Organelle Membrane Extensions in Mammalian Cells
Abstract
Simple Summary
Abstract
1. Introduction
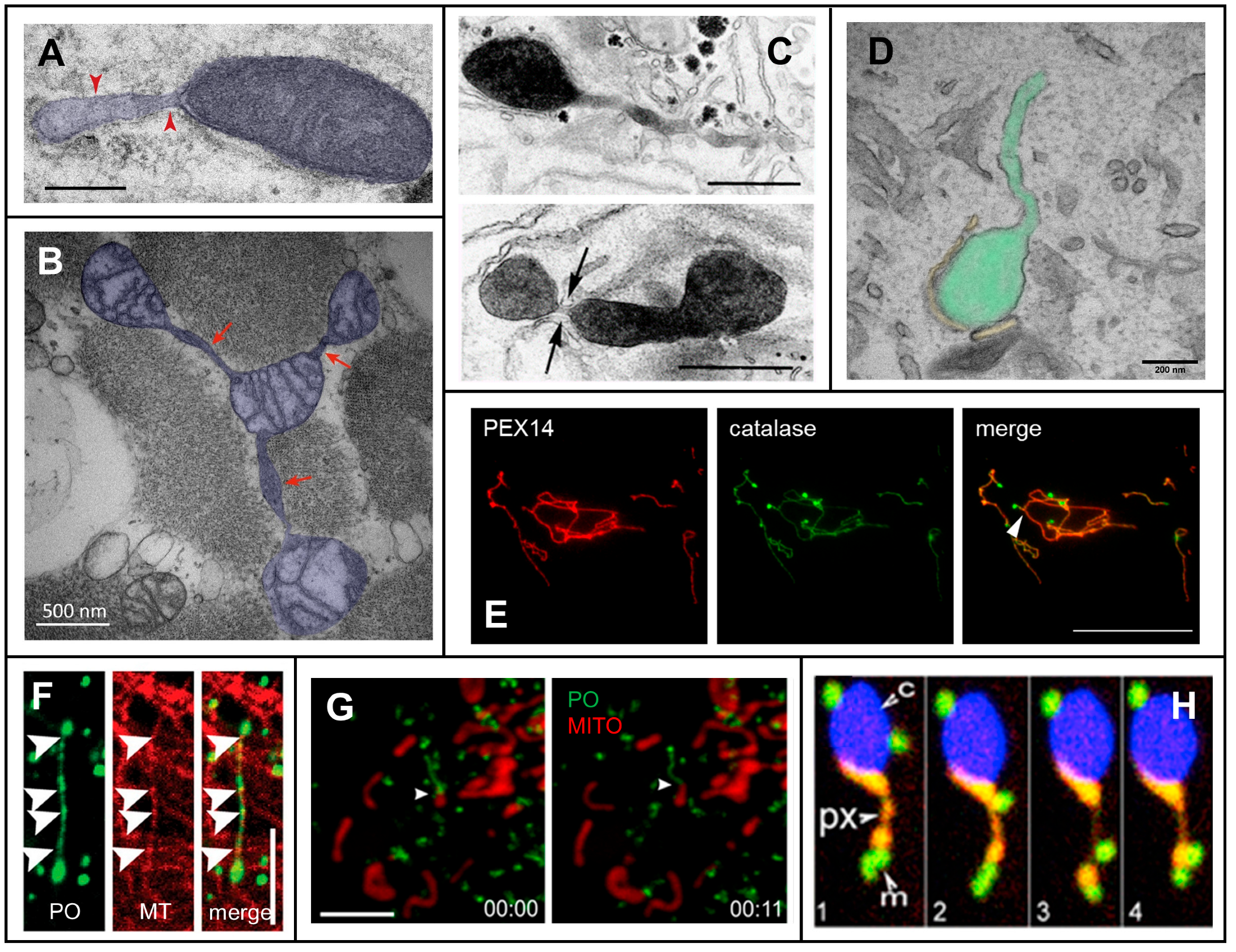
2. Mechanisms of Membrane Protrusion Formation
2.1. Dynamic Tubulation of Mitochondria
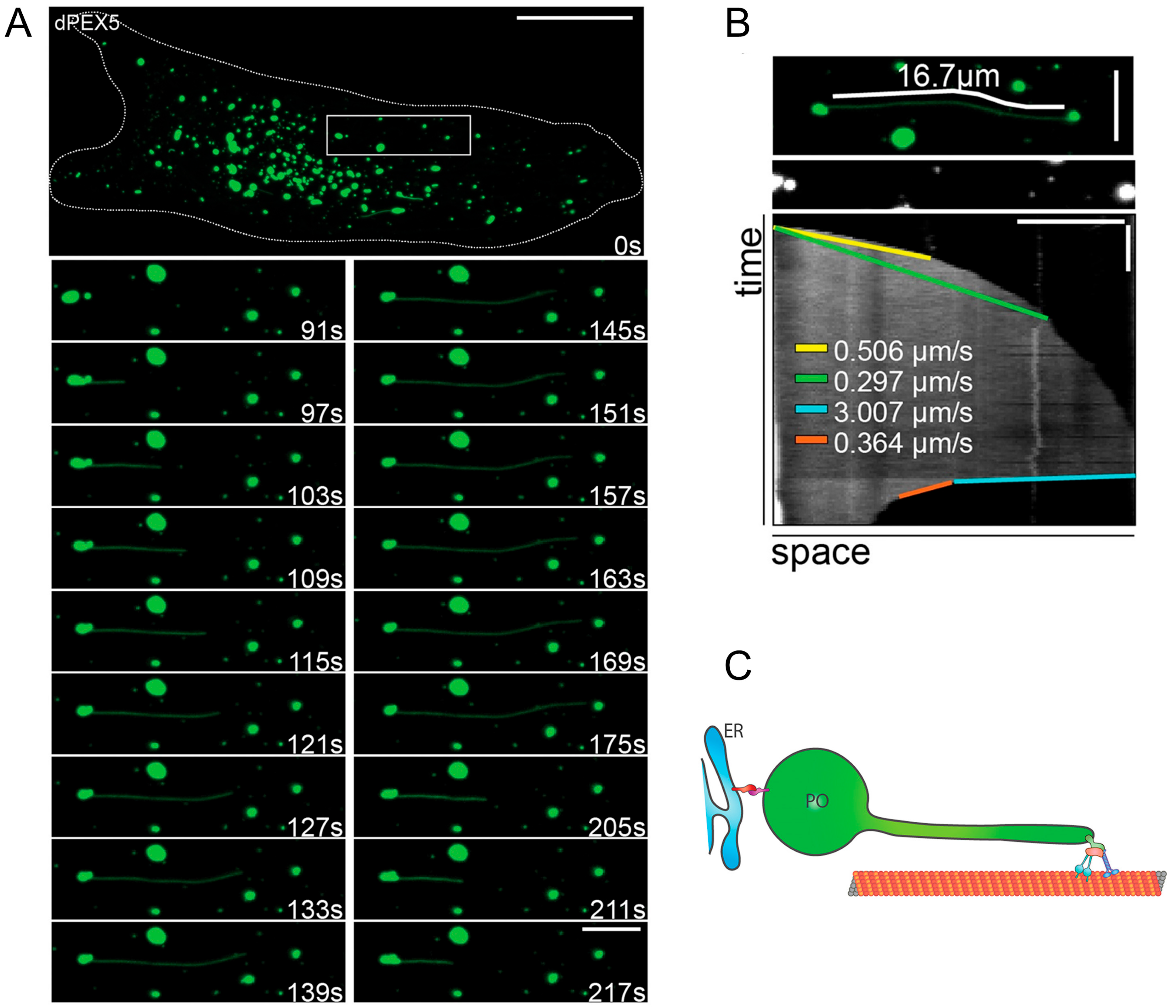
2.2. Peroxisomal Membrane Protrusions
2.2.1. Peroxisomal Shaping Proteins
2.2.2. Peroxisome-ER Interaction, Tethers and Lipid Flow
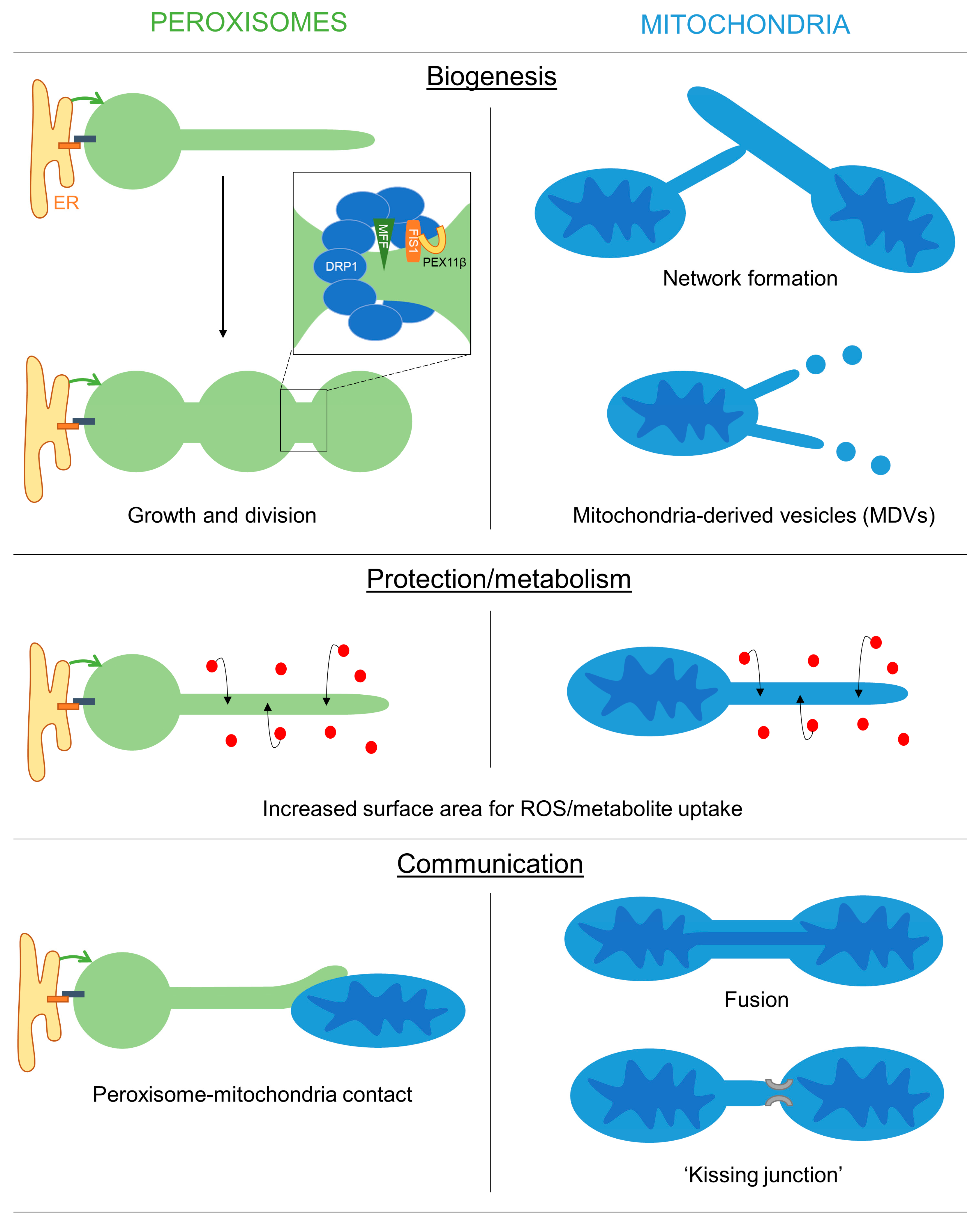
3. Possible Functions of Organelle Membrane Extensions
3.1. A Role for Membrane Protrusions in Organelle Biogenesis and Dynamics
3.2. A Protective Role for Organelle Membrane Protrusions
3.3. Organelle Protrusions, Communication and Metabolic Exchange
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
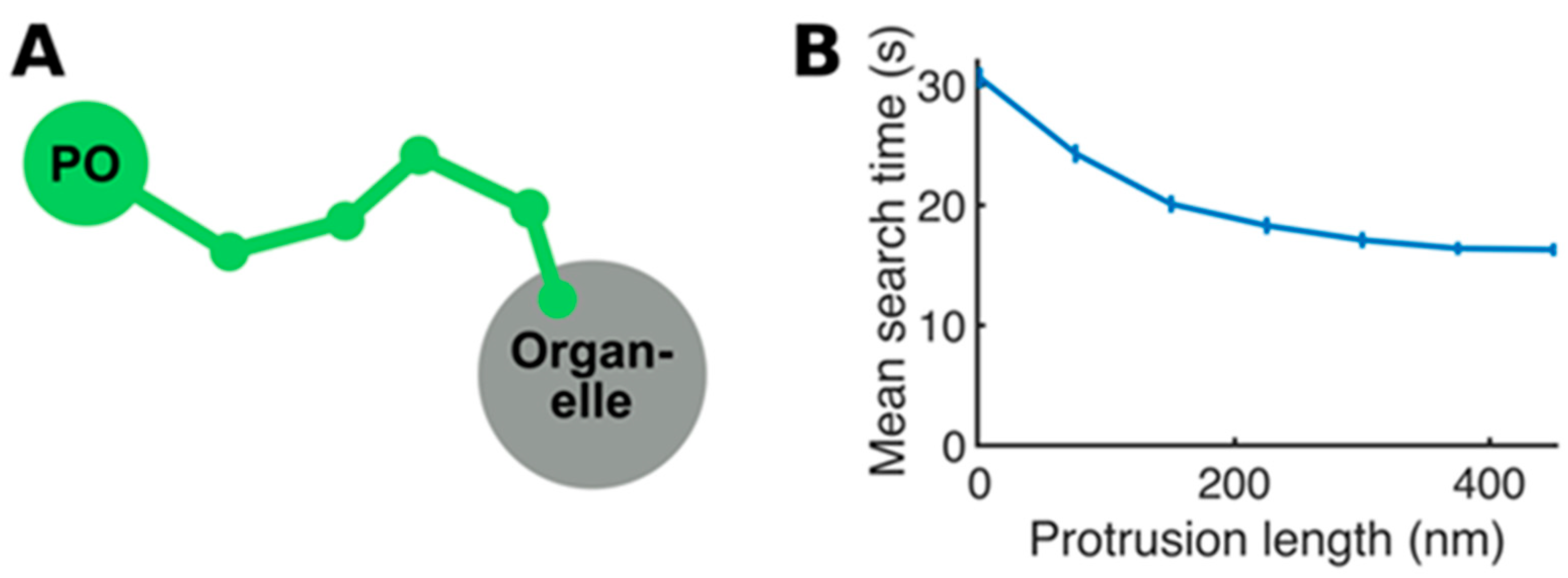
Appendix A
Details of the Mathematical Model
References
- Grainger, F.; James, D.W. Mitochondrial Extensions Associated with Microtubules in Outgrowing Processes from Chick Spinal Cord In Vitro. J. Cell Sci. 1969, 4, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Fahimi, H.D. Three-Dimensional Reconstruction of a Peroxisomal Reticulum in Regenerating Rat Liver: Evidence of Interconnections between Heterogeneous Segments. J. Cell Biol. 1987, 105, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Köhler, R.H.; Hanson, M.R. Plastid Tubules of Higher Plants Are Tissue-Specific and Developmentally Regulated. J. Cell Sci. 2000, 113, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Köhler, R.H.; Cao, J.; Zipfel, W.R.; Webb, W.W.; Hanson, M.R. Exchange of Protein Molecules through Connections between Higher Plant Plastids. Science 1997, 276, 2039–2042. [Google Scholar] [CrossRef] [PubMed]
- Jedd, G.; Chua, N.-H. Visualization of Peroxisomes in Living Plant Cells Reveals Acto-Myosin-Dependent Cytoplasmic Streaming and Peroxisome Budding. Plant Cell Physiol. 2002, 43, 384–392. [Google Scholar] [CrossRef]
- Cutler, S.R.; Ehrhardt, D.W.; Griffitts, J.S.; Somerville, C.R. Random GFP::CDNA Fusions Enable Visualization of Subcellular Structures in Cells of Arabidopsis at a High Frequency. Proc. Natl. Acad. Sci. USA 2000, 97, 3718–3723. [Google Scholar] [CrossRef]
- Scott, I.; Sparkes, I.A.; Logan, D.C. The Missing Link: Inter-Organellar Connections in Mitochondria and Peroxisomes? Trends Plant Sci. 2007, 12, 380–383. [Google Scholar] [CrossRef]
- Logan, D.C.; Scott, I.; Tobin, A.K. ADL2a, like ADL2b, Is Involved in the Control of Higher Plant Mitochondrial Morphology. J. Exp. Bot. 2004, 55, 783–785. [Google Scholar] [CrossRef]
- Mathur, J. Organelle Extensions in Plant Cells. Plant Physiol. 2021, 185, 593–607. [Google Scholar] [CrossRef]
- Wiltshire, E.J.; Collings, D.A. New Dynamics in an Old Friend: Dynamic Tubular Vacuoles Radiate through the Cortical Cytoplasm of Red Onion Epidermal Cells. Plant Cell Physiol. 2009, 50, 1826–1839. [Google Scholar] [CrossRef]
- Mathur, J.; Mammone, A.; Barton, K.A. Organelle Extensions in Plant Cells. J. Integr. Plant Biol. 2012, 54, 851–867. [Google Scholar] [CrossRef]
