Nectin Family Ligands Trigger Immune Effector Functions in Health and Autoimmunity
Simple Summary
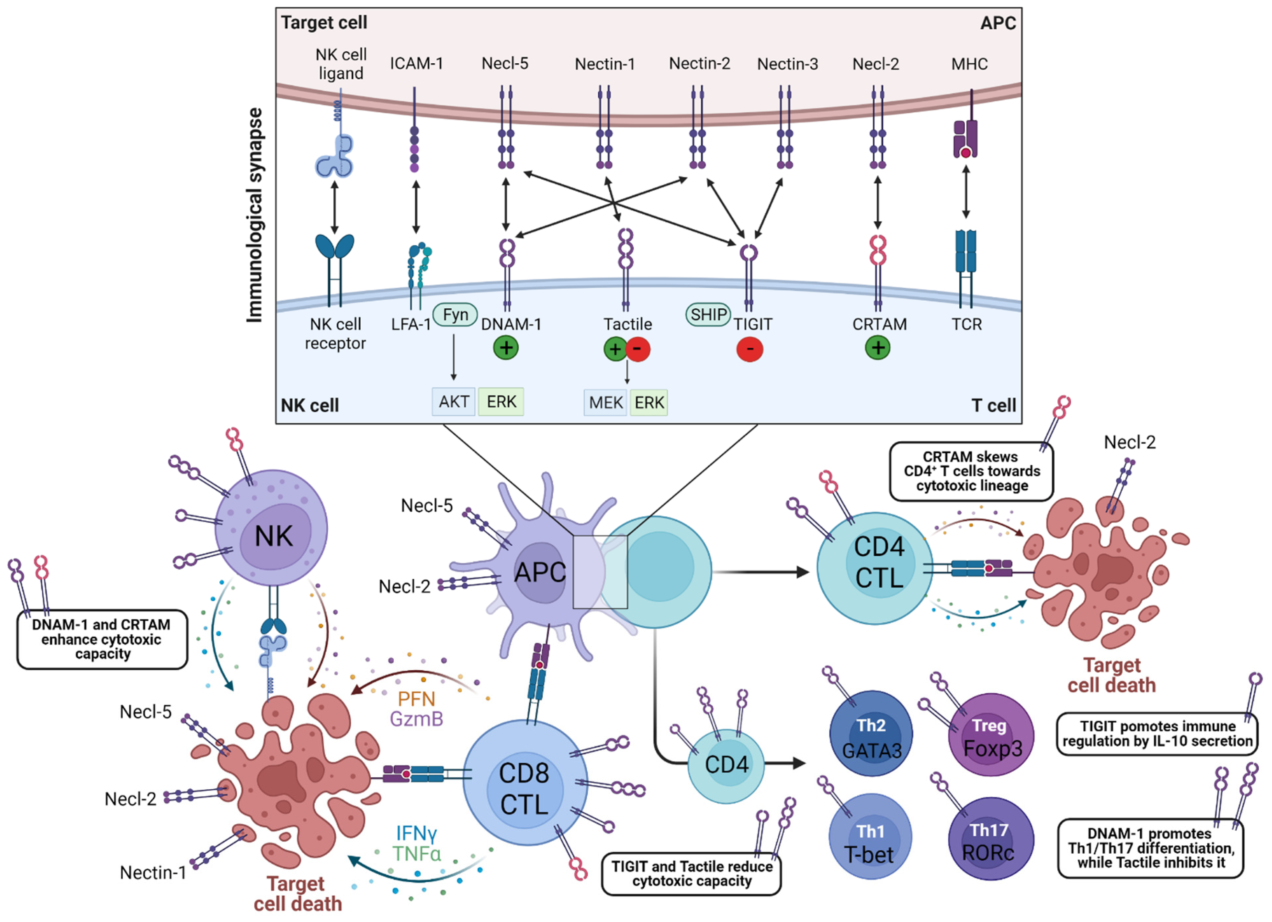
Abstract
1. Introduction
2. Immune-Cell Expression
| T Lymphocytes | B Lymphocytes | NK Cells | Monocytes Macrophages | Ref. | |
|---|---|---|---|---|---|
| DNAM-1 (CD226) | + | + | + | + | [12,13,15,16,17,40,41] |
| CRTAM (CD355) | + | - | + | - | [30,31,33] |
| Tactile (CD96) | + | +/- | + | + | [12,13,14,55,56] |
| TIGIT | + | + | + | - | [12,26,28] |
| Nectin-1 (CD111, HVEC, PRR1, PVRL1, HIgR) | - | ND | - | ND | [36,39] |
| Nectin-2 (CD112, HVEB, PRR2, PVRL2, PVRR2) | - | - | - | + | [36,40] |
| Nectin-3 (CD113, PPR3, PRR3, PVRL3, PVRR3) | + | ND | + | + * | [36,39] |
| Nectin-4 (PVRL4, PRR4) | - | - | ND | - | [36,37,40] |
| Necl-1 (CADM3, SYNCAM3, IGSF4B, TSLL1) | ND | ND | ND | ND | / |
| Necl-2 (CADM1, SYNCAM1, IGSF4, IGSF4A, TSLC1, RA175) | +/- | - | ND | - | [52,53,54] |
| Necl-3 (CADM2, SYNCAM2, IGSF4D) | ND | ND | ND | ND | / |
| Necl-4 (CADM4, SYNCAM4, IGSF4C, TSLL2) | ND | ND | ND | ND | / |
| Necl-5 (CD155, HVED, PVR, PVS, TAGE4) | + | + | ND | + | [40,41,44,57] |
3. Effector Functions
3.1. DNAM-1/CD226
| Disease | Dysregulation | Dysfunction | Ref. |
|---|---|---|---|
| Multiple sclerosis | DNAM-1 ↘ on NK cells | Defective regulation of autoreactive T cells | [19,67] |
| DNAM-1 ↗ on Tregs | Inducing a pro-inflammatory phenotype and reduced suppressive capacity | [20,44,62,63,64] | |
| TIGIT ↘ on B cells | Defective regulation of Th cell balance | [28,71] | |
| Systemic sclerosis | DNAM-1 ↗ on CD8+ CTLs | Increased cytokine production and cytotoxicity | [17] |
| DNAM-1 ↘ on NK cells | Defective regulation of autoreactive T cells | [21] | |
| Rheumatoid arthritis | DNAM-1 ↗ on NK cells | Increased cytotoxicity towards synovial fibroblasts | [68] |
| DNAM-1 ↗ on CD4+ CTLs | Increased cytokine production and cytotoxicity | [69] | |
| CRTAM ↗ in synovium | Hub gene for diagnosis and therapy | [72] | |
| Type 1 diabetes | CRTAM ≈ on NK T cells and CD8+ CTLs | Targeted cytotoxicity of CD8+ CTLs towards Necl-2+ pancreatic islet cells | [32,73,74,75] |
| Atopic dermatitis | TIGIT ↗/↘ on CD4+ T cells | Retrains inflammation by inhibiting cell proliferation; negatively correlated with disease severity | [27] |
| Psoriasis | TIGIT ↘ on CD4+ T cells | Reduced regulation of cell proliferation and cytokine production; negatively correlated with disease severity | [29] |
| Ankylosing spondylitis | Tactile ↘ on CD4+ T cells | Inducing a pro-inflammatory phenotype and cytokine production | [18] |
| Viral infections | CRTAM ↗ on CD8+ CTLs | Increased cytotoxicity towards infected cells | [53,73,75] |
| Tactile ↘ on CD8+ CTLs | Immunosenescent phenotype in persistent HIV infection | [76,77] |
3.2. CRTAM/CD355
3.3. Tactile/CD96
3.4. TIGIT
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gérard, A.; Cope, A.P.; Kemper, C.; Alon, R.; Köchl, R. LFA-1 in T cell priming, differentiation, and effector functions. Trends Immunol. 2021, 42, 706–722. [Google Scholar] [CrossRef] [PubMed]
- Molon, B.; Liboni, C.; Viola, A. CD28 and chemokine receptors: Signalling amplifiers at the immunological synapse. Front. Immunol. 2022, 13, 938004. [Google Scholar] [CrossRef] [PubMed]
- Engels, N.; Wienands, J. The signaling tool box for tyrosine-based costimulation of lymphocytes. Curr. Opin. Immunol. 2011, 23, 324–329. [Google Scholar] [CrossRef]
- Kumar, S. Natural killer cell cytotoxicity and its regulation by inhibitory receptors. Immunology 2018, 154, 383–393. [Google Scholar] [CrossRef]
- Takai, Y.; Irie, K.; Shimizu, K.; Sakisaka, T.; Ikeda, W. Nectins and nectin-like molecules: Roles in cell adhesion, migration, and polarization. Cancer Sci. 2003, 94, 655–667. [Google Scholar] [CrossRef]
- Deuss, F.A.; Watson, G.M.; Goodall, K.J.; Leece, I.; Chatterjee, S.; Fu, Z.; Thaysen-Andersen, M.; Andrews, D.M.; Rossjohn, J.; Berry, R. Structural basis for the recognition of nectin-like protein-5 by the human-activating immune receptor, DNAM-1. J. Biol. Chem. 2019, 294, 12534–12546. [Google Scholar] [CrossRef]
