Rediscovery of Remarkably Rare Anaerobic Tentaculiferous Ciliate Genera Legendrea and Dactylochlamys (Ciliophora: Litostomatea)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Morphological Characterization
2.3. DNA Extraction, Amplification, and Sequencing
2.4. Phylogenetic Analyses
3. Results
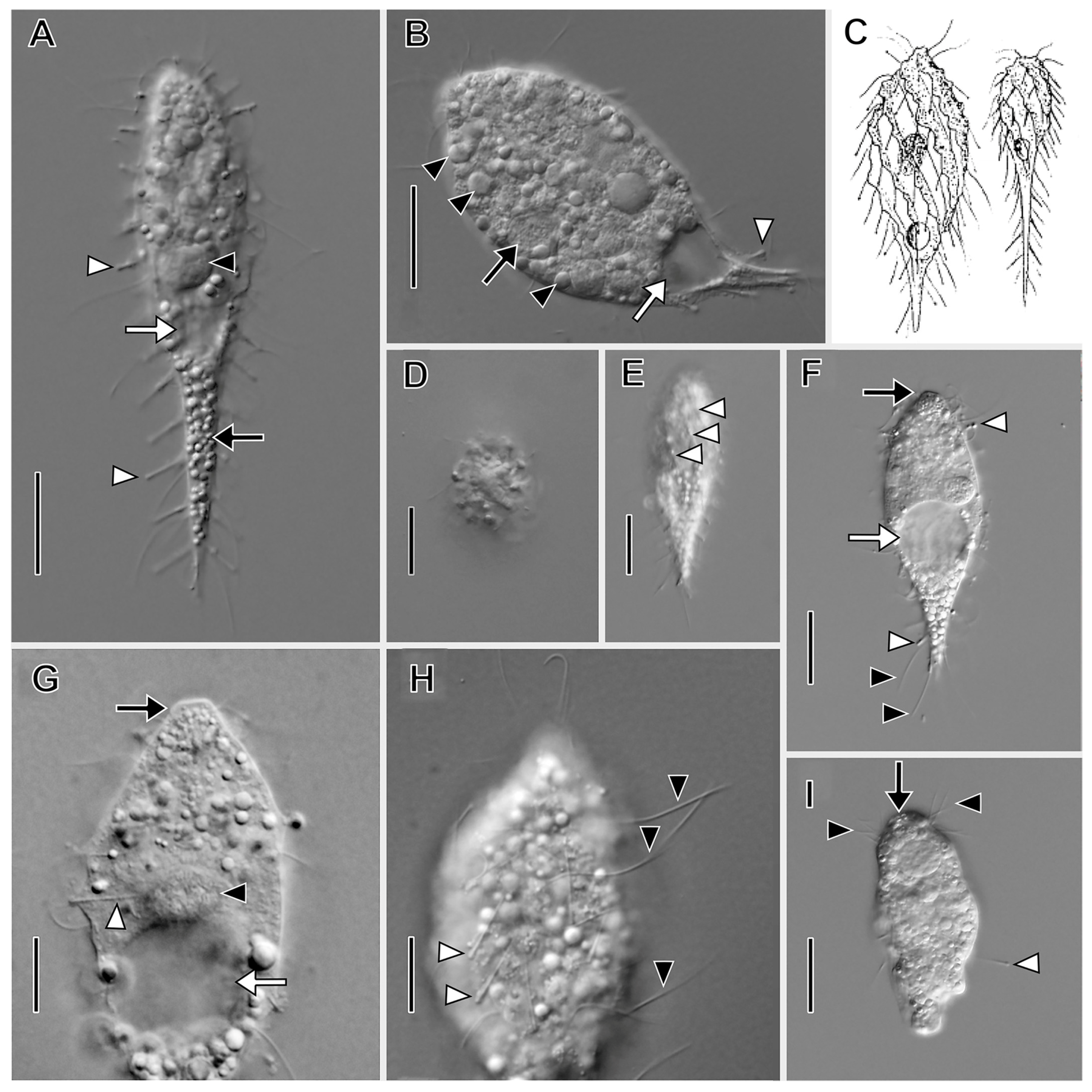
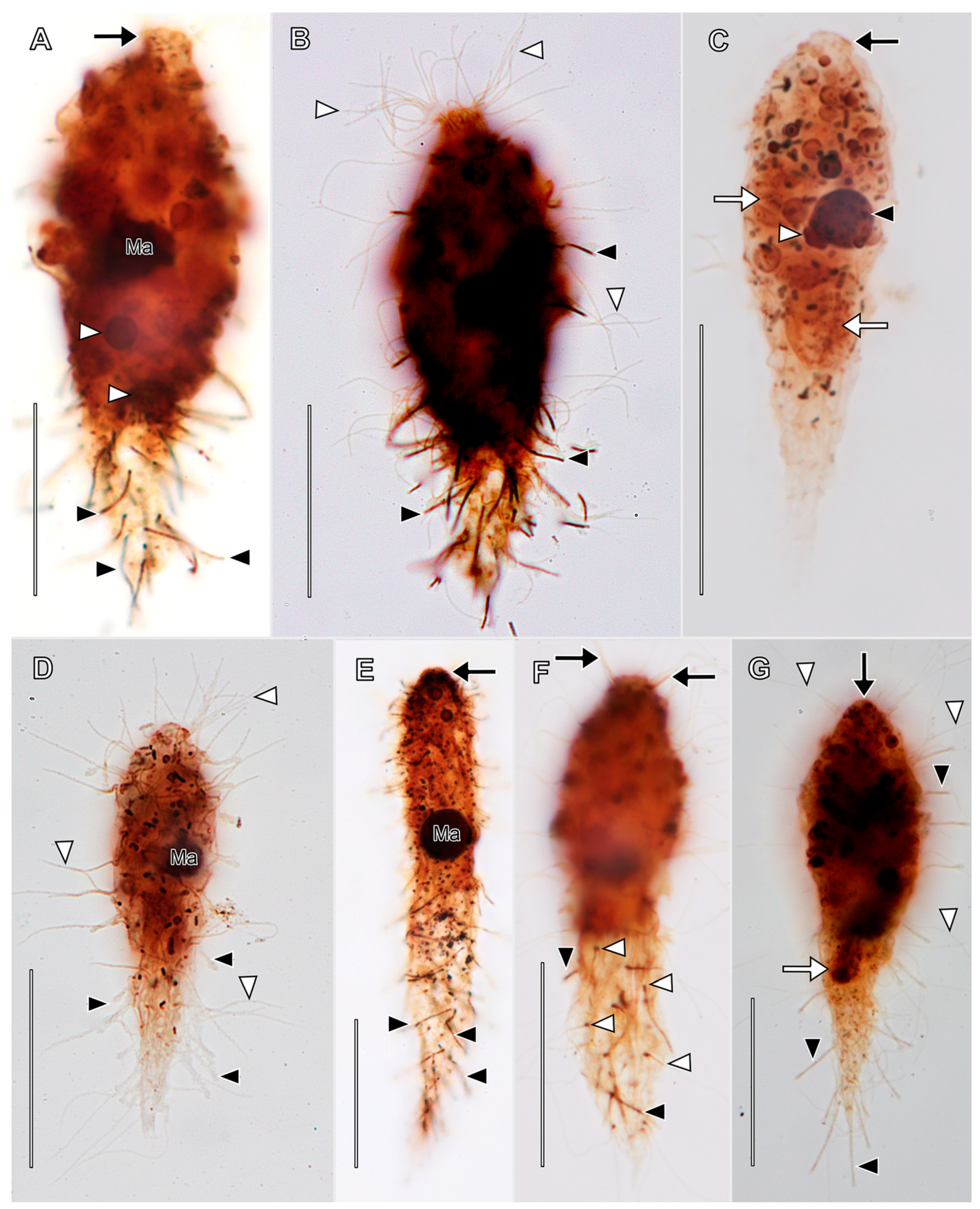
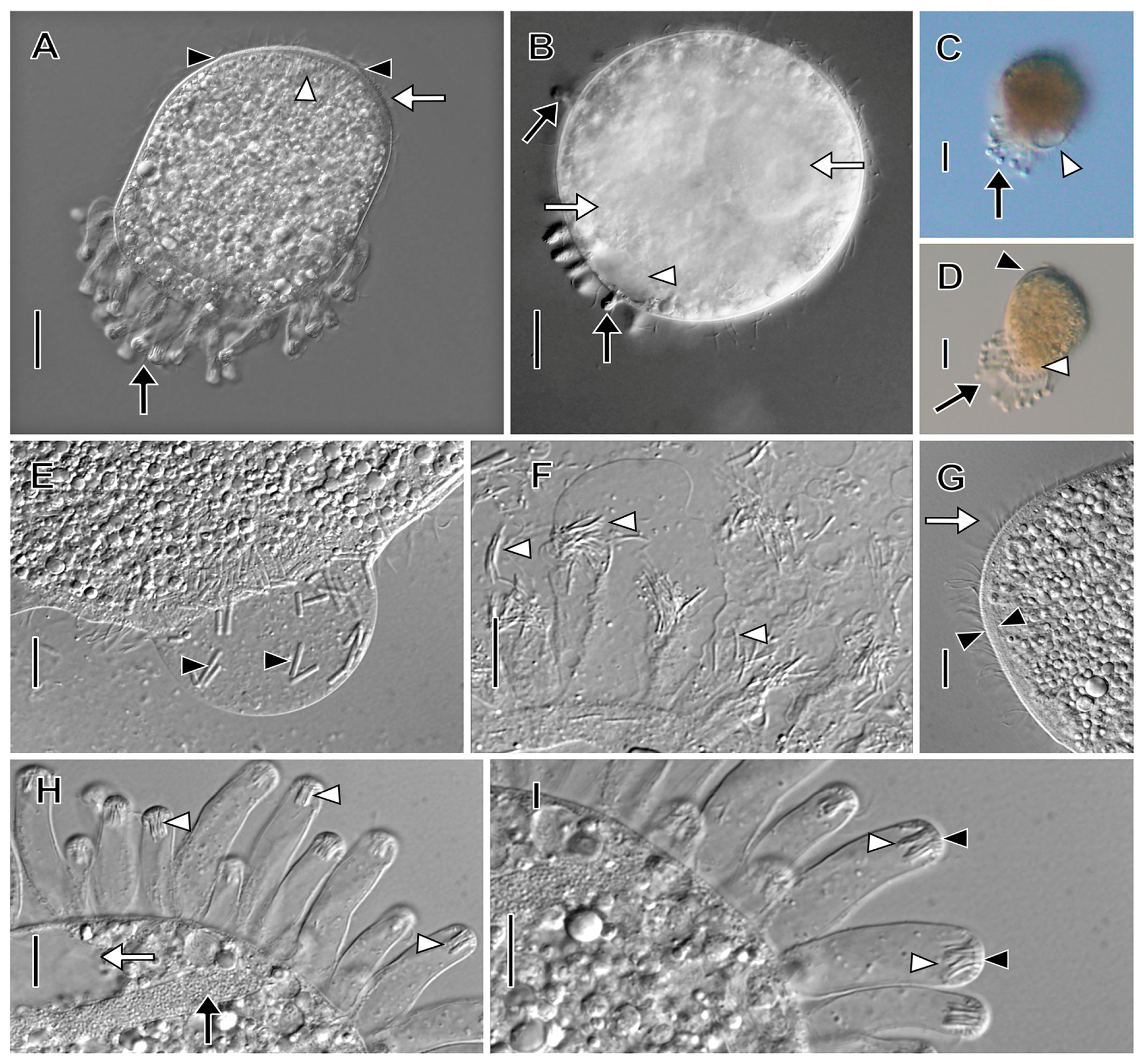
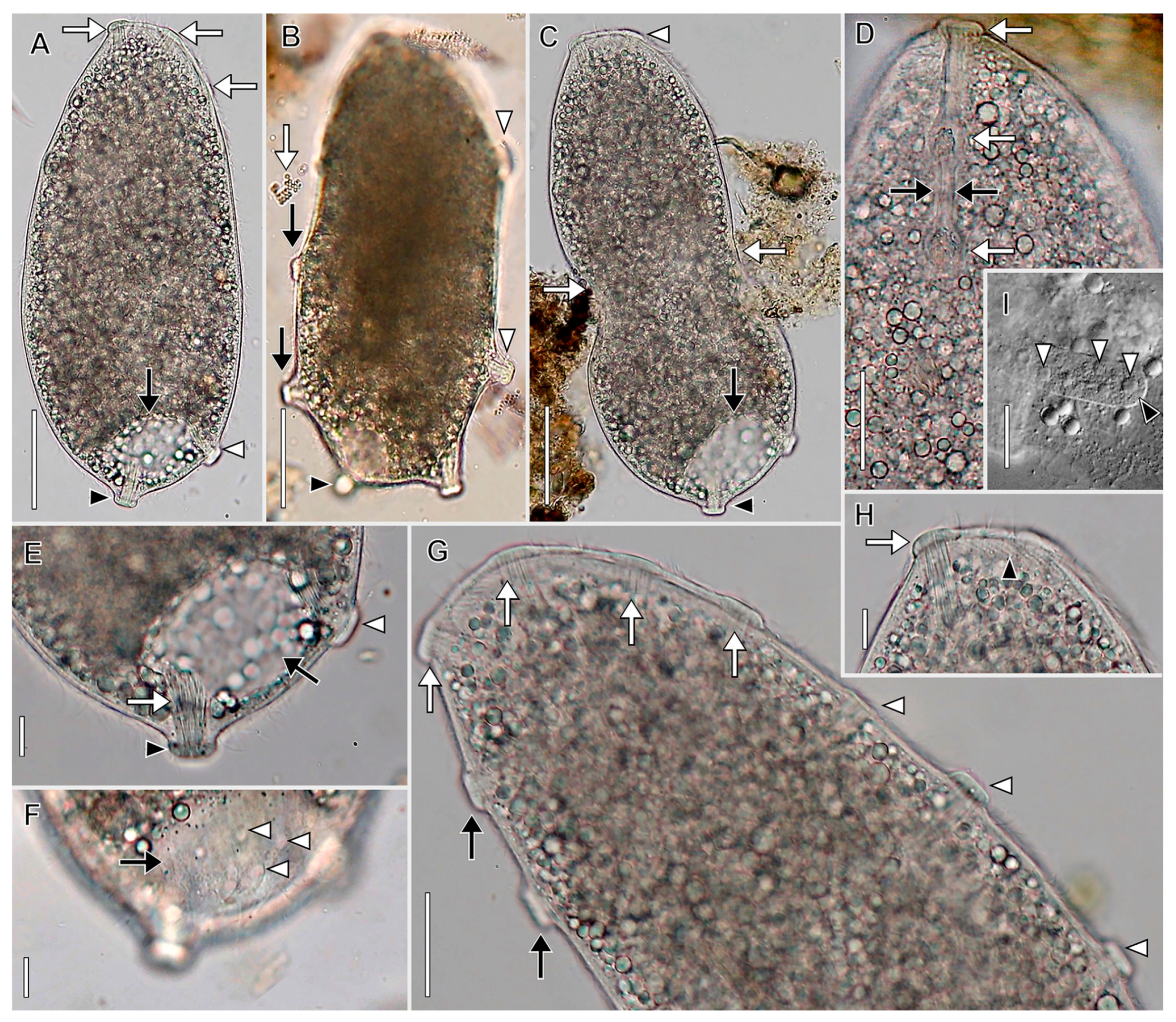
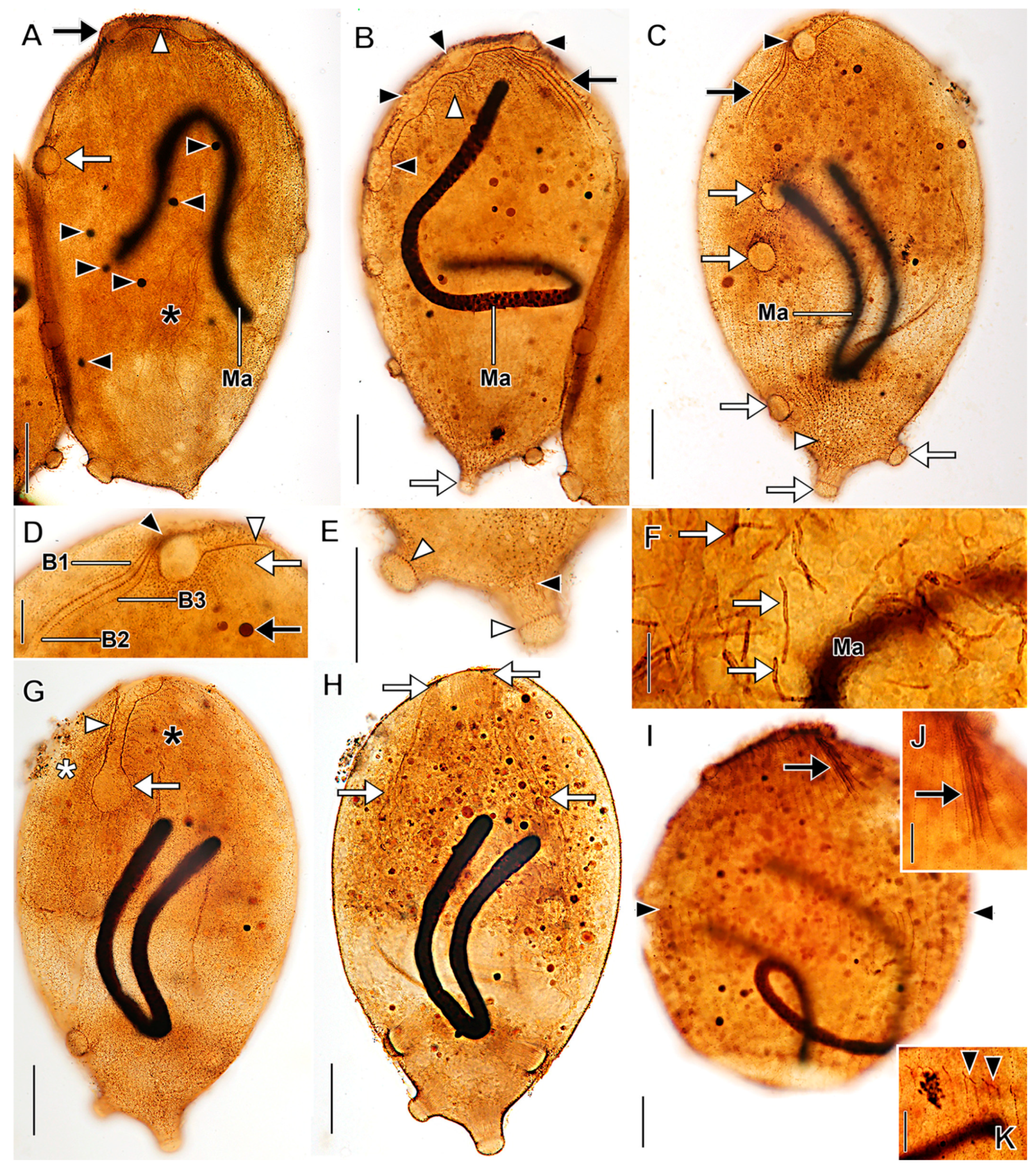
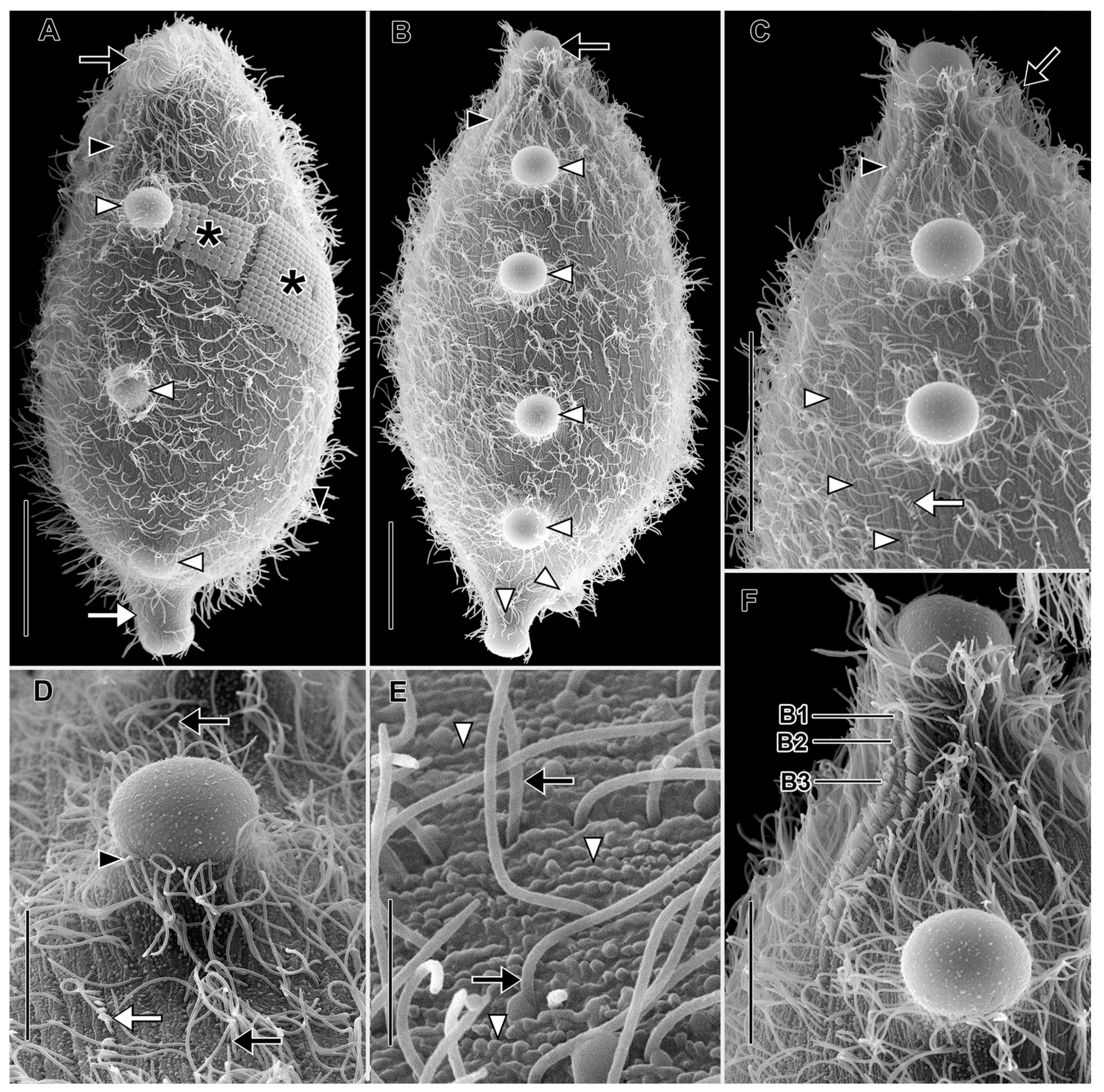
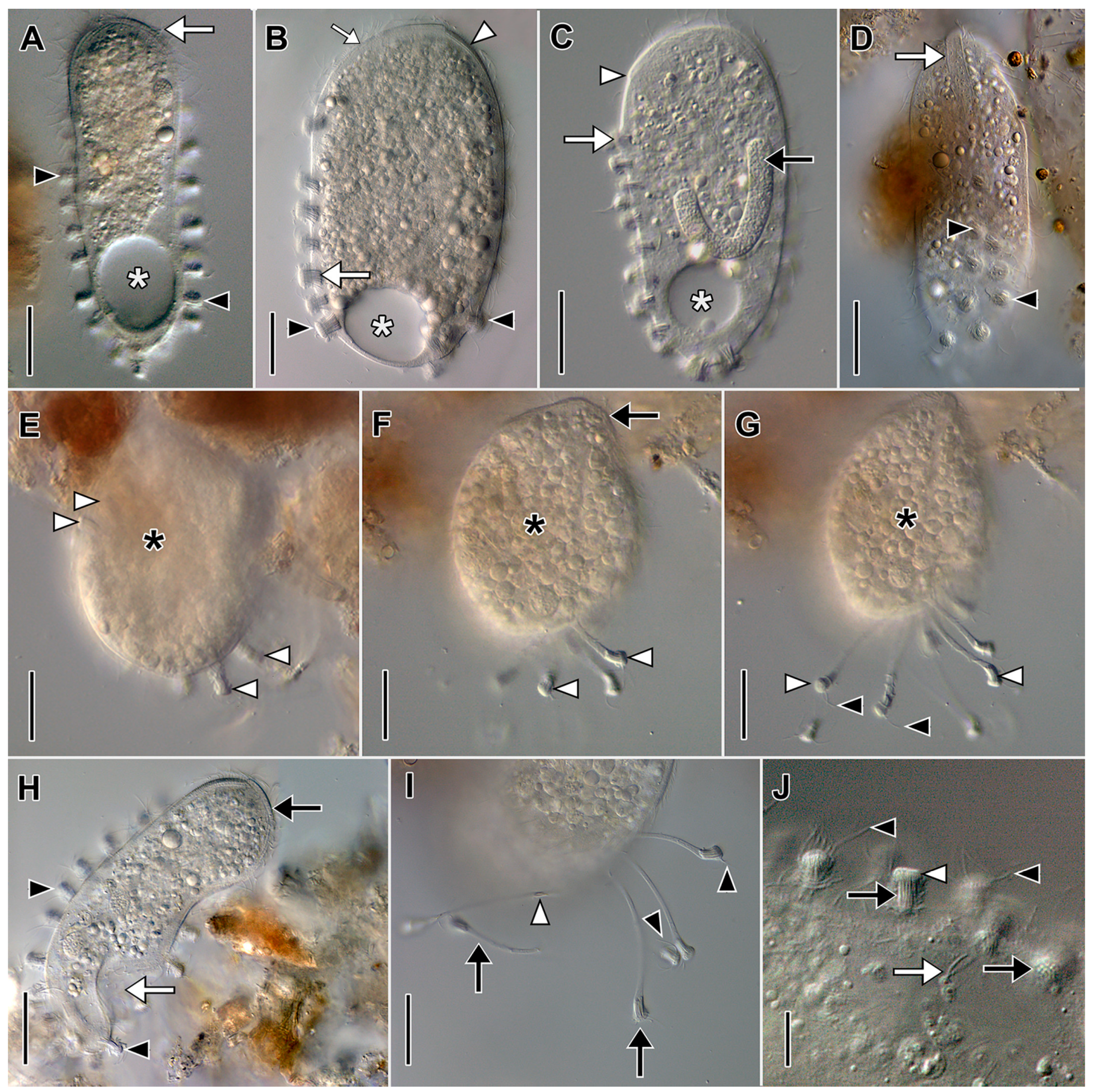
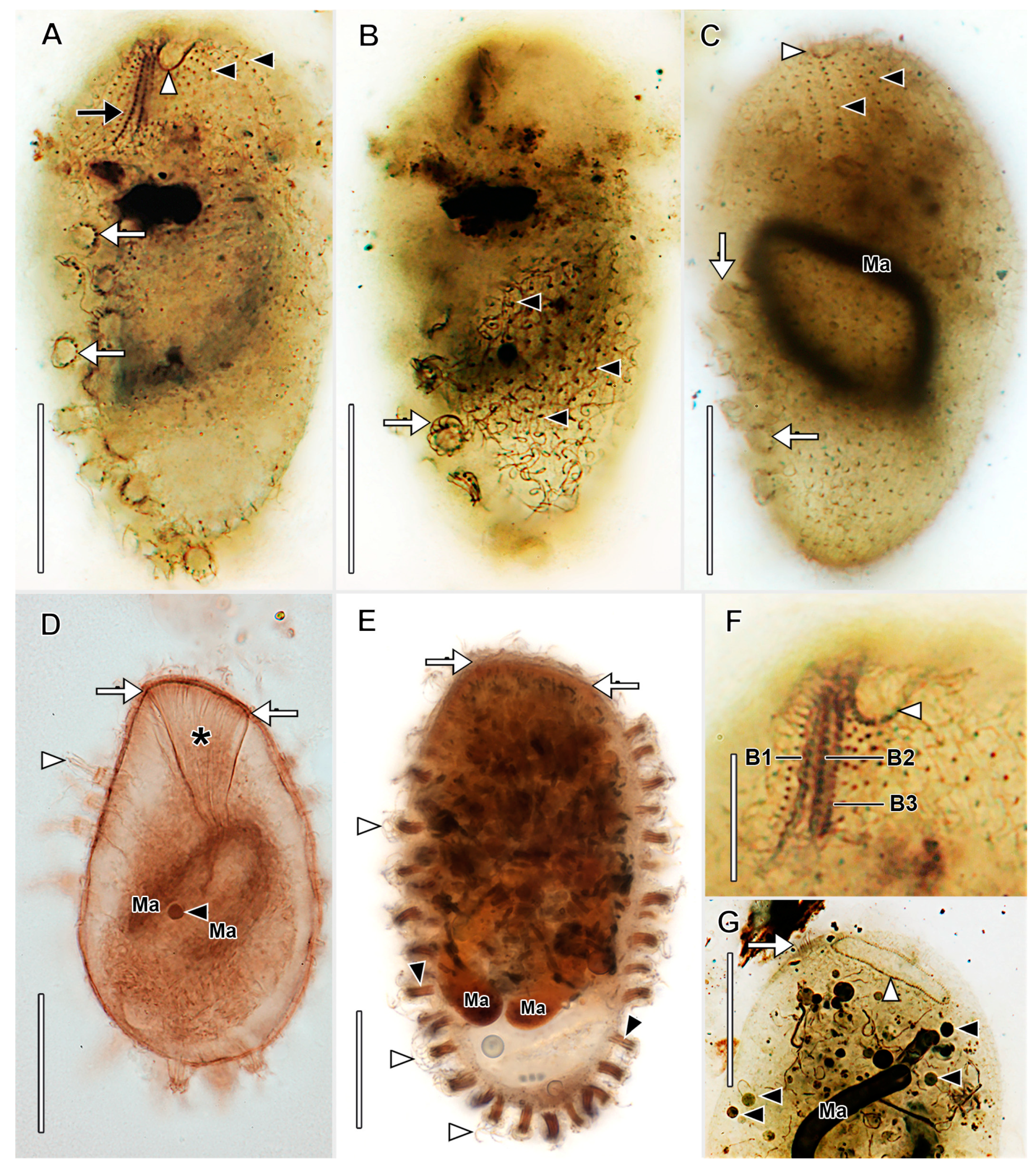
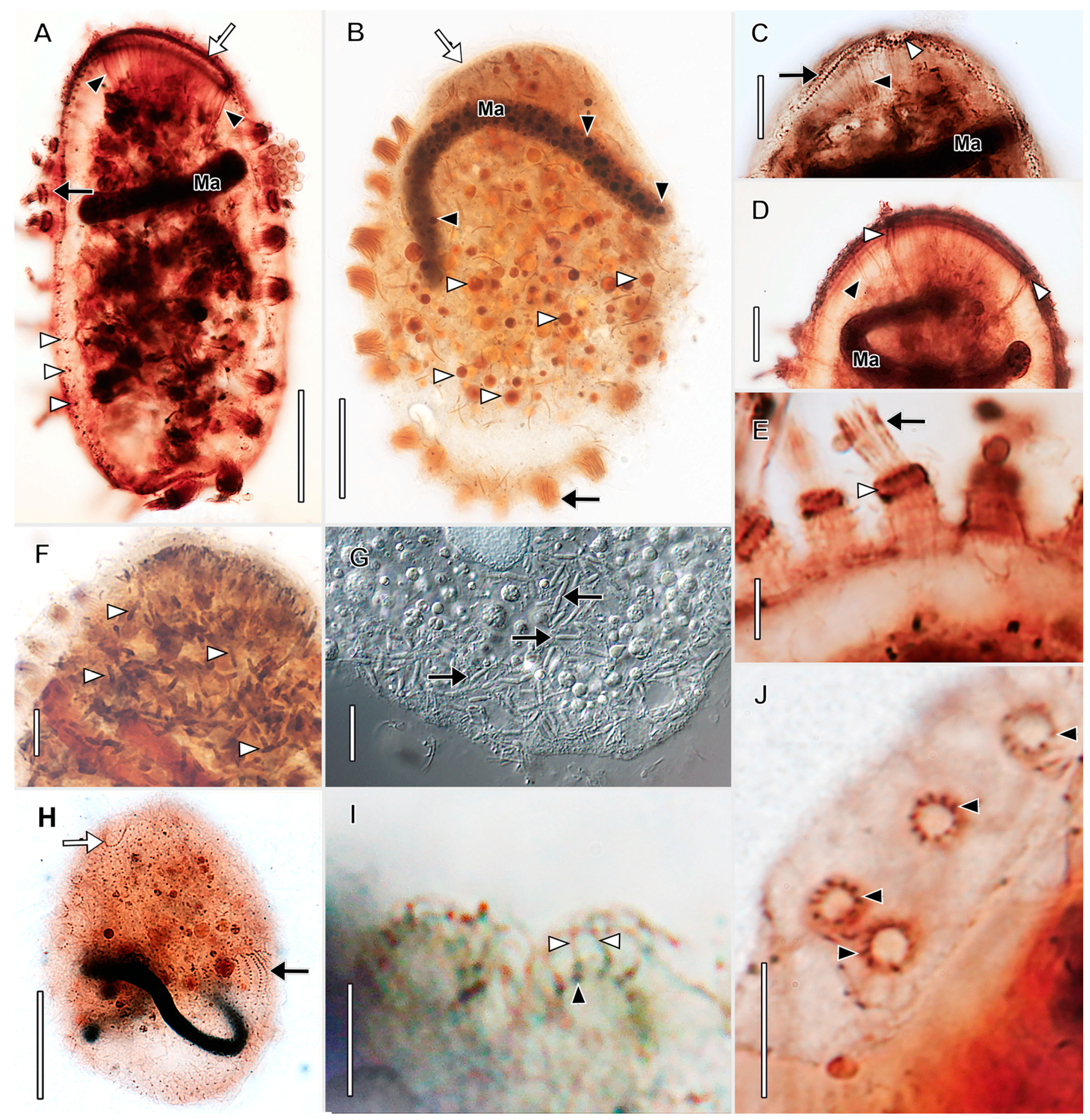
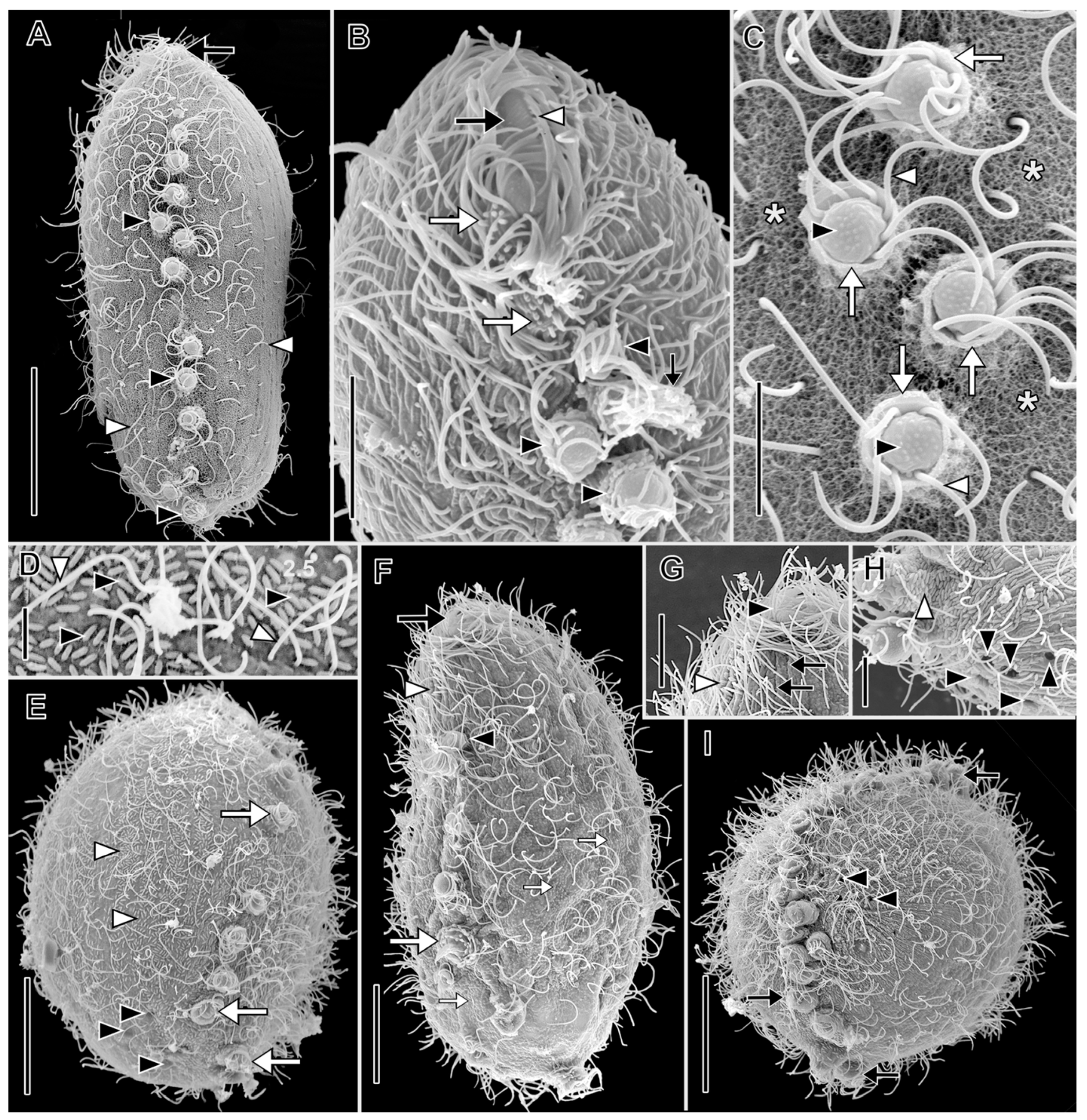
3.1. Morphological Descriptions and Diagnoses
3.1.1. Dactylochlamys Pisciformis Lauterborn, 1901
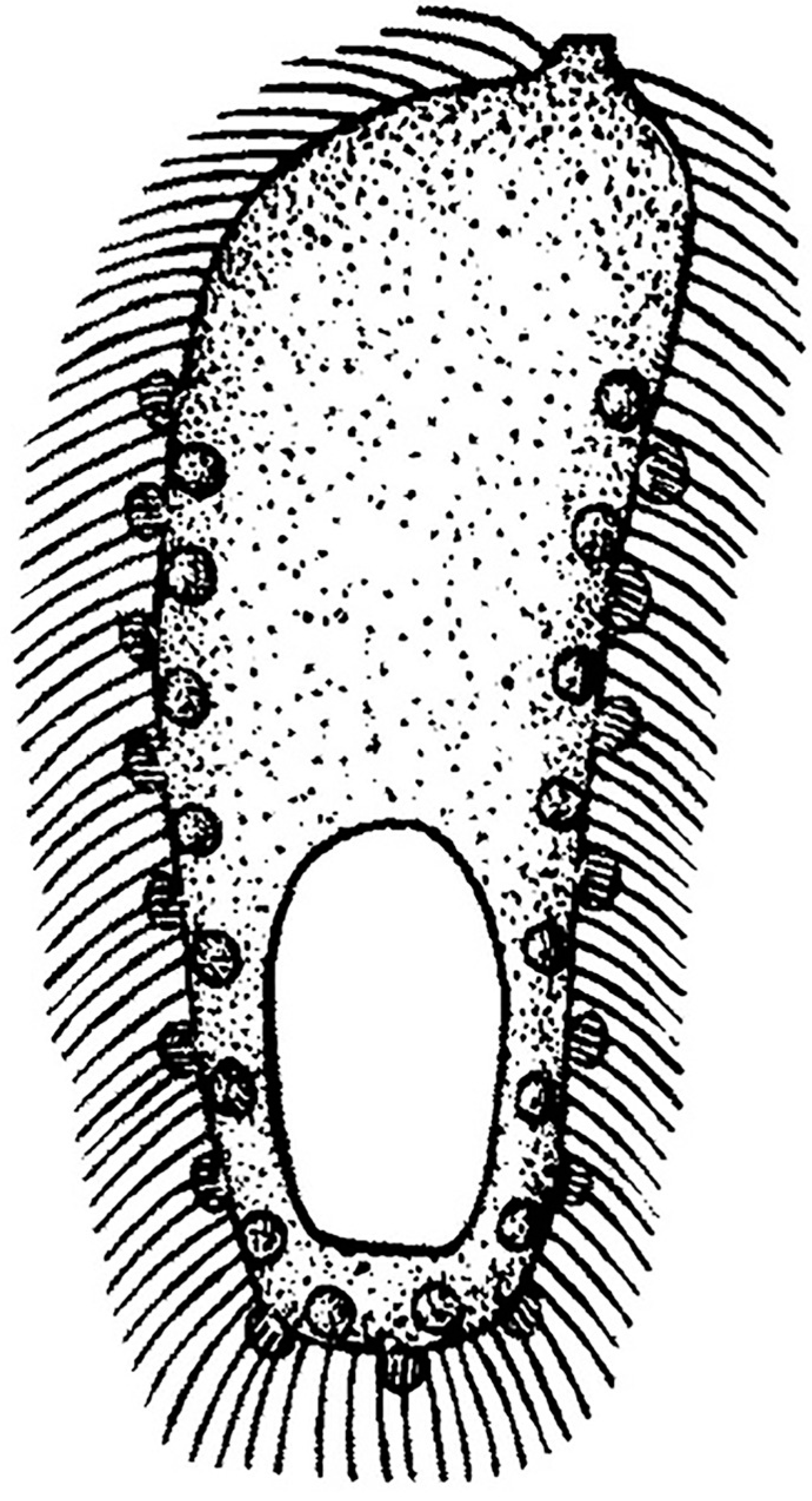
3.1.2. Legendrea Fauré-Fremiet, 1908
- Legendrea loyezae Fauré-Fremiet, 1908
- Legendrea pespelicani Penard, 1922
- Legendrea ornata (Stokes, 1887) Penard, 1914
- Original combination, Holophrya ornata Stokes, 1887
3.2. Molecular Data and Phylogenetic Analysis
4. Discussion
4.1. Morphological Comparison of Legendrea Species and Similar Species
4.2. Remarks on the Rarity and Ecology of Dactylochlamys and Legendrea
4.3. Phylogenetic Analysis and the Relationship of Tentaculiferous Ciliates
4.4. Putative Prokaryotic Endosymbionts
4.5. The Role of Citizen Science in Ciliatology
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lynn, D.H. The Ciliated Protozoa: Characterization, Classification, and Guide to the Literature; Springer: Dordrecht, Germany, 2008. [Google Scholar]
- Foissner, W.; Xu, K. Monograph of the Spathidiida (Ciliophora, Haptoria). Monogr. Biol. 2007, 81, 1–485. [Google Scholar]
