Simple Summary
Oxidative stress is recognized as one of the pathogenetic mechanisms underpinning insulin resistance, the hallmark of type 2 diabetes. Although oxidative stress can be elicited by unhealthy dietary patterns rich in long-chain saturated fatty acids and sugars, it may be exacerbated in genetically predisposed individuals carrying single nucleotide polymorphisms which dampen the activity of proteins responsible for maintaining redox balance, that is, the balance between oxygen and nitrogen reactive species production and their detoxification. In light of this, this literature review aims at providing an overview of the potential role of microalgae as a source of nutraceuticals able to improve insulin sensitivity by tackling oxidative stress, particularly in the aforementioned individuals. Microalgae represent a source of phenolic compounds, carotenoids, vitamins, and omega-3 polyunsaturated fatty acids which are able to counter oxidative stress, by either modulating intracellular pathways related to inflammation and oxidative stress or by acting as direct reactive oxygen species scavengers. Thus, microalgae represent a promising tool for precise nutritional interventions to tackle oxidative stress and improve insulin sensitivity. Nevertheless, further studies are warranted to confirm whether their supplementation in genetically predisposed individuals may be sufficient to restore redox balance and counter insulin resistance.
Abstract
Microalgae represent a growing innovative source of nutraceuticals such as carotenoids and phenolic compound which are naturally present within these single-celled organisms or can be induced in response to specific growth conditions. The presence of the unfavourable allelic variant in genes involved in the control of oxidative stress, due to one or more SNPs in gene encoding protein involved in the regulation of redox balance, can lead to pathological conditions such as insulin resistance, which, in turn, is directly involved in the pathogenesis of type 2 diabetes mellitus. In this review we provide an overview of the main SNPs in antioxidant genes involved in the promotion of insulin resistance with a focus on the potential role of microalgae-derived antioxidant molecules as novel nutritional tools to mitigate oxidative stress and improve insulin sensitivity.
1. Introduction
In genetics, different variability sources have been reported, but among these, single nucleotide polymorphisms (SNPs) are prominent as they represent around 90% of human genetic variation. To be defined as SNPs, the polymorphic alleles arising from the substitution of a single base have to be present in more than 1% of the population. SNPs can occur within the coding sequence of a gene, an intronic or intergenic region, and can consequently affect the amino acid sequence of the resulting protein or modulate the expression of the gene itself, respectively [1]. However, this is not always the case, with not all nucleotide variations leading to an amino acid substitution in the protein encoded by the SNPs affected gene.
On the contrary, however, in some cases, SNPs can act as important predictors of the response to certain specific drugs, susceptibility to certain environmental stressors such as toxins, and the development of some diseases, including metabolic diseases [2]. In this regard, a wide variety of SNPs have been associated with the susceptibility to develop metabolic syndrome (MetS) and type 2 diabetes mellitus (T2DM) [3], which are both underlain by insulin resistance (IR). Insulin resistance is referred to as a blunted response of insulin tissue targets to this hormone leading to impaired glycaemic control, dyslipidemia along with an increased risk of developing cardiovascular disease and certain types of cancer [4,5]. In this regard, insulin resistance impairs glucose homeostasis by negatively affecting glucose uptake and metabolism in metabolically active tissues, namely the skeletal muscle, the adipose tissue, and the liver [6]. In the skeletal muscle, insulin resistance hampers GLUT-4-dependent glucose uptake leading to an impairment in glucose oxidative metabolism and non-oxidative glucose disposal [7]. The same holds true for the adipose tissue, where defective insulin signaling impairs glucose uptake. This occurs in concert with a loss of inhibition of lipolysis leading to an increase in circulating free-fatty acids which further deteriorates insulin sensitivity by promoting lipotoxic lipid accumulation in metabolically active tissues like the liver and the skeletal muscle [8]. Finally, in the liver, the ability of insulin to lower hepatic glucose production is compromised, further fueling circulating hyperglycaemia [9].
To date, a wide array of SNPs has been identified as genetic susceptibility factors for IR, including SNPs in genes encoding proteins directly involved in the insulin signaling pathway such as insulin receptor and in genes responsible for the modulation of insulin sensitivity as described in the next paragraphs. Additionally, other SNPs are able to trigger pathogenetic mechanisms directly involved in hampering insulin signaling, such as oxidative stress [10]. In this regard, several SNPs have been identified in genes encoding enzymes with antioxidant activity and therefore responsible for the maintenance of redox balance [11,12,13].