- Deuss, F.A.; Watson, G.M.; Fu, Z.; Rossjohn, J.; Berry, R. Structural Basis for CD96 Immune Receptor Recognition of Nectin-like Protein-5, CD155. Structure 2019, 27, 219–228.e213. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, N.; Lu, Y.; Davidson, D.; Colonna, M.; Veillette, A. DNAM-1 controls NK cell activation via an ITT-like motif. J. Exp. Med. 2015, 212, 2165–2182. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, H.; Li, M.; Hu, D.; Li, C.; Ge, B.; Jin, B.; Fan, Z. Recruitment of Grb2 and SHIP1 by the ITT-like motif of TIGIT suppresses granule polarization and cytotoxicity of NK cells. Cell Death Differ. 2013, 20, 456–464. [Google Scholar] [CrossRef]
- Meyer, D.; Seth, S.; Albrecht, J.; Maier, M.K.; du Pasquier, L.; Ravens, I.; Dreyer, L.; Burger, R.; Gramatzki, M.; Schwinzer, R.; et al. CD96 interaction with CD155 via its first Ig-like domain is modulated by alternative splicing or mutations in distal Ig-like domains. J. Biol. Chem. 2009, 284, 2235–2244. [Google Scholar] [CrossRef]
- Yeh, J.H.; Sidhu, S.S.; Chan, A.C. Regulation of a late phase of T cell polarity and effector functions by Crtam. Cell 2008, 132, 846–859. [Google Scholar] [CrossRef]
- Lepletier, A.; Lutzky, V.P.; Mittal, D.; Stannard, K.; Watkins, T.S.; Ratnatunga, C.N.; Smith, C.; McGuire, H.M.; Kemp, R.A.; Mukhopadhyay, P.; et al. The immune checkpoint CD96 defines a distinct lymphocyte phenotype and is highly expressed on tumor-infiltrating T cells. Immunol. Cell Biol. 2019, 97, 152–164. [Google Scholar] [CrossRef]
- Manes, T.D.; Pober, J.S. Identification of endothelial cell junctional proteins and lymphocyte receptors involved in transendothelial migration of human effector memory CD4+ T cells. J. Immunol. 2011, 186, 1763–1768. [Google Scholar] [CrossRef]
- Chiang, E.Y.; de Almeida, P.E.; de Almeida Nagata, D.E.; Bowles, K.H.; Du, X.; Chitre, A.S.; Banta, K.L.; Kwon, Y.; McKenzie, B.; Mittman, S.; et al. CD96 functions as a co-stimulatory receptor to enhance CD8+ T cell activation and effector responses. Eur. J. Immunol. 2020, 6, 891–902. [Google Scholar] [CrossRef]
- Shibuya, A.; Campbell, D.; Hannum, C.; Yssel, H.; Franz-Bacon, K.; McClanahan, T.; Kitamura, T.; Nicholl, J.; Sutherland, G.R.; Lanier, L.L.; et al. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity 1996, 4, 573–581. [Google Scholar] [CrossRef]
- Nagayama-Hasegawa, Y.; Honda, S.I.; Shibuya, A.; Shibuya, K. Expression and function of DNAM-1 on human B-lineage cells. Cytometry B Clin. Cytom. 2020, 98, 368–374. [Google Scholar] [CrossRef]
- Ayano, M.; Tsukamoto, H.; Kohno, K.; Ueda, N.; Tanaka, A.; Mitoma, H.; Akahoshi, M.; Arinobu, Y.; Niiro, H.; Horiuchi, T.; et al. Increased CD226 Expression on CD8+ T Cells Is Associated with Upregulated Cytokine Production and Endothelial Cell Injury in Patients with Systemic Sclerosis. J. Immunol. 2015, 195, 892–900. [Google Scholar] [CrossRef]
- Wu, F.; Yang, H.; Xu, X.; Ren, C.; Zheng, Y.; Zhang, H.; Cai, B.; Qiu, R.; Ren, W.; Quan, R. CD96 Downregulation Promotes the Immune Response of CD4 T Cells and Associates with Ankylosing Spondylitis. Biomed. Res. Int. 2022, 2022, 3946754. [Google Scholar] [CrossRef]
- Gross, C.C.; Schulte-Mecklenbeck, A.; Runzi, A.; Kuhlmann, T.; Posevitz-Fejfar, A.; Schwab, N.; Schneider-Hohendorf, T.; Herich, S.; Held, K.; Konjevic, M.; et al. Impaired NK-mediated regulation of T-cell activity in multiple sclerosis is reconstituted by IL-2 receptor modulation. Proc. Natl. Acad. Sci. USA 2016, 113, E2973–E2982. [Google Scholar] [CrossRef]
- Tapia-Maltos, M.A.; Treviño-Frenk, I.; García-González, H.B.; Rosetti, M.; Barriga-Maldonado, V.; Morales-Ramírez, F.; López-Hernández, D.C.; Rosetti, F.; Crispín, J.C. Identification of regulatory T cell molecules associated with severity of multiple sclerosis. Mult. Scler. 2021, 27, 1695–1705. [Google Scholar] [CrossRef]
- Puxeddu, I.; Bongiorni, F.; Chimenti, D.; Bombardieri, S.; Moretta, A.; Bottino, C.; Migliorini, P. Cell surface expression of activating receptors and co-receptors on peripheral blood NK cells in systemic autoimmune diseases. Scand. J. Rheumatol. 2012, 41, 298–304. [Google Scholar] [CrossRef]