- Castro, I.G.; Richards, D.M.; Metz, J.; Costello, J.L.; Passmore, J.B.; Schrader, T.A.; Gouveia, A.; Ribeiro, D.; Schrader, M. A Role for Mitochondrial Rho GTPase 1 (MIRO1) in Motility and Membrane Dynamics of Peroxisomes. Traffic 2018, 19, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Schrader, M.; Fahimi, H.D. Growth and Division of Peroxisomes. Int Rev. Cytol. 2006, 255, 237–290. [Google Scholar] [CrossRef] [PubMed]
- Bowes, T.; Gupta, R.S. Novel Mitochondrial Extensions Provide Evidence for a Link between Microtubule-Directed Movement and Mitochondrial Fission. Biochem. Biophys. Res. Commun. 2008, 376, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Lavorato, M.; Formenti, F.; Franzini-Armstrong, C. The Structural Basis for Intermitochondrial Communications Is Fundamentally Different in Cardiac and Skeletal Muscle. Exp. Physiol. 2020, 105, 606–612. [Google Scholar] [CrossRef]
- Vincent, A.E.; Turnbull, D.M.; Eisner, V.; Hajnóczky, G.; Picard, M. Mitochondrial Nanotunnels. Trends Cell Biol. 2017, 27, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.J.; Eren, E.; Petralia, R.S.; Gu, J.W.; Wang, Y.-X.; Kapogiannis, D. Mitochondrial Protrusions in Neuronal Cells. iScience 2020, 23, 101514. [Google Scholar] [CrossRef]
- Passmore, J.B.; Carmichael, R.E.; Schrader, T.A.; Godinho, L.F.; Ferdinandusse, S.; Lismont, C.; Wang, Y.; Hacker, C.; Islinger, M.; Fransen, M.; et al. Mitochondrial Fission Factor (MFF) Is a Critical Regulator of Peroxisome Maturation. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118709. [Google Scholar] [CrossRef]
- Kustatscher, G.; Grabowski, P.; Schrader, T.A.; Passmore, J.B.; Schrader, M.; Rappsilber, J. Co-Regulation Map of the Human Proteome Enables Identification of Protein Functions. Nat. Biotechnol. 2019, 37, 1361–1371. [Google Scholar] [CrossRef]
- Jaipargas, E.-A.; Mathur, N.; Bou Daher, F.; Wasteneys, G.O.; Mathur, J. High Light Intensity Leads to Increased Peroxule-Mitochondria Interactions in Plants. Front. Cell Dev. Biol. 2016, 4, 6. [Google Scholar] [CrossRef]
- Dubey, G.P.; Ben-Yehuda, S. Intercellular Nanotubes Mediate Bacterial Communication. Cell 2011, 144, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Torralba, D.; Baixauli, F.; Sánchez-Madrid, F. Mitochondria Know No Boundaries: Mechanisms and Functions of Intercellular Mitochondrial Transfer. Front. Cell Dev. Biol. 2016, 4, 107. [Google Scholar] [CrossRef] [PubMed]
- Mattes, B.; Scholpp, S. Emerging Role of Contact-Mediated Cell Communication in Tissue Development and Diseases. Histochem. Cell Biol. 2018, 150, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Costello, J.L.; Passmore, J.B.; Islinger, M.; Schrader, M. Multi-Localized Proteins: The Peroxisome-Mitochondria Connection. In Proteomics of Peroxisomes; Subcellular Biochemistry 89; Springer: Singapore, 2018; pp. 383–415. [Google Scholar] [CrossRef]
- Wang, C.; Du, W.; Su, Q.P.; Zhu, M.; Feng, P.; Li, Y.; Zhou, Y.; Mi, N.; Zhu, Y.; Jiang, D.; et al. Dynamic Tubulation of Mitochondria Drives Mitochondrial Network Formation. Cell Res. 2015, 25, 1108–1120. [Google Scholar] [CrossRef] [PubMed]