- Cedrola, F.; Senra, M.V.X.; Rossi, M.F.; Fregulia, P.; D’Agosto, M.; Dias, R.J.P. Trichostomatid ciliates (Alveolata, Ciliophora, Trichostomatia) systematics and diversity: Past, present and future. Front. Microbiol. 2020, 10, 2967. [Google Scholar] [CrossRef]
- Foissner, W.; Berger, H.; Schaumburg, H. Identification and Ecology of Limnetic Plankton Ciliates; Informationsberichte des Bayerischen Landesamtes für Wasserwirtschaft: Deggendorf, Germany, 1999. [Google Scholar]
- Kreutz, M.; Foissner, W. The Sphagnum ponds of Simmelried in Germany: A biodiversity hot-spot for microscopic organisms. Protozool. Monogr. 2006, 3, 1–267. [Google Scholar]
- Jiang, L.; Wang, C.; Zhuang, W.; Li, S.; Hu, X. Taxonomy, phylogeny, and geographical distribution of the little-known Helicoprorodon multinucleatum Dragesco, 1960 (Ciliophora, Haptorida) and key to species within the genus. Eur. J. Protistol. 2021, 78, 125769. [Google Scholar] [CrossRef] [PubMed]
- Penard, E. Un curieux infusoire Legendrea bellerophon. Rev. Suisse Zool. 1914, 22, 407–432. [Google Scholar]
- Weiss, J.; Andreou, D.; Esteban, G.F. The Extraordinarily rare ciliate Legendrea loyezae Fauré-Fremiet, 1908 (Haptoria, Ciliophora). Protist 2022, 173, 125912. [Google Scholar] [CrossRef] [PubMed]
- Penard, E. Études sur les Infusoires d’Eau Douce; George & Cie: Geneve, Swizerland, 1922. [Google Scholar]
- Kahl, A. Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria) I. Allgemeiner Teil und Prostomata. Die Tierwelt Dtschl. Und Der Angrenz. Meerest. Nach Ihren Merkmalen Und Nach Ihrer Lebensw. 1930, 18, 1–186. [Google Scholar]
- Foissner, W.; Xu, K.; Kreutz, M. The Apertospathulidae, a new family of haptorid ciliates (Protozoa, Ciliophora). J. Eukaryot. Microbiol. 2005, 52, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Bourland, W.A.; Wendell, L.; Hampikian, G. Morphologic and molecular description of Metopus fuscus Kahl from North America and new rDNA sequences from seven metopids (Armophorea, Metopidae). Eur. J. Protistol. 2014, 50, 213–230. [Google Scholar] [CrossRef]
- Dieckmann, J. An improved protargol impregnation for ciliates yielding reproducible results. Eur. J. Protistol. 1995, 31, 372–382. [Google Scholar] [CrossRef]
- Foissner, W. An update of ‘basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa’. Int. J. Syst. Evol. Microbiol. 2014, 64, 271–292. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Bourland, W.A.; Song, W. Protargol synthesis: An in-house protocol. J. Eukaryot. Microbiol. 2013, 60, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Strüder-Kypke, M.C.; Lynn, D.H. Sequence analyses of the small subunit rRNA gene confirm the paraphyly of oligotrich ciliates sensu lato and support the monophyly of the subclasses Oligotrichia and Choreotrichia (Ciliophora, Spirotrichea). J. Zool. 2003, 260, 87–97. [Google Scholar] [CrossRef]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 7, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Jerome, C.A.; Lynn, D.H.; Simon, E.M. Description of Tetrahymena empidokyrea n. sp., a new species in the Tetrahymena pyriformis sibling species complex (Ciliophora, Oligohymenophorea), and an assessment of its phylogenetic position using small-subunit rRNA sequences. Can. J. Zool. 1996, 74, 1898–1906. [Google Scholar] [CrossRef]
- Miao, M.; Warren, A.; Song, W.; Wang, S.; Shang, H.; Chen, Z. Analysis of the internal transcribed spacer 2 (ITS2) region of scuticociliates and related taxa (Ciliophora, Oligohymenophorea) to infer their evolution and phylogeny. Protist 2008, 159, 519–533. [Google Scholar] [CrossRef]
- Pawlowski, J. Introduction to the molecular systematics of foraminifera. Micropaleontology 2000, 46, 1–12. Available online: http://www.jstor.org/stable/1486176 (accessed on 22 December 2022).
- van Hoek, A.H.; van Alen, T.A.; Sprakel, V.S.; Leunissen, J.A.; Brigge, T.; Vogels, G.D.; Hackstein, J.H. Multiple acquisition of methanogenic archaeal symbionts by anaerobic ciliates. Mol. Biol. Evol. 2000, 17, 251–258. [Google Scholar] [CrossRef]
- Irbis, C.; Ushida, K. Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J. Gen. Appl. Microbiol. 2004, 50, 203–212. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Gentekaki, E.; Kolisko, M.; Boscaro, V.; Bright, K.J.; Dini, F.; Di Giuseppe, G.; Gong, Y.; Miceli, C.; Modeo, L.; Molestina, R.E.; et al. Large-scale phylogenomic analysis reveals the phylogenetic position of the problematic taxon Protocruzia and unravels the deep phylogenetic affinities of the ciliate lineages. Mol. Phylogenet. Evol. 2014, 78, 36–42. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Flouri, T.; Izquierdo-Carrasco, F.; Darriba, D.; Aberer, A.J.; Nguyen, L.T.; Minh, B.Q.; Von Haeseler, A.; Stamatakis, A. The phylogenetic likelihood library. Syst. Biol. 2015, 64, 356–362. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Warren, D.L.; Geneva, A.J.; Lanfear, R. RWTY (R We There Yet): An R package for examining convergence of Bayesian phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Lauterborn, R. Die “sapropelische“ Lebewelt. Zool. Anz. 1901, 24, 50–55. [Google Scholar]
- Wetzel, A. Der Faulschlamm und seine ziliaten Leitformen. Z. Morphol. Ökol. Tiere 1928, 13, 179–328. [Google Scholar] [CrossRef]
- Fauré-Fremiet, E. Sur deux Infusoires nouveaux de la famille des Trachelidae. Bull. Soc. Zool. Fr. 1908, 33, 13–16. [Google Scholar]
- Stokes, A.C. Notices of new fresh-water infusoria. Proc. Am. Philos. Soc. 1887, 24, 244–255. [Google Scholar]
- Babko, R.V.; Kuzmina, T.M. Spatial distribution of ciliates (Protista, Ciliophora) in the river Bityza (Dnieper basin). Vestnik Zoologii 1999, 33, 83–89. [Google Scholar]