- Bai, L.; Jiang, J.; Li, H.; Zhang, R. Role of CD226 Rs763361 Polymorphism in Susceptibility to Multiple Autoimmune Diseases. Immunol. Investig. 2020, 49, 926–942. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, X.; Leng, S.; Xu, Q.; Sheng, Z.; Zhang, Y.; Yu, J.; Feng, Q.; Hou, M.; Peng, J.; et al. Immune Checkpoint-Related Gene Polymorphisms Are Associated With Primary Immune Thrombocytopenia. Front. Immunol. 2020, 11, 615941. [Google Scholar] [CrossRef]
- Reinards, T.H.; Albers, H.M.; Brinkman, D.M.; Kamphuis, S.S.; van Rossum, M.A.; Girschick, H.J.; Wouters, C.; Hoppenreijs, E.P.; Saurenmann, R.K.; Hinks, A.; et al. CD226 (DNAM-1) is associated with susceptibility to juvenile idiopathic arthritis. Ann. Rheum. Dis. 2015, 74, 2193–2198. [Google Scholar] [CrossRef]
- Saevarsdottir, S.; Olafsdottir, T.A.; Ivarsdottir, E.V.; Halldorsson, G.H.; Gunnarsdottir, K.; Sigurdsson, A.; Johannesson, A.; Sigurdsson, J.K.; Juliusdottir, T.; Lund, S.H.; et al. FLT3 stop mutation increases FLT3 ligand level and risk of autoimmune thyroid disease. Nature 2020, 584, 619–623. [Google Scholar] [CrossRef]
- Yu, X.; Harden, K.; Gonzalez, L.C.; Francesco, M.; Chiang, E.; Irving, B.; Tom, I.; Ivelja, S.; Refino, C.J.; Clark, H.; et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2009, 10, 48–57. [Google Scholar] [CrossRef]
- Kurita, M.; Yoshihara, Y.; Ishiuji, Y.; Chihara, M.; Ishiji, T.; Asahina, A.; Yanaba, K. Expression of T-cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain on CD4+ T cells in patients with atopic dermatitis. J. Dermatol. 2019, 46, 37–42. [Google Scholar] [CrossRef]
- Hasan, M.M.; Nair, S.S.; O’Leary, J.G.; Thompson-Snipes, L.; Nyarige, V.; Wang, J.; Park, W.; Stegall, M.; Heilman, R.; Klintmalm, G.B.; et al. Implication of TIGIT(+) human memory B cells in immune regulation. Nat. Commun. 2021, 12, 1534. [Google Scholar] [CrossRef]
- Wang, F.F.; Wang, Y.; Wang, L.; Wang, T.S.; Bai, Y.P. TIGIT expression levels on CD4+ T cells are correlated with disease severity in patients with psoriasis. Clin. Exp. Dermatol. 2018, 43, 675–682. [Google Scholar] [CrossRef]
- Patino-Lopez, G.; Hevezi, P.; Lee, J.; Willhite, D.; Verge, G.M.; Lechner, S.M.; Ortiz-Navarrete, V.; Zlotnik, A. Human class-I restricted T cell associated molecule is highly expressed in the cerebellum and is a marker for activated NKT and CD8+ T lymphocytes. J. Neuroimmunol. 2006, 171, 145–155. [Google Scholar] [CrossRef]
- Boles, K.S.; Barchet, W.; Diacovo, T.; Cella, M.; Colonna, M. The tumor suppressor TSLC1/NECL-2 triggers NK-cell and CD8+ T-cell responses through the cell-surface receptor CRTAM. Blood 2005, 106, 779–786. [Google Scholar] [CrossRef]
- Beristain-Covarrubias, N.; Canche-Pool, E.B.; Ramirez-Velazquez, C.; Barragan-Galvez, J.C.; Gomez-Diaz, R.A.; Ortiz-Navarrete, V. Class I-Restricted T Cell-Associated Molecule Is a Marker for IFN-γ-Producing iNKT Cells in Healthy Subjects and Patients with Type 1 Diabetes. J. Interferon. Cytokine Res. 2017, 37, 39–49. [Google Scholar] [CrossRef]
- Ramirez-Velazquez, C.; Beristain-Covarrubias, N.; Guido-Bayardo, L.; Ortiz-Navarrete, V. Peripheral blood T cells and neutrophils from asthma patients express class-I MHC-restricted T cell-associated molecule. Allergy Asthma Clin. Immunol. 2014, 10, 46. [Google Scholar] [CrossRef][Green Version]
- Takeuchi, A.; Badr Mel, S.; Miyauchi, K.; Ishihara, C.; Onishi, R.; Guo, Z.; Sasaki, Y.; Ike, H.; Takumi, A.; Tsuji, N.M.; et al. CRTAM determines the CD4+ cytotoxic T lymphocyte lineage. J. Exp. Med. 2016, 213, 123–138. [Google Scholar] [CrossRef]
- Patil, V.S.; Madrigal, A.; Schmiedel, B.J.; Clarke, J.; O’Rourke, P.; de Silva, A.D.; Harris, E.; Peters, B.; Seumois, G.; Weiskopf, D.; et al. Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci. Immunol. 2018, 3, eaan8664. [Google Scholar] [CrossRef]
- Devilard, E.; Xerri, L.; Dubreuil, P.; Lopez, M.; Reymond, N. Nectin-3 (CD113) interacts with Nectin-2 (CD112) to promote lymphocyte transendothelial migration. PLoS ONE 2013, 8, e77424. [Google Scholar] [CrossRef]