- Lavorato, M.; Iyer, V.R.; Dewight, W.; Cupo, R.R.; Debattisti, V.; Gomez, L.; De la Fuente, S.; Zhao, Y.-T.; Valdivia, H.H.; Hajnóczky, G.; et al. Increased Mitochondrial Nanotunneling Activity, Induced by Calcium Imbalance, Affects Intermitochondrial Matrix Exchanges. Proc. Natl. Acad. Sci. USA 2017, 114, E849–E858. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Sun, L.; Ji, S.; Zhao, T.; Zhang, W.; Xu, J.; Zhang, J.; Wang, Y.; Wang, X.; Franzini-Armstrong, C.; et al. Kissing and Nanotunneling Mediate Intermitochondrial Communication in the Heart. Proc. Natl. Acad. Sci. USA 2013, 110, 2846–2851. [Google Scholar] [CrossRef]
- Eisner, V.; Cupo, R.R.; Gao, E.; Csordás, G.; Slovinsky, W.S.; Paillard, M.; Cheng, L.; Ibetti, J.; Chen, S.R.W.; Chuprun, J.K.; et al. Mitochondrial Fusion Dynamics Is Robust in the Heart and Depends on Calcium Oscillations and Contractile Activity. Proc. Natl. Acad. Sci. USA 2017, 114, E859–E868. [Google Scholar] [CrossRef]
- Vincent, A.E.; Ng, Y.S.; White, K.; Davey, T.; Mannella, C.; Falkous, G.; Feeney, C.; Schaefer, A.M.; McFarland, R.; Gorman, G.S.; et al. The Spectrum of Mitochondrial Ultrastructural Defects in Mitochondrial Myopathy. Sci. Rep. 2016, 6, 30610. [Google Scholar] [CrossRef]
- Vincent, A.E.; White, K.; Davey, T.; Philips, J.; Ogden, R.T.; Lawless, C.; Warren, C.; Hall, M.G.; Ng, Y.S.; Falkous, G.; et al. Quantitative 3D Mapping of the Human Skeletal Muscle Mitochondrial Network. Cell Rep. 2019, 26, 996–1009.e4. [Google Scholar] [CrossRef]
- Chung, D.J.; Madison, G.P.; Aponte, A.M.; Singh, K.; Li, Y.; Pirooznia, M.; Bleck, C.K.E.; Darmani, N.A.; Balaban, R.S. Metabolic Design in a Mammalian Model of Extreme Metabolism, the North American Least Shrew (Cryptotis parva). J. Physiol. 2022, 600, 547–567. [Google Scholar] [CrossRef]
- Boardman, N.T.; Trani, G.; Scalabrin, M.; Romanello, V.; Wüst, R.C.I. Intra-Cellular to Inter-Organ Mitochondrial Communication in Striated Muscle in Health and Disease. Endocr. Rev. 2023. [Google Scholar] [CrossRef] [PubMed]
- Morcillo, P.; Cordero, H.; Ijomone, O.M.; Ayodele, A.; Bornhorst, J.; Gunther, L.; Macaluso, F.P.; Bowman, A.B.; Aschner, M. Defective Mitochondrial Dynamics Underlie Manganese-Induced Neurotoxicity. Mol. Neurobiol. 2021, 58, 3270–3289. [Google Scholar] [CrossRef] [PubMed]
- Quintana-Cabrera, R.; Scorrano, L. Determinants and Outcomes of Mitochondrial Dynamics. Mol. Cell 2023, 83, 857–876. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Guo, Y.; Xue, B.; Shi, P.; Chen, Y.; Su, Q.P.; Hao, H.; Zhao, S.; Wu, C.; Yu, L.; et al. ER-Mitochondria Contacts Promote MtDNA Nucleoids Active Transportation via Mitochondrial Dynamic Tubulation. Nat. Commun. 2020, 11, 4471. [Google Scholar] [CrossRef]
- König, T.; Nolte, H.; Aaltonen, M.J.; Tatsuta, T.; Krols, M.; Stroh, T.; Langer, T.; McBride, H.M. MIROs and DRP1 Drive Mitochondrial-Derived Vesicle Biogenesis and Promote Quality Control. Nat. Cell Biol. 2021, 23, 1271–1286. [Google Scholar] [CrossRef]
- Neuspiel, M.; Schauss, A.C.; Braschi, E.; Zunino, R.; Rippstein, P.; Rachubinski, R.A.; Andrade-Navarro, M.A.; McBride, H.M. Cargo-Selected Transport from the Mitochondria to Peroxisomes Is Mediated by Vesicular Carriers. Curr. Biol. 2008, 18, 102–108. [Google Scholar] [CrossRef]