- Lackey, J.B. The Plankton Algae and Protozoa of Two Tennessee Rivers. Am. Midl. Nat. 1942, 27, 191–202. [Google Scholar] [CrossRef]
- Strayer, D. The benthic micrometazoans of Mirror Lake, New Hampshire. Arch. Hydrobiol. 1985, 72, 287–426. [Google Scholar]
- Vd’ačný, P.; Bourland, W.A.; Orsi, W.; Epstein, S.S.; Foissner, W. Genealogical analyses of multiple loci of litostomatean ciliates (Protista, Ciliophora, Litostomatea). Mol. Phylogenet. Evol. 2012, 65, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Vďačný, P.; Breiner, H.W.; Yashchenko, V.; Dunthorn, M.; Stoeck, T.; Foissner, W. The chaos prevails: Molecular phylogeny of the Haptoria (Ciliophora, Litostomatea). Protist 2014, 165, 93–111. [Google Scholar] [CrossRef] [PubMed]
- Jankowski, A.W. Phylum Ciliophora Doflein, 1901. In Protista. Part 2, Handbook on Zoology; Alimov, A.F., Ed.; Russian Academy of Sciences, Zoological Institut: St. Petersburg, Russia, 2007; pp. 371–993. [Google Scholar]
- Corliss, J.O. The Ciliated Protozoa. Characterization, Classification and Guide to the Literature; Pergamon Press: New York, NY, USA, 1979. [Google Scholar]
- Jang, S.W.; Vďačný, P.; Shazib, S.U.A.; Shin, M.K. Linking morphology and molecules: Integrative taxonomy of spathidiids (Protista: Ciliophora: Litostomatea) from Korea. J. Nat. Hist. 2017, 51, 1–36. [Google Scholar] [CrossRef]
- Rajter, Ľ.; Vďačný, P. Rapid radiation, gradual extinction and parallel evolution challenge generic classification of spathidiid ciliates (Protista, Ciliophora). Zool. Scr. 2015, 45, 200–223. [Google Scholar] [CrossRef]
- Ylidiz, I. Morphology and phylogeny of Apertospathula oktemae n. sp. (Ciliophora, Haptoria, Spathidiida) from Lake Van, Turkey. Eur. J. Protistol. 2018, 66, 1–8. [Google Scholar] [CrossRef]
- Holt, P.; Corliss, J.O. Pattern variability in microtubular arrays associated with the tentacles of Actinobolina (Ciliatea: Gymnostomatida). J. Cell Biol. 1973, 58, 213–219. [Google Scholar] [CrossRef]
- Lasek-Nesselquist, E.; Johnson, M.D. A phylogenomic approach to clarifying the relationship of Mesodinium within the Ciliophora: A sase Study in the complexity of mixed-species transcriptome analyses. Genome Biol. Evol. 2019, 11, 3218–3232. [Google Scholar] [CrossRef]
- Lindholm, T.; Lindroosand, P.; Mörk, A.C. Ultrastructure of the photosynthetic ciliate Mesodinium rubrum. Biosystems 1988, 21, 141–149. [Google Scholar] [CrossRef]
- Foissner, W.; Foissner, I. Fine structure and systematic position of Enchelyomorpha vermicularis (Smith, 1899) Kahl, 1930, an anaerobic ciliate (Protozoa, Ciliophora) from domestic sewage. Acta Protozool. 1995, 34, 21–34. [Google Scholar]
- Rudzinska, M.A. The fine structure and function of the tentacle in Tokophrya infusionum. J. Cell Biol. 1965, 25, 459–477. [Google Scholar] [CrossRef]
- Rudzinska, M.A. Do Suctoria really feed by suction? Bioscience 1973, 23, 87–94. [Google Scholar] [CrossRef]
- Greening, C.; Ahmed, F.H.; Mohamed, A.E.; Lee, B.M.; Pandey, G.; Warden, A.C.; Scott, C.; Oakeshott, J.G.; Taylor, M.C.; Jackson, C.J. Physiology, biochemistry, and applications of F420- and F0- dependent redox reactions. Microbiol. Mol. Biol. Rev. 2016, 80, 451–493. [Google Scholar] [CrossRef]
- Boxma, B.; de Graaf, R.M.; van der Staay, G.W.; van Alen, T.A.; Ricard, G.; Gabaldón, T.; van Hoek, A.H.; Moon-van der Staay, S.Y.; Koopman, W.J.; van Hellemond, J.J.; et al. An anaerobic mitochondrion that produces hydrogen. Nature 2015, 434, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Fenchel, T.; Finlay, B.J. The biology of free-living anaerobic ciliates. Eur. J. Protistol. 1991, 26, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Rotterová, J.; Salomaki, E.; Pánek, T.; Bourland, W.; Žihala, D.; Táborský, P.; Edgcomb, V.P.; Beinart, R.A.; Kolísko, M.; Čepička, I. Genomics of new ciliate lineages provides insight into the evolution of obligate anaerobiosis. Cur. Biol. 2020, 30, 2037–2050.e6. [Google Scholar] [CrossRef] [PubMed]
- Kuever, J. The Family Syntrophaceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 281–288. [Google Scholar]
- Bass, D.; Boenigk, J. Everything is everywhere: A twenty-first century reconstruction with respect to protists. In Biogeography of Microscopic Organisms: Is Everything Small Everywhere? Fontaneto, D., Ed.; Cambridge University Press: Cambridge, UK, 2011; pp. 88–110. [Google Scholar]
- Fenchel, T.; Finlay, B.J.; Esteban, G.F. Cosmopolitan metapopulations? Protist 2019, 170, 314–318. [Google Scholar] [CrossRef]
- Foissner, W. Protist diversity and distribution: Some basic considerations. Biodivers. Conserv. 2008, 17, 235–242. [Google Scholar] [CrossRef]
- Agnarsson, I.; Kuntner, M. Taxonomy in a changing world: Seeking solutions for a science in crisis. Syst. Biol. 2007, 56, 531–539. [Google Scholar] [CrossRef]
- Engel, M.S.; Ceríaco, L.M.; Daniel, G.M.; Dellapé, P.M.; Löbl, I.; Marinov, M.; Reis, R.E.; Young, M.T.; Dubois, A.; Agarwal, I.; et al. The taxonomic impediment: A shortage of taxonomists, not the lack of technical approaches. Zool. J. Linn. Soc. 2021, 193, 381–387. [Google Scholar] [CrossRef]
- Acorn, J. Amateurs and abandoned science. Am. Entomol. 2009, 55, 127–128. [Google Scholar] [CrossRef]
- Fischer, E.E.; Cobb, N.S.; Kawahara, A.Y.; Zaspel, J.M.; Cognato, A.I. Decline of amateur Lepidoptera collectors threatens the future of specimen-based research. Bioscience 2021, 71, 396–404. [Google Scholar] [CrossRef]