- Fabre-Lafay, S.; Monville, F.; Garrido-Urbani, S.; Berruyer-Pouyet, C.; Ginestier, C.; Reymond, N.; Finetti, P.; Sauvan, R.; Adelaide, J.; Geneix, J.; et al. Nectin-4 is a new histological and serological tumor associated marker for breast cancer. BMC Cancer 2007, 7, 73. [Google Scholar] [CrossRef]
- Zhu, Y.; Paniccia, A.; Schulick, A.C.; Chen, W.; Koenig, M.R.; Byers, J.T.; Yao, S.; Bevers, S.; Edil, B.H. Identification of CD112R as a novel checkpoint for human T cells. J. Exp. Med. 2016, 213, 167–176. [Google Scholar] [CrossRef]
- Holmes, V.M.; Maluquer de Motes, C.; Richards, P.T.; Roldan, J.; Bhargava, A.K.; Orange, J.S.; Krummenacher, C. Interaction between nectin-1 and the human natural killer cell receptor CD96. PLoS ONE 2019, 14, e0212443. [Google Scholar] [CrossRef]
- Reymond, N.; Imbert, A.M.; Devilard, E.; Fabre, S.; Chabannon, C.; Xerri, L.; Farnarier, C.; Cantoni, C.; Bottino, C.; Moretta, A.; et al. DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J. Exp. Med. 2004, 199, 1331–1341. [Google Scholar] [CrossRef]
- Sullivan, D.P.; Seidman, M.A.; Muller, W.A. Poliovirus receptor (CD155) regulates a step in transendothelial migration between PECAM and CD99. Am. J. Pathol. 2013, 182, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Reymond, N.; Borg, J.P.; Lecocq, E.; Adelaide, J.; Campadelli-Fiume, G.; Dubreuil, P.; Lopez, M. Human nectin3/PRR3: A novel member of the PVR/PRR/nectin family that interacts with afadin. Gene 2000, 255, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Lopez, M.; Cocchi, F.; Avitabile, E.; Leclerc, A.; Adelaide, J.; Campadelli-Fiume, G.; Dubreuil, P. Novel, soluble isoform of the herpes simplex virus (HSV) receptor nectin1 (or PRR1-HIgR-HveC) modulates positively and negatively susceptibility to HSV infection. J. Virol. 2001, 75, 5684–5691. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lozano, E.; Dominguez-Villar, M.; Kuchroo, V.; Hafler, D.A. The TIGIT/CD226 axis regulates human T cell function. J. Immunol. 2012, 188, 3869–3875. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.J.; Andrews, D.M.; Smyth, M.J. Receptors that interact with nectin and nectin-like proteins in the immunosurveillance and immunotherapy of cancer. Curr. Opin. Immunol. 2012, 24, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zheng, Q.; Xin, N.; Wang, W.; Zhao, C. CD155, an onco-immunologic molecule in human tumors. Cancer Sci. 2017, 108, 1934–1938. [Google Scholar] [CrossRef]
- Molfetta, R.; Milito, N.D.; Zitti, B.; Lecce, M.; Fionda, C.; Cippitelli, M.; Santoni, A.; Paolini, R. The Ubiquitin-proteasome pathway regulates Nectin2/CD112 expression and impairs NK cell recognition and killing. Eur. J. Immunol. 2019, 49, 873–883. [Google Scholar] [CrossRef]
- Sasaki, H.; Nishikata, I.; Shiraga, T.; Akamatsu, E.; Fukami, T.; Hidaka, T.; Kubuki, Y.; Okayama, A.; Hamada, K.; Okabe, H.; et al. Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute-type adult T-cell leukemia. Blood 2005, 105, 1204–1213. [Google Scholar] [CrossRef]
- Belaaloui, G.; Imbert, A.M.; Bardin, F.; Tonnelle, C.; Dubreuil, P.; Lopez, M.; Chabannon, C. Functional characterization of human CD34+ cells that express low or high levels of the membrane antigen CD111 (nectin 1). Leukemia 2003, 17, 1137–1145. [Google Scholar] [CrossRef]
- Stamm, H.; Klingler, F.; Grossjohann, E.M.; Muschhammer, J.; Vettorazzi, E.; Heuser, M.; Mock, U.; Thol, F.; Vohwinkel, G.; Latuske, E.; et al. Immune checkpoints PVR and PVRL2 are prognostic markers in AML and their blockade represents a new therapeutic option. Oncogene 2018, 37, 5269–5280. [Google Scholar] [CrossRef]
- Wang, X.; Xing, Z.; Chen, H.; Yang, H.; Wang, Q.; Xing, T. High expression of nectin-1 indicates a poor prognosis and promotes metastasis in hepatocellular carcinoma. Front. Oncol. 2022, 12, 953529. [Google Scholar] [CrossRef]
- Tatsumi, K.; Taatjes, D.J.; Wadsworth, M.P.; Bouchard, B.A.; Bovill, E.G. Cell adhesion molecule 1 (CADM1) is ubiquitously present in the endothelium and smooth muscle cells of the human macro- and micro-vasculature. Histochem. Cell Biol. 2012, 138, 815–820. [Google Scholar] [CrossRef]
- Manivannan, K.; Rowan, A.G.; Tanaka, Y.; Taylor, G.P.; Bangham, C.R. CADM1/TSLC1 Identifies HTLV-1-Infected Cells and Determines Their Susceptibility to CTL-Mediated Lysis. PLoS Pathog. 2016, 12, e1005560. [Google Scholar] [CrossRef]
- Kim, H.R.; Jeon, B.H.; Lee, H.S.; Im, S.H.; Araki, M.; Araki, K.; Yamamura, K.; Choi, S.C.; Park, D.S.; Jun, C.D. IGSF4 is a novel TCR ζ-chain-interacting protein that enhances TCR-mediated signaling. J. Exp. Med. 2011, 208, 2545–2560. [Google Scholar] [CrossRef]
- Fuchs, A.; Cella, M.; Giurisato, E.; Shaw, A.S.; Colonna, M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J. Immunol. 2004, 172, 3994–3998. [Google Scholar] [CrossRef]
- Hoffmann, J.; Fišer, K.; Liebetrau, C.; Staubach, N.; Kost, D.; Voss, S.; Heiden, A.Z.; Dörr, O.; Lipps, C.; Nef, H.M.; et al. High-Content Immunophenotyping and Hierarchical Clustering Reveal Sources of Heterogeneity and New Surface Markers of Human Blood Monocyte Subsets. Thromb. Haemost. 2020, 120, 141–155. [Google Scholar] [CrossRef]
- Lange, R.; Peng, X.; Wimmer, E.; Lipp, M.; Bernhardt, G. The poliovirus receptor CD155 mediates cell-to-matrix contacts by specifically binding to vitronectin. Virology 2001, 285, 218–227. [Google Scholar] [CrossRef]
- Ramsbottom, K.M.; Hawkins, E.D.; Shimoni, R.; McGrath, M.; Chan, C.J.; Russell, S.M.; Smyth, M.J.; Oliaro, J. Cutting edge: DNAX accessory molecule 1-deficient CD8+ T cells display immunological synapse defects that impair antitumor immunity. J. Immunol. 2014, 192, 553–557. [Google Scholar] [CrossRef]
- Hou, S.; Ge, K.; Zheng, X.; Wei, H.; Sun, R.; Tian, Z. CD226 protein is involved in immune synapse formation and triggers Natural Killer (NK) cell activation via its first extracellular domain. J. Biol. Chem. 2014, 289, 6969–6977. [Google Scholar] [CrossRef]
- Shibuya, K.; Shirakawa, J.; Kameyama, T.; Honda, S.; Tahara-Hanaoka, S.; Miyamoto, A.; Onodera, M.; Sumida, T.; Nakauchi, H.; Miyoshi, H.; et al. CD226 (DNAM-1) is involved in lymphocyte function-associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J. Exp. Med. 2003, 198, 1829–1839. [Google Scholar] [CrossRef]
- Enqvist, M.; Ask, E.H.; Forslund, E.; Carlsten, M.; Abrahamsen, G.; Béziat, V.; Andersson, S.; Schaffer, M.; Spurkland, A.; Bryceson, Y.; et al. Coordinated expression of DNAM-1 and LFA-1 in educated NK cells. J. Immunol. 2015, 194, 4518–4527. [Google Scholar] [CrossRef]
- Lozano, E.; Joller, N.; Cao, Y.; Kuchroo, V.K.; Hafler, D.A. The CD226/CD155 interaction regulates the proinflammatory (Th1/Th17)/anti-inflammatory (Th2) balance in humans. J. Immunol. 2013, 191, 3673–3680. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zeng, H.; Zhang, Y.; Chen, K.; Zhang, C.; Song, C.; Fang, L.; Xu, Z.; Yang, K.; Jin, B.; et al. CD226 ligation protects against EAE by promoting IL-10 expression via regulation of CD4+ T cell differentiation. Oncotarget 2016, 7, 19251–19264. [Google Scholar] [CrossRef] [PubMed]
- Murata, R.; Kinoshita, S.; Matsuda, K.; Kawaguchi, A.; Shibuya, A.; Shibuya, K. G307S DNAM-1 Mutation Exacerbates Autoimmune Encephalomyelitis via Enhancing CD4+T Cell Activation. J. Immunol. 2022, 12, 2304–2312. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liang, S.; Jin, J.; Fang, L.; Ma, Q.; Wang, X.; Song, Y.; Chen, L. CD226 attenuates Treg suppressive capacity via CTLA-4 and TIGIT during EAE. Immunol. Res. 2019, 67, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Stannard, K.A.; Lemoine, S.; Waterhouse, N.J.; Vari, F.; Chatenoud, L.; Gandhi, M.K.; Martinet, L.; Smyth, M.J.; Guillerey, C. Human peripheral blood DNAM-1(neg) NK cells are a terminally differentiated subset with limited effector functions. Blood Adv. 2019, 3, 1681–1694. [Google Scholar] [CrossRef]
- Ott, M.; Avendano-Guzman, E.; Ullrich, E.; Dreyer, C.; Strauss, J.; Harden, M.; Schon, M.; Schon, M.P.; Bernhardt, G.; Stadelmann, C.; et al. Laquinimod, a prototypic quinoline-3-carboxamide and aryl hydrocarbon receptor agonist, utilizes a CD155-mediated natural killer/dendritic cell interaction to suppress CNS autoimmunity. J. Neuroinflamm. 2019, 16, 49. [Google Scholar] [CrossRef]
- Nielsen, N.; Pascal, V.; Fasth, A.E.; Sundström, Y.; Galsgaard, E.D.; Ahern, D.; Andersen, M.; Baslund, B.; Bartels, E.M.; Bliddal, H.; et al. Balance between activating NKG2D, DNAM-1, NKp44 and NKp46 and inhibitory CD94/NKG2A receptors determine natural killer degranulation towards rheumatoid arthritis synovial fibroblasts. Immunology 2014, 142, 581–593. [Google Scholar] [CrossRef]
- Fasth, A.E.; Björkström, N.K.; Anthoni, M.; Malmberg, K.J.; Malmström, V. Activating NK-cell receptors co-stimulate CD4+CD28− T cells in patients with rheumatoid arthritis. Eur. J. Immunol. 2010, 40, 378–387. [Google Scholar] [CrossRef]
- Elhai, M.; Chiocchia, G.; Marchiol, C.; Lager, F.; Renault, G.; Colonna, M.; Bernhardt, G.; Allanore, Y.; Avouac, J. Targeting CD226/DNAX accessory molecule-1 (DNAM-1) in collagen-induced arthritis mouse models. J. Inflamm. 2015, 12, 9. [Google Scholar] [CrossRef]
- Asashima, H.; Axisa, P.P.; Pham, T.H.G.; Longbrake, E.E.; Ruff, W.E.; Lele, N.; Cohen, I.; Raddassi, K.; Sumida, T.S.; Hafler, D.A. Impaired TIGIT expression on B cells drives circulating follicular helper T cell expansion in multiple sclerosis. J. Clin. Investig. 2022, 132. [Google Scholar] [CrossRef]
- He, X.; Yin, J.; Yu, M.; Wang, H.; Qiu, J.; Wang, A.; He, X.; Wu, X. Identification and Validation of Hub Genes for Predicting Treatment Targets and Immune Landscape in Rheumatoid Arthritis. Biomed. Res. Int. 2022, 2022, 8023779. [Google Scholar] [CrossRef]
- Fuchs, Y.F.; Sharma, V.; Eugster, A.; Kraus, G.; Morgenstern, R.; Dahl, A.; Reinhardt, S.; Petzold, A.; Lindner, A.; Löbel, D.; et al. Gene Expression-Based Identification of Antigen-Responsive CD8+ T Cells on a Single-Cell Level. Front. Immunol. 2019, 10, 2568. [Google Scholar] [CrossRef]
- Sona, C.; Yeh, Y.T.; Patsalos, A.; Halasz, L.; Yan, X.; Kononenko, N.L.; Nagy, L.; Poy, M.N. Evidence of islet CADM1-mediated immune cell interactions during human type 1 diabetes. JCI Insight 2022, 7. [Google Scholar] [CrossRef]
- Takeuchi, A.; Itoh, Y.; Takumi, A.; Ishihara, C.; Arase, N.; Yokosuka, T.; Koseki, H.; Yamasaki, S.; Takai, Y.; Miyoshi, J.; et al. CRTAM confers late-stage activation of CD8+ T cells to regulate retention within lymph node. J. Immunol. 2009, 183, 4220–4228. [Google Scholar] [CrossRef]
- Eriksson, E.M.; Keh, C.E.; Deeks, S.G.; Martin, J.N.; Hecht, F.M.; Nixon, D.F. Differential expression of CD96 surface molecule represents CD8+ T cells with dissimilar effector function during HIV-1 infection. PLoS ONE 2012, 7, e51696. [Google Scholar] [CrossRef]
- Bunet, R.; Nayrac, M.; Ramani, H.; Sylla, M.; Durand, M.; Chartrand-Lefebvre, C.; Routy, J.P.; Landay, A.L.; Gauchat, J.F.; Chomont, N.; et al. Loss of CD96 Expression as a Marker of HIV-Specific CD8+ T-Cell Differentiation and Dysfunction. Front. Immunol. 2021, 12, 673061. [Google Scholar] [CrossRef]
- Medina-Contreras, O.; Soldevila, G.; Patiño-Lopez, G.; Canche-Pool, E.; Valle-Rios, R.; Ortiz-Navarrete, V. Role of CRTAM during mouse early T lymphocytes development. Dev. Comp. Immunol. 2010, 34, 196–202. [Google Scholar] [CrossRef]
- Galibert, L.; Diemer, G.S.; Liu, Z.; Johnson, R.S.; Smith, J.L.; Walzer, T.; Comeau, M.R.; Rauch, C.T.; Wolfson, M.F.; Sorensen, R.A.; et al. Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for class-I-restricted T-cell-associated molecule. J. Biol. Chem. 2005, 280, 21955–21964. [Google Scholar] [CrossRef]
- Rojas-Marquez, C.; Valle-Rios, R.; Lopez-Bayghen, E.; Ortiz-Navarrete, V. CRTAM is negatively regulated by ZEB1 in T cells. Mol. Immunol. 2015, 66, 290–298. [Google Scholar] [CrossRef]
- Valle-Rios, R.; Patiño-Lopez, G.; Medina-Contreras, O.; Canche-Pool, E.; Recillas-Targa, F.; Lopez-Bayghen, E.; Zlotnik, A.; Ortiz-Navarrete, V. Characterization of CRTAM gene promoter: AP-1 transcription factor control its expression in human T CD8 lymphocytes. Mol. Immunol. 2009, 46, 3379–3387. [Google Scholar] [CrossRef] [PubMed]
- Giangreco, A.; Hoste, E.; Takai, Y.; Rosewell, I.; Watt, F.M. Epidermal Cadm1 expression promotes autoimmune alopecia via enhanced T cell adhesion and cytotoxicity. J. Immunol. 2012, 188, 1514–1522. [Google Scholar] [CrossRef]
- Arase, N.; Takeuchi, A.; Unno, M.; Hirano, S.; Yokosuka, T.; Arase, H.; Saito, T. Heterotypic interaction of CRTAM with Necl2 induces cell adhesion on activated NK cells and CD8+ T cells. Int. Immunol. 2005, 17, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Barragan, L.; Cortez, V.S.; Wang, Q.; McDonald, K.G.; Chai, J.N.; Di Luccia, B.; Gilfillan, S.; Hsieh, C.S.; Newberry, R.D.; Sibley, L.D.; et al. CRTAM Protects Against Intestinal Dysbiosis During Pathogenic Parasitic Infection by Enabling Th17 Maturation. Front. Immunol. 2019, 10, 1423. [Google Scholar] [CrossRef] [PubMed]
- Cortez, V.S.; Cervantes-Barragan, L.; Song, C.; Gilfillan, S.; McDonald, K.G.; Tussiwand, R.; Edelson, B.T.; Murakami, Y.; Murphy, K.M.; Newberry, R.D.; et al. CRTAM controls residency of gut CD4+CD8+ T cells in the steady state and maintenance of gut CD4+ Th17 during parasitic infection. J. Exp. Med. 2014, 211, 623–633. [Google Scholar] [CrossRef]
- Perez-Lopez, A.; Nuccio, S.P.; Ushach, I.; Edwards, R.A.; Pahu, R.; Silva, S.; Zlotnik, A.; Raffatellu, M. CRTAM Shapes the Gut Microbiota and Enhances the Severity of Infection. J. Immunol. 2019, 203, 532–543. [Google Scholar] [CrossRef]
- Catros, V.; Dessarthe, B.; Thedrez, A.; Toutirais, O. [Nectins and nectin-like receptors DNAM-1 and CRTAM: New ways for tumor escape]. Med. Sci. 2014, 30, 537–543. [Google Scholar] [CrossRef]
- Ramirez-Ramirez, D.; Padilla-Castaneda, S.; Galan-Enriquez, C.S.; Vadillo, E.; Prieto-Chavez, J.L.; Jimenez-Hernandez, E.; Vilchis-Ordonez, A.; Sandoval, A.; Balandran, J.C.; Perez-Tapia, S.M.; et al. CRTAM(+) NK cells endowed with suppressor properties arise in leukemic bone marrow. J. Leukoc. Biol. 2019, 105, 999–1013. [Google Scholar] [CrossRef]
- Dessarthe, B.; Thedrez, A.; Latouche, J.B.; Cabillic, F.; Drouet, A.; Daniel, P.; de La Pintière, C.T.; Catros, V.; Toutirais, O. CRTAM receptor engagement by Necl-2 on tumor cells triggers cell death of activated Vγ9Vδ2 T cells. J. Immunol. 2013, 190, 4868–4876. [Google Scholar] [CrossRef]
- Oh-Oka, K.; Abe, F.; Shibuya, A.; Shibuya, K. CD96 Blockade Ameliorates Imiquimod-Induced Psoriasis-like Dermatitis via Suppression of IL-17A Production by Dermal γδ T Cells. J. Immunol. 2022, 12, 2313–2321. [Google Scholar] [CrossRef]
- Mittal, D.; Lepletier, A.; Madore, J.; Aguilera, A.R.; Stannard, K.; Blake, S.J.; Whitehall, V.L.J.; Liu, C.; Bettington, M.L.; Takeda, K.; et al. CD96 Is an Immune Checkpoint That Regulates CD8+ T-cell Antitumor Function. Cancer Immunol. Res. 2019, 7, 559–571. [Google Scholar] [CrossRef]
- Stanko, K.; Iwert, C.; Appelt, C.; Vogt, K.; Schumann, J.; Strunk, F.J.; Ahrlich, S.; Schlickeiser, S.; Romagnani, C.; Jürchott, K.; et al. CD96 expression determines the inflammatory potential of IL-9-producing Th9 cells. Proc. Natl. Acad. Sci. USA 2018, 115, E2940–E2949. [Google Scholar] [CrossRef]
- Whelan, S.; Ophir, E.; Kotturi, M.F.; Levy, O.; Ganguly, S.; Leung, L.; Vaknin, I.; Kumar, S.; Dassa, L.; Hansen, K.; et al. PVRIG and PVRL2 Are Induced in Cancer and Inhibit CD8+ T-cell Function. Cancer Immunol. Res. 2019, 7, 257–268. [Google Scholar] [CrossRef]
- Aterido, A.; Palau, N.; Domènech, E.; Nos Mateu, P.; Gutiérrez, A.; Gomollón, F.; Mendoza, J.L.; Garcia-Planella, E.; Barreiro-de Acosta, M.; Muñoz, F.; et al. Genetic association between CD96 locus and immunogenicity to anti-TNF therapy in Crohn’s disease. Pharmacogenomics J. 2019, 19, 547–555. [Google Scholar] [CrossRef]
- Ho, C.H.; Silva, A.A.; Tomita, B.; Weng, H.Y.; Ho, I.C. Differential impacts of TNFα inhibitors on the transcriptome of Th cells. Arthritis Res. Ther. 2021, 23, 199. [Google Scholar] [CrossRef]
- Chan, C.J.; Martinet, L.; Gilfillan, S.; Souza-Fonseca-Guimaraes, F.; Chow, M.T.; Town, L.; Ritchie, D.S.; Colonna, M.; Andrews, D.M.; Smyth, M.J. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat. Immunol. 2014, 15, 431–438. [Google Scholar] [CrossRef]
- Zhao, X.; Feng, X.; Liu, P.; Ye, J.; Tao, R.; Li, R.; Shen, B.; Zhang, X.; Wang, X.; Zhao, D. Abnormal expression of CD96 on natural killer cell in peripheral blood of patients with chronic obstructive pulmonary disease. Clin. Respir. J. 2022, 16, 546–554. [Google Scholar] [CrossRef]
- Johnston, R.J.; Comps-Agrar, L.; Hackney, J.; Yu, X.; Huseni, M.; Yang, Y.; Park, S.; Javinal, V.; Chiu, H.; Irving, B.; et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8+ T cell effector function. Cancer Cell 2014, 26, 923–937. [Google Scholar] [CrossRef]
- Joller, N.; Hafler, J.P.; Brynedal, B.; Kassam, N.; Spoerl, S.; Levin, S.D.; Sharpe, A.H.; Kuchroo, V.K. Cutting edge: TIGIT has T cell-intrinsic inhibitory functions. J. Immunol. 2011, 186, 1338–1342. [Google Scholar] [CrossRef]
- Zhao, W.; Dong, Y.; Wu, C.; Ma, Y.; Jin, Y.; Ji, Y. TIGIT overexpression diminishes the function of CD4 T cells and ameliorates the severity of rheumatoid arthritis in mouse models. Exp. Cell Res. 2016, 340, 132–138. [Google Scholar] [CrossRef]
- Sato, K.; Yamashita-Kanemaru, Y.; Abe, F.; Murata, R.; Nakamura-Shinya, Y.; Kanemaru, K.; Muratani, M.; Veillette, A.; Goto, M.; Ito, M.; et al. DNAM-1 regulates Foxp3 expression in regulatory T cells by interfering with TIGIT under inflammatory conditions. Proc. Natl. Acad. Sci. USA 2021, 118, e2021309118. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xia, P.; Du, Y.; Liu, S.; Huang, G.; Chen, J.; Zhang, H.; Hou, N.; Cheng, X.; Zhou, L.; et al. T-cell immunoglobulin and ITIM domain (TIGIT) receptor/poliovirus receptor (PVR) ligand engagement suppresses interferon-gamma production of natural killer cells via beta-arrestin 2-mediated negative signaling. J. Biol. Chem. 2014, 289, 17647–17657. [Google Scholar] [CrossRef] [PubMed]
- Stanietsky, N.; Simic, H.; Arapovic, J.; Toporik, A.; Levy, O.; Novik, A.; Levine, Z.; Beiman, M.; Dassa, L.; Achdout, H.; et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 17858–17863. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, A.; Liu, X.; Han, S.; Sun, Y.; Zhang, J.; Guo, L.; Zhang, Y. Blocking TIGIT/CD155 signalling reverses CD8+ T cell exhaustion and enhances the antitumor activity in cervical cancer. J. Transl. Med. 2022, 20, 280. [Google Scholar] [CrossRef]
- Maas, R.J.; Hoogstad-van Evert, J.S.; Van der Meer, J.M.; Mekers, V.; Rezaeifard, S.; Korman, A.J.; de Jonge, P.K.; Cany, J.; Woestenenk, R.; Schaap, N.P.; et al. TIGIT blockade enhances functionality of peritoneal NK cells with altered expression of DNAM-1/TIGIT/CD96 checkpoint molecules in ovarian cancer. Oncoimmunology 2020, 9, 1843247. [Google Scholar] [CrossRef]
- McKinney, E.F.; Lee, J.C.; Jayne, D.R.; Lyons, P.A.; Smith, K.G. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 2015, 523, 612–616. [Google Scholar] [CrossRef]
- Blazkova, J.; Huiting, E.D.; Boddapati, A.K.; Shi, V.; Whitehead, E.J.; Justement, J.S.; Nordstrom, J.L.; Moir, S.; Lack, J.; Chun, T.W. Correlation Between TIGIT Expression on CD8+ T Cells and Higher Cytotoxic Capacity. J. Infect. Dis. 2021, 224, 1599–1604. [Google Scholar] [CrossRef]
- Collier, J.L.; Weiss, S.A.; Pauken, K.E.; Sen, D.R.; Sharpe, A.H. Not-so-opposite ends of the spectrum: CD8+ T cell dysfunction across chronic infection, cancer and autoimmunity. Nat. Immunol. 2021, 22, 809–819. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hermans, D.; van Beers, L.; Broux, B. Nectin Family Ligands Trigger Immune Effector Functions in Health and Autoimmunity. Biology 2023, 12, 452. https://doi.org/10.3390/biology12030452
Hermans D, van Beers L, Broux B. Nectin Family Ligands Trigger Immune Effector Functions in Health and Autoimmunity. Biology. 2023; 12(3):452. https://doi.org/10.3390/biology12030452
Chicago/Turabian StyleHermans, Doryssa, Lisa van Beers, and Bieke Broux. 2023. "Nectin Family Ligands Trigger Immune Effector Functions in Health and Autoimmunity" Biology 12, no. 3: 452. https://doi.org/10.3390/biology12030452
APA StyleHermans, D., van Beers, L., & Broux, B. (2023). Nectin Family Ligands Trigger Immune Effector Functions in Health and Autoimmunity. Biology, 12(3), 452. https://doi.org/10.3390/biology12030452