In particular, aberrant antioxidant genes lead to an imbalance in the cellular redox status underpinned by an increase in reactive oxygen species (ROS) and a decrease in their detoxification. The resulting redox imbalance leads to the disruption of the insulin signaling pathway at different levels, paving the way for the development of IR [14,15].
ROS are reactive molecules due to the presence of at least an unpaired electron in the outermost orbital [16], occur within the cell, and also represent the product of environmental factors such as active and passive smoking, ultraviolet exposure, and psychophysical stress. ROS are unstable molecules that, in order to return to their balanced state, subtract electrons from other nearby atoms, generating new unstable molecules in a chain reaction [17]. In physiological concentrations, ROS have fundamental roles including regulation of gene expression, intracellular communication, defence of the organism from pathogens, and stimulation of immune functions, but the lack of balance can cause damage to cellular components including proteins, lipids, and DNA [18,19,20].
Dietary patterns rich in fat and refined carbohydrates are key in fostering obesity and its comorbidities by triggering inflammation, lipotoxicity, and oxidative stress in metabolically active tissues as well as the hypothalamus [7,21,22]. Considering the deleterious impact of unhealthy dietary patters on oxidative stress, this phenomenon may be exacerbated in individuals carrying SNPs in genes encoding antioxidant proteins, thereby further fuelling insulin resistance [23].
In contrast, following a healthy dietary pattern, like the Mediterranean diet, rich in antioxidant molecules such as carotenoids, vitamins, phenolic compounds, and mono- as well as polyunsaturated fatty acids, coupled with adequate physical activity are essential factors to preserve insulin sensitivity and also by improving redox balance [7,24,25].
In this context, microalgae may represent a novel nutritional tool to counter oxidative stress, particularly due to the fact that these single-celled organisms provide a unique combination of antioxidant molecules [26].
Several microalgae species have already been approved by the major food regulatory authorities for human consumption, and more species are in the process of being approved. Although microalgae also include cyanobacteria, it was chosen not to mention them as they have some limitations in the production of molecules such as carotenoids and vitamins, thus becoming less attractive than green microalgae in regards to their use as functional foods [27,28]. In addition, although it is established that seaweeds and plants approved for human consumption can accumulate the same antioxidant molecules produced in green microalgae, the latter are also inducible and can therefore be used as bioreactors for the synthesis, and bioaccumulation, of specific molecules in response to particular stress stimuli [29]. In addition, microalgae can be grown in tightly controlled environments with the possibility to constantly monitor culture conditions such as temperature, pH, salinity, nutrient concentration, and illumination. This prevents the microalgae cultures from being influenced by environmental conditions as well as chemical and biological contaminations [30]. All these peculiarities make microalgae an excellent novel food and a source of nutraceuticals with multiple applications. In addition, despite their nutritional value, also in terms of antioxidant potential, there are no reports in the literature dissecting the relationship between microalgae antioxidant molecules, oxidative stress, and the effects on insulin sensitivity.
Herein we provide an overview on the main SNPs in antioxidant genes driving insulin resistance and the possible use of microalgae as a source of antioxidant molecules to improve oxidative stress and possibly improve insulin sensitivity, partially compensating the functional impairment induced by SNPs.
2. SNPs Promoting IR
Several studies provided evidence that SNPs are associated with cardio–metabolic disorders including obesity, T2DM, and cardiovascular disease [31]. Variants in more than 50 and 80 loci were, respectively, found to be linked with obesity and T2DM [32], and occur in genes that regulate glucose homeostasis, insulin signaling, and energy balance [33]. The mere presence of SNPs may increase the risk of developing metabolic diseases but is not sufficient for their occurrence. Indeed, in the absence of highly penetrant mutations, metabolic disease arises as a consequence of a complex interaction between intrinsic biological factors, including SNPs, and an obesogenic environment characterized by the consumption of highly palatable energy dense foods and physical inactivity. A pivotal metabolic aberration shared between the cardio–metabolic comorbidities linked with obesity is represented by IR [34]. Thus, considering the association between SNPs and impaired cardio–metabolic health, it is not surprising that they may also contribute to the pathogenesis of IR [35]. In this regard, SNPs in genes encoding peroxisome proliferator-activated receptor gamma (PPARγ), insