRef]
- He, M.; Chiang, H.H.; Luo, H.; Zheng, Z.; Qiao, Q.; Wang, L.; Tan, M.; Ohkubo, R.; Mu, W.C.; Zhao, S.; et al. An Acetylation Switch of the NLRP3 Inflammasome Regulates Aging-Associated Chronic Inflammation and Insulin Resistance. Cell Metab. 2020, 31, 580–591.e5. [Google Scholar] [CrossRef]
- Capece, D.; Verzella, D.; Flati, I.; Arboretto, P.; Cornice, J.; Franzoso, G. NF-kappaB: Blending metabolism, immunity, and inflammation. Trends Immunol. 2022, 43, 757–775. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.G.; Hayden, M.S.; Ghosh, S. NF-kappaB, inflammation, and metabolic disease. Cell Metab. 2011, 13, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Mauro, C.; Leow, S.C.; Anso, E.; Rocha, S.; Thotakura, A.K.; Tornatore, L.; Moretti, M.; De Smaele, E.; Beg, A.A.; Tergaonkar, V.; et al. NF-kappaB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat. Cell. Biol. 2011, 13, 1272–1279. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, K.; Araki, K.; Tobiume, K.; Tanaka, N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol. 2008, 10, 611–618. [Google Scholar] [CrossRef]
- Johnson, R.F.; Witzel, I.I.; Perkins, N.D. p53-dependent regulation of mitochondrial energy production by the RelA subunit of NF-kappaB. Cancer Res. 2011, 71, 5588–5597. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Yang, X.D.; Lamb, A.; Chen, L.F. Posttranslational modifications of NF-kappaB: Another layer of regulation for NF-kappaB signaling pathway. Cell Signal. 2010, 22, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Stark, G.R. NF-kappaB: Regulation by Methylation. Cancer Res. 2015, 75, 3692–3695. [Google Scholar] [CrossRef]
- Zhang, T.; Park, K.A.; Li, Y.; Byun, H.S.; Jeon, J.; Lee, Y.; Hong, J.H.; Kim, J.M.; Huang, S.M.; Choi, S.W.; et al. PHF20 regulates NF-kappaB signalling by disrupting recruitment of PP2A to p65. Nat. Commun. 2013, 4, 2062. [Google Scholar] [CrossRef]
- Milanovic, M.; Kracht, M.; Schmitz, M.L. The cytokine-induced conformational switch of nuclear factor kappaB p65 is mediated by p65 phosphorylation. Biochem. J. 2014, 457, 401–413. [Google Scholar] [CrossRef]
- Chen, L.F.; Williams, S.A.; Mu, Y.; Nakano, H.; Duerr, J.M.; Buckbinder, L.; Greene, W.C. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol. Cell Biol. 2005, 25, 7966–7975. [Google Scholar] [CrossRef]
- Lawrence, T.; Bebien, M.; Liu, G.Y.; Nizet, V.; Karin, M. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature 2005, 434, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.F.; Mu, Y.; Greene, W.C. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002, 21, 6539–6548. [Google Scholar] [CrossRef] [PubMed]
- Buerki, C.; Rothgiesser, K.M.; Valovka, T.; Owen, H.R.; Rehrauer, H.; Fey, M.; Lane, W.S.; Hottiger, M.O. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 2008, 36, 1665–1680. [Google Scholar] [CrossRef]
- Bonizzi, G.; Karin, M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004, 25, 280–288. [Google Scholar] [CrossRef]
- Infantino, V.; Iacobazzi, V.; Palmieri, F.; Menga, A. ATP-citrate lyase is essential for macrophage inflammatory response. Biochem. Biophys. Res. Commun. 2013, 440, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Iacobazzi, V.; Infantino, V. Citrate--new functions for an old metabolite. Biol. Chem. 2014, 395, 387–399. [Google Scholar] [CrossRef]
- Santarsiero, A.; Convertini, P.; Vassallo, A.; Santoro, V.; Todisco, S.; Iacobazzi, D.; Fondufe-Mittendorf, Y.; Martelli, G.; de Oliveira, M.R.; Montanaro, R.; et al. Phenolic Compounds of Red Wine Aglianico del Vulture Modulate the Functional Activity of Macrophages via Inhibition of NF-kappaB and the Citrate Pathway. Oxidative Med. Cell. Longev. 2021, 2021, 5533793. [Google Scholar] [CrossRef]
- Santarsiero, A.; Onzo, A.; Pascale, R.; Acquavia, M.A.; Coviello, M.; Convertini, P.; Todisco, S.; Marsico, M.; Pifano, C.; Iannece, P.; et al. Pistacia lentiscus Hydrosol: Untargeted Metabolomic Analysis and Anti-Inflammatory Activity Mediated by NF-kappaB and the Citrate Pathway. Oxidative Med. Cell. Longev. 2020, 2020, 4264815. [Google Scholar] [CrossRef]
- Salminen, A.; Hyttinen, J.M.; Kaarniranta, K. AMP-activated protein kinase inhibits NF-kappaB signaling and inflammation: Impact on healthspan and lifespan. J. Mol. Med. 2011, 89, 667–676. [Google Scholar] [CrossRef]
- Yeung, F.; Hoberg, J.E.; Ramsey, C.S.; Keller, M.D.; Jones, D.R.; Frye, R.A.; Mayo, M.W. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004, 23, 2369–2380. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhou, X.; Liu, Y.; Tan, S.; Li, Y. The Role of Sirtuin-1 in Immune Response and Systemic Lupus Erythematosus. Front. Immunol. 2021, 12, 632383. [Google Scholar] [CrossRef]