- Yamashita, A.; Fujimoto, M.; Katayama, K.; Yamaoka, S.; Tsutsumi, N.; Arimura, S.-I. Formation of Mitochondrial Outer Membrane Derived Protrusions and Vesicles in Arabidopsis thaliana. PLoS ONE 2016, 11, e0146717. [Google Scholar] [CrossRef]
- Zhang, L.; Trushin, S.; Christensen, T.A.; Bachmeier, B.V.; Gateno, B.; Schroeder, A.; Yao, J.; Itoh, K.; Sesaki, H.; Poon, W.W.; et al. Altered Brain Energetics Induces Mitochondrial Fission Arrest in Alzheimer’s Disease. Sci. Rep. 2016, 6, 18725. [Google Scholar] [CrossRef]
- Bharti, P.; Schliebs, W.; Schievelbusch, T.; Neuhaus, A.; David, C.; Kock, K.; Herrmann, C.; Meyer, H.E.; Wiese, S.; Warscheid, B.; et al. PEX14 Is Required for Microtubule-Based Peroxisome Motility in Human Cells. J. Cell Sci. 2011, 124, 1759–1768. [Google Scholar] [CrossRef]
- Theiss, C.; Neuhaus, A.; Schliebs, W.; Erdmann, R. TubStain: A Universal Peptide-Tool to Label Microtubules. Histochem. Cell Biol. 2012, 138, 531–540. [Google Scholar] [CrossRef]
- Barros-Barbosa, A.; Ferreira, M.J.; Rodrigues, T.A.; Pedrosa, A.G.; Grou, C.P.; Pinto, M.P.; Fransen, M.; Francisco, T.; Azevedo, J.E. Membrane Topologies of PEX13 and PEX14 Provide New Insights on the Mechanism of Protein Import into Peroxisomes. FEBS J. 2019, 286, 205–222. [Google Scholar] [CrossRef]
- Roux, A.; Cappello, G.; Cartaud, J.; Prost, J.; Goud, B.; Bassereau, P. A Minimal System Allowing Tubulation with Molecular Motors Pulling on Giant Liposomes. Proc. Natl. Acad. Sci. USA 2002, 99, 5394–5399. [Google Scholar] [CrossRef] [PubMed]
- Koster, G.; VanDuijn, M.; Hofs, B.; Dogterom, M. Membrane Tube Formation from Giant Vesicles by Dynamic Association of Motor Proteins. Proc. Natl. Acad. Sci. USA 2003, 100, 15583–15588. [Google Scholar] [CrossRef] [PubMed]
- Waterham, H.R.; Ferdinandusse, S.; Wanders, R.J.A. Human Disorders of Peroxisome Metabolism and Biogenesis. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, R.E.; Schrader, M. Determinants of Peroxisome Membrane Dynamics. Front. Physiol. 2022, 13, 834411. [Google Scholar] [CrossRef]
- Opaliński, Ł.; Kiel, J.A.K.W.; Williams, C.; Veenhuis, M.; van der Klei, I.J. Membrane Curvature during Peroxisome Fission Requires Pex11. EMBO J. 2011, 30, 5–16. [Google Scholar] [CrossRef]
- Itoyama, A.; Honsho, M.; Abe, Y.; Moser, A.; Yoshida, Y.; Fujiki, Y.; Gould, S.J. Docosahexaenoic Acid Mediates Peroxisomal Elongation, a Prerequisite for Peroxisome Division. J. Cell Sci. 2012, 125, 589–602. [Google Scholar] [CrossRef]
- Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Sanz-Fernández, M.; Hu, J.; Sandalio, L.M. Peroxisomes Extend Peroxules in a Fast Response to Stress via a Reactive Oxygen Species-Mediated Induction of the Peroxin PEX11a. Plant Physiol. 2016, 171, 1665–1674. [Google Scholar] [CrossRef]
- Williams, C.; Opalinski, L.; Landgraf, C.; Costello, J.; Schrader, M.; Krikken, A.M.; Knoops, K.; Kram, A.M.; Volkmer, R.; van der Klei, I.J. The Membrane Remodeling Protein Pex11p Activates the GTPase Dnm1p during Peroxisomal Fission. Proc. Natl. Acad. Sci. USA 2015, 112, 6377–6382. [Google Scholar] [CrossRef]