- Gura, T. Citizen science: Amateur experts. Nature 2013, 496, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Bergerot, B. The Citizen Science Paradox. Land 2022, 11, 1151. [Google Scholar] [CrossRef]
- Foissner, W.; Wenzel, F. Life and legacy of an outstanding ciliate taxonomist, Alfred Kahl (1877–1946), including a facsimile of his forgotten monograph from 1943. Acta Protozool. 2004, 43, 3–69. [Google Scholar]
- Santaoja, M. Social media in learning on nature: Case Finnish amateur mycologists. Horiz. Int. J. Learn. Futures 2022, 30, 122–130. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pomahač, O.; Méndez-Sánchez, D.; Poláková, K.; Müller, M.; Solito, M.-M.; Bourland, W.A.; Čepička, I. Rediscovery of Remarkably Rare Anaerobic Tentaculiferous Ciliate Genera Legendrea and Dactylochlamys (Ciliophora: Litostomatea). Biology 2023, 12, 707. https://doi.org/10.3390/biology12050707
Pomahač O, Méndez-Sánchez D, Poláková K, Müller M, Solito M-M, Bourland WA, Čepička I. Rediscovery of Remarkably Rare Anaerobic Tentaculiferous Ciliate Genera Legendrea and Dactylochlamys (Ciliophora: Litostomatea). Biology. 2023; 12(5):707. https://doi.org/10.3390/biology12050707
Chicago/Turabian StylePomahač, Ondřej, Daniel Méndez-Sánchez, Kateřina Poláková, Michael Müller, Michel-Marie Solito, William A. Bourland, and Ivan Čepička. 2023. "Rediscovery of Remarkably Rare Anaerobic Tentaculiferous Ciliate Genera Legendrea and Dactylochlamys (Ciliophora: Litostomatea)" Biology 12, no. 5: 707. https://doi.org/10.3390/biology12050707
APA StylePomahač, O., Méndez-Sánchez, D., Poláková, K., Müller, M., Solito, M.-M., Bourland, W. A., & Čepička, I. (2023). Rediscovery of Remarkably Rare Anaerobic Tentaculiferous Ciliate Genera Legendrea and Dactylochlamys (Ciliophora: Litostomatea). Biology, 12(5), 707. https://doi.org/10.3390/biology12050707