- D’Ignazio, L.; Bandarra, D.; Rocha, S. NF-kappaB and HIF crosstalk in immune responses. FEBS J. 2016, 283, 413–424. [Google Scholar] [CrossRef]
- Bandarra, D.; Biddlestone, J.; Mudie, S.; Muller, H.A.; Rocha, S. HIF-1alpha restricts NF-kappaB-dependent gene expression to control innate immunity signals. Dis. Model. Mech. 2015, 8, 169–181. [Google Scholar] [PubMed]
- Todisco, S.; Santarsiero, A.; Convertini, P.; De Stefano, G.; Gilio, M.; Iacobazzi, V.; Infantino, V. PPAR Alpha as a Metabolic Modulator of the Liver: Role in the Pathogenesis of Nonalcoholic Steatohepatitis (NASH). Biology 2022, 11, 792. [Google Scholar] [CrossRef] [PubMed]
- Pontis, S.; Ribeiro, A.; Sasso, O.; Piomelli, D. Macrophage-derived lipid agonists of PPAR-alpha as intrinsic controllers of inflammation. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 7–14. [Google Scholar] [CrossRef]
- Cheang, W.S.; Tian, X.Y.; Wong, W.T.; Huang, Y. The peroxisome proliferator-activated receptors in cardiovascular diseases: Experimental benefits and clinical challenges. Br. J. Pharm. 2015, 172, 5512–5522. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Tian, R.; Zhang, X.; Yao, Q.; Chen, Q.; Liu, B.; Liao, L.; Gong, Y.; Yang, H.; Wang, D. PPARgamma inhibits small airway remodeling through mediating the polarization homeostasis of alveolar macrophages in COPD. Clin. Immunol. 2023, 250, 109293. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.J.; Liu, Z.G. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Schieber, M.; Chandel, N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef]
- Chandel, N.S.; Trzyna, W.C.; McClintock, D.S.; Schumacker, P.T. Role of oxidants in NF-kappa B activation and TNF-alpha gene transcription induced by hypoxia and endotoxin. J. Immunol. 2000, 165, 1013–1021. [Google Scholar] [CrossRef]
- Djavaheri-Mergny, M.; Javelaud, D.; Wietzerbin, J.; Besancon, F. NF-kappaB activation prevents apoptotic oxidative stress via an increase of both thioredoxin and MnSOD levels in TNFalpha-treated Ewing sarcoma cells. FEBS Lett. 2004, 578, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Hu, J.; Ketterer, B.; Taylor, J.B. The organization of the human GSTP1-1 gene promoter and its response to retinoic acid and cellular redox status. Biochem. J. 1996, 313 Pt 1, 155–161. [Google Scholar] [CrossRef]
- Schreiber, J.; Jenner, R.G.; Murray, H.L.; Gerber, G.K.; Gifford, D.K.; Young, R.A. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc. Natl. Acad. Sci. USA 2006, 103, 5899–5904. [Google Scholar] [CrossRef] [PubMed]
- Nagy, C.; Haschemi, A. Time and Demand are Two Critical Dimensions of Immunometabolism: The Process of Macrophage Activation and the Pentose Phosphate Pathway. Front. Immunol. 2015, 6, 164. [Google Scholar] [CrossRef]
- Infantino, V.; Convertini, P.; Cucci, L.; Panaro, M.A.; Di Noia, M.A.; Calvello, R.; Palmieri, F.; Iacobazzi, V. The mitochondrial citrate carrier: A new player in inflammation. Biochem. J. 2011, 438, 433–436. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A. A critical role for citrate metabolism in LPS signalling. Biochem. J. 2011, 438, e5–e6. [Google Scholar] [CrossRef]
- Mills, E.L.; Kelly, B.; Logan, A.; Costa, A.S.H.; Varma, M.; Bryant, C.E.; Tourlomousis, P.; Dabritz, J.H.M.; Gottlieb, E.; Latorre, I.; et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016, 167, 457–470.e13. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, R.; Gu, H.; Zhang, E.; Qu, J.; Cao, W.; Huang, X.; Yan, H.; He, J.; Cai, Z. Metabolic reprogramming in macrophage responses. Biomark. Res. 2021, 9, 1. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacobazzi, D.; Convertini, P.; Todisco, S.; Santarsiero, A.; Iacobazzi, V.; Infantino, V. New Insights into NF-κB Signaling in Innate Immunity: Focus on Immunometabolic Crosstalks. Biology 2023, 12, 776. https://doi.org/10.3390/biology12060776
Iacobazzi D, Convertini P, Todisco S, Santarsiero A, Iacobazzi V, Infantino V. New Insights into NF-κB Signaling in Innate Immunity: Focus on Immunometabolic Crosstalks. Biology. 2023; 12(6):776. https://doi.org/10.3390/biology12060776
Chicago/Turabian StyleIacobazzi, Dominga, Paolo Convertini, Simona Todisco, Anna Santarsiero, Vito Iacobazzi, and Vittoria Infantino. 2023. "New Insights into NF-κB Signaling in Innate Immunity: Focus on Immunometabolic Crosstalks" Biology 12, no. 6: 776. https://doi.org/10.3390/biology12060776
APA StyleIacobazzi, D., Convertini, P., Todisco, S., Santarsiero, A., Iacobazzi, V., & Infantino, V. (2023). New Insights into NF-κB Signaling in Innate Immunity: Focus on Immunometabolic Crosstalks. Biology, 12(6), 776. https://doi.org/10.3390/biology12060776