- Carmichael, R.E.; Islinger, M.; Schrader, M. Fission Impossible (?)—New Insights into Disorders of Peroxisome Dynamics. Cells 2022, 11, 1922. [Google Scholar] [CrossRef]
- Motley, A.M.; Hettema, E.H. Yeast Peroxisomes Multiply by Growth and Division. J. Cell Biol. 2007, 178, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Nagotu, S.; Saraya, R.; Otzen, M.; Veenhuis, M.; van der Klei, I.J. Peroxisome Proliferation in Hansenula Polymorpha Requires Dnm1p Which Mediates Fission but Not de Novo Formation. Biochim. Biophys. Acta 2008, 1783, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Knoblach, B.; Rachubinski, R.A. How Peroxisomes Partition between Cells. A Story of Yeast, Mammals and Filamentous Fungi. Curr. Opin. Cell Biol. 2016, 41, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Schrader, T.A.; Carmichael, R.E.; Islinger, M.; Costello, J.L.; Hacker, C.; Bonekamp, N.A.; Weishaupt, J.H.; Andersen, P.M.; Schrader, M. PEX11β and FIS1 Cooperate in Peroxisome Division Independent of Mitochondrial Fission Factor. J. Cell Sci. 2022, 135, 259924. [Google Scholar] [CrossRef]
- Otera, H.; Wang, C.; Cleland, M.M.; Setoguchi, K.; Yokota, S.; Youle, R.J.; Mihara, K. Mff Is an Essential Factor for Mitochondrial Recruitment of Drp1 during Mitochondrial Fission in Mammalian Cells. J. Cell Biol. 2010, 191, 1141–1158. [Google Scholar] [CrossRef]
- Koch, A.; Schneider, G.; Lüers, G.H.; Schrader, M. Peroxisome Elongation and Constriction but Not Fission Can Occur Independently of Dynamin-like Protein 1. J. Cell Sci. 2004, 117, 3995–4006. [Google Scholar] [CrossRef]
- Costello, J.L.; Castro, I.G.; Hacker, C.; Schrader, T.A.; Metz, J.; Zeuschner, D.; Azadi, A.S.; Godinho, L.F.; Costina, V.; Findeisen, P.; et al. ACBD5 and VAPB Mediate Membrane Associations between Peroxisomes and the ER. J. Cell Biol. 2017, 216, 331–342. [Google Scholar] [CrossRef]
- Hua, R.; Cheng, D.; Coyaud, É.; Freeman, S.; Di Pietro, E.; Wang, Y.; Vissa, A.; Yip, C.M.; Fairn, G.D.; Braverman, N.; et al. VAPs and ACBD5 Tether Peroxisomes to the ER for Peroxisome Maintenance and Lipid Homeostasis. J. Cell Biol. 2017, 216, 367–377. [Google Scholar] [CrossRef]
- Kors, S.; Hacker, C.; Bolton, C.; Maier, R.; Reimann, L.; Kitchener, E.J.A.; Warscheid, B.; Costello, J.L.; Schrader, M. Regulating Peroxisome-ER Contacts via the ACBD5-VAPB Tether by FFAT Motif Phosphorylation and GSK3β. J. Cell Biol. 2022, 221, e202003143. [Google Scholar] [CrossRef]
- Bishop, A.; Kamoshita, M.; Passmore, J.B.; Hacker, C.; Schrader, T.A.; Waterham, H.R.; Costello, J.L.; Schrader, M. Fluorescent Tools to Analyse Peroxisome-ER Interactions in Mammalian Cells. Contact 2019, 2. [Google Scholar] [CrossRef]
- Guillén-Samander, A.; Leonzino, M.; Hanna, M.G.; Tang, N.; Shen, H.; De Camilli, P. VPS13D Bridges the ER to Mitochondria and Peroxisomes via Miro. J. Cell Biol. 2021, 220, e202010004. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, H.A.; Wang, C.; Kanfer, G.; Shah, H.V.; Velayos-Baeza, A.; Dulovic-Mahlow, M.; Brüggemann, N.; Anding, A.; Baehrecke, E.H.; Maric, D.; et al. VPS13D Promotes Peroxisome Biogenesis. J. Cell Biol. 2021, 220, e202001188. [Google Scholar] [CrossRef] [PubMed]
- Dziurdzik, S.K.; Conibear, E. The Vps13 Family of Lipid Transporters and Its Role at Membrane Contact Sites. Int. J. Mol. Sci. 2021, 22, 2905. [Google Scholar] [CrossRef] [PubMed]
- Neuman, S.D.; Levine, T.P.; Bashirullah, A. A Novel Superfamily of Bridge-like Lipid Transfer Proteins. Trends Cell Biol. 2022, 32, 962–974. [Google Scholar] [CrossRef]
- Ferdinandusse, S.; Falkenberg, K.D.; Koster, J.; Mooyer, P.A.; Jones, R.; van Roermund, C.W.T.; Pizzino, A.; Schrader, M.; Wanders, R.J.A.; Vanderver, A.; et al. ACBD5 Deficiency Causes a Defect in Peroxisomal Very Long-Chain Fatty Acid Metabolism. J. Med. Genet. 2017, 54, 330–337. [Google Scholar] [CrossRef]
- Yagita, Y.; Shinohara, K.; Abe, Y.; Nakagawa, K.; Al-Owain, M.; Alkuraya, F.S.; Fujiki, Y. Deficiency of a Retinal Dystrophy Protein, Acyl-CoA Binding Domain-Containing 5 (ACBD5), Impairs Peroxisomal β-Oxidation of Very-Long-Chain Fatty Acids. J. Biol. Chem. 2017, 292, 691–705. [Google Scholar] [CrossRef]
- Schrader, M.; Thiemann, M.; Fahimi, H.D. Peroxisomal Motility and Interaction With Microtubules. Microsc. Res. Tech. 2003, 61, 171–178. [Google Scholar] [CrossRef]
- Schrader, M.; Burkhardt, J.K.; Baumgart, E.; Lüers, G.H.; Spring, H.; Völkl, A.; Fahimi, H.D. Interaction of Microtubules with Peroxisomes. Tubular and Spherical Peroxisomes in HepG2 Cells and Their Alteractions Induced by Microtubule-Active Drugs. Eur. J. Cell Biol. 1996, 69, 24–35. [Google Scholar]
- Passmore, J.B.; Pinho, S.; Gomez-Lazaro, M.; Schrader, M. The Respiratory Chain Inhibitor Rotenone Affects Peroxisomal Dynamics via Its Microtubule-Destabilising Activity. Histochem. Cell Biol. 2017, 148, 331–341. [Google Scholar] [CrossRef]
- Azadi, A.S.; Carmichael, R.E.; Kovacs, W.J.; Koster, J.; Kors, S.; Waterham, H.R.; Schrader, M. A Functional SMAD2/3 Binding Site in the PEX11β Promoter Identifies a Role for TGFβ in Peroxisome Proliferation in Humans. Front. Cell Dev. Biol. 2020, 8, 577637. [Google Scholar] [CrossRef]
- Sinclair, A.M.; Trobacher, C.P.; Mathur, N.; Greenwood, J.S.; Mathur, J. Peroxule Extension over ER-Defined Paths Constitutes a Rapid Subcellular Response to Hydroxyl Stress. Plant J. 2009, 59, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Barton, K.A.; Jaipargas, E.-A.; Griffiths, N.; Mathur, J. Live Imaging of Peroxisomes and Peroxules in Plants. In Molecular Machines Involved in Peroxisome Biogenesis and Maintenance; Brocard, C., Hartig, A., Eds.; Springer: Vienna, Austria, 2014; pp. 233–253. ISBN 978-3-7091-1788-0. [Google Scholar]
- Mathur, J.; Shaikh, A.; Mathur, N. Peroxisome Mitochondria Inter-Relations in Plants. In Proteomics of Peroxisomes; Subcellular Biochemistry 89; Springer: Singapore, 2018; pp. 417–433. [Google Scholar] [CrossRef]
- Barton, K.; Mathur, N.; Mathur, J. Simultaneous Live-Imaging of Peroxisomes and the ER in Plant Cells Suggests Contiguity but No Luminal Continuity between the Two Organelles. Front. Physiol. 2013, 4, 196. [Google Scholar] [CrossRef] [PubMed]
- Purves, W.K.; Sadava, D.E.; Orians, G.H.; Heller, H.C. Life: The Science of Biology; Freeman, W.H., Ed.; Macmillan: New York, NY, USA, 2004; ISBN 9780716798569. [Google Scholar]
- Hamilton, N.; Kerr, M.C.; Burrage, K.; Teasdale, R.D. Analyzing Real-Time Video Microscopy: The Dynamics and Geometry of Vesicles and Tubules in Endocytosis. In Current Protocols in Cell Biology; Wiley: Hoboken, NJ, USA, 2007; Chapter 14, Unit 4.16. [Google Scholar] [CrossRef]
- Schrader, M.; Wodopia, R.; Fahimi, H.D. Induction of Tubular Peroxisomes by UV Irradiation and Reactive Oxygen Species in HepG2 Cells. J. Histochem. Cytochem. 1999, 47, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Van Veldhoven, P.P.; Brees, C.; Rubio, N.; Nordgren, M.; Apanasets, O.; Kunze, M.; Baes, M.; Agostinis, P.; Fransen, M. Mitochondria Are Targets for Peroxisome-Derived Oxidative Stress in Cultured Mammalian Cells. Free Radic. Biol. Med. 2013, 65, 882–894. [Google Scholar] [CrossRef] [PubMed]
- Mattiazzi Ušaj, M.; Brložnik, M.; Kaferle, P.; Žitnik, M.; Wolinski, H.; Leitner, F.; Kohlwein, S.D.; Zupan, B.; Petrovič, U. Genome-Wide Localization Study of Yeast Pex11 Identifies Peroxisome–Mitochondria Interactions through the ERMES Complex. J. Mol. Biol. 2015, 427, 2072–2087. [Google Scholar] [CrossRef] [PubMed]
- Shai, N.; Yifrach, E.; van Roermund, C.W.T.; Cohen, N.; Bibi, C.; IJlst, L.; Cavellini, L.; Meurisse, J.; Schuster, R.; Zada, L.; et al. Systematic Mapping of Contact Sites Reveals Tethers and a Function for the Peroxisome-Mitochondria Contact. Nat. Commun. 2018, 9, 1761. [Google Scholar] [CrossRef]
- Binns, D.; Januszewski, T.; Chen, Y.; Hill, J.; Markin, V.S.; Zhao, Y.; Gilpin, C.; Chapman, K.D.; Anderson, R.G.W.; Goodman, J.M. An Intimate Collaboration between Peroxisomes and Lipid Bodies. J. Cell Biol. 2006, 173, 719–731. [Google Scholar] [CrossRef]
- Kalutsky, M.A.; Galimzyanov, T.R.; Molotkovsky, R.J. A Model of Lipid Monolayer-Bilayer Fusion of Lipid Droplets and Peroxisomes. Membranes 2022, 12, 992. [Google Scholar] [CrossRef]
- Thazar-Poulot, N.; Miquel, M.; Fobis-Loisy, I.; Gaude, T. Peroxisome Extensions Deliver the Arabidopsis SDP1 Lipase to Oil Bodies. Proc. Natl. Acad. Sci. USA 2015, 112, 4158–4163. [Google Scholar] [CrossRef]
- Silva, B.S.C.; DiGiovanni, L.; Kumar, R.; Carmichael, R.E.; Kim, P.K.; Schrader, M. Maintaining Social Contacts: The Physiological Relevance of Organelle Interactions. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118800. [Google Scholar] [CrossRef]
- Bonekamp, N.A.; Sampaio, P.; de Abreu, F.V.; Lüers, G.H.; Schrader, M. Transient Complex Interactions of Mammalian Peroxisomes without Exchange of Matrix or Membrane Marker Proteins. Traffic 2012, 13, 960–978. [Google Scholar] [CrossRef] [PubMed]
- Huybrechts, S.J.; Van Veldhoven, P.P.; Brees, C.; Mannaerts, G.P.; Los, G.V.; Fransen, M. Peroxisome Dynamics in Cultured Mammalian Cells. Traffic 2009, 10, 1722–1733. [Google Scholar] [CrossRef] [PubMed]
- Schrader, M.; King, S.J.; Stroh, T.A.; Schroer, T.A. Real Time Imaging Reveals a Peroxisomal Reticulum in Living Cells. J. Cell Sci. 2000, 113, 3663–3671. [Google Scholar] [CrossRef] [PubMed]
- Chornyi, S.; IJlst, L.; van Roermund, C.W.T.; Wanders, R.J.A.; Waterham, H.R. Peroxisomal Metabolite and Cofactor Transport in Humans. Front. Cell Dev. Biol. 2020, 8, 613892. [Google Scholar] [CrossRef]
- Klar, T.A.; Jakobs, S.; Dyba, M.; Egner, A.; Hell, S.W. Fluorescence Microscopy with Diffraction Resolution Barrier Broken by Stimulated Emission. Proc. Natl. Acad. Sci. USA 2000, 97, 8206–8210. [Google Scholar] [CrossRef]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-Diffraction-Limit Imaging by Stochastic Optical Reconstruction Microscopy (STORM). Nat. Methods 2006, 3, 793–795. [Google Scholar] [CrossRef]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef]
- Heilemann, M.; van de Linde, S.; Schüttpelz, M.; Kasper, R.; Seefeldt, B.; Mukherjee, A.; Tinnefeld, P.; Sauer, M. Subdiffraction-Resolution Fluorescence Imaging with Conventional Fluorescent Probes. Angew. Chem. Int. Ed. Engl. 2008, 47, 6172–6176. [Google Scholar] [CrossRef]
- Van de Linde, S.; Löschberger, A.; Klein, T.; Heidbreder, M.; Wolter, S.; Heilemann, M.; Sauer, M. Direct Stochastic Optical Reconstruction Microscopy with Standard Fluorescent Probes. Nat. Protoc. 2011, 6, 991–1009. [Google Scholar] [CrossRef]
- Legant, W.R.; Shao, L.; Grimm, J.B.; Brown, T.A.; Milkie, D.E.; Avants, B.B.; Lavis, L.D.; Betzig, E. High-Density Three-Dimensional Localization Microscopy across Large Volumes. Nat. Methods 2016, 13, 359–365. [Google Scholar] [CrossRef]
- Spahn, C.K.; Glaesmann, M.; Grimm, J.B.; Ayala, A.X.; Lavis, L.D.; Heilemann, M. A Toolbox for Multiplexed Super-Resolution Imaging of the E. Coli Nucleoid and Membrane Using Novel PAINT Labels. Sci. Rep. 2018, 8, 14768. [Google Scholar] [CrossRef] [PubMed]
- Hoboth, P.; Šebesta, O.; Sztacho, M.; Castano, E.; Hozák, P. Dual-Color DSTORM Imaging and ThunderSTORM Image Reconstruction and Analysis to Study the Spatial Organization of the Nuclear Phosphatidylinositol Phosphates. MethodsX 2021, 8, 101372. [Google Scholar] [CrossRef] [PubMed]
- Hoboth, P.; Šebesta, O.; Hozák, P. How Single-Molecule Localization Microscopy Expanded Our Mechanistic Understanding of RNA Polymerase II Transcription. Int. J. Mol. Sci. 2021, 22, 6694. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carmichael, R.E.; Richards, D.M.; Fahimi, H.D.; Schrader, M. Organelle Membrane Extensions in Mammalian Cells. Biology 2023, 12, 664. https://doi.org/10.3390/biology12050664
Carmichael RE, Richards DM, Fahimi HD, Schrader M. Organelle Membrane Extensions in Mammalian Cells. Biology. 2023; 12(5):664. https://doi.org/10.3390/biology12050664
Chicago/Turabian StyleCarmichael, Ruth E., David M. Richards, H. Dariush Fahimi, and Michael Schrader. 2023. "Organelle Membrane Extensions in Mammalian Cells" Biology 12, no. 5: 664. https://doi.org/10.3390/biology12050664
APA StyleCarmichael, R. E., Richards, D. M., Fahimi, H. D., & Schrader, M. (2023). Organelle Membrane Extensions in Mammalian Cells. Biology, 12(5), 664. https://doi.org/10.3390/biology12050664




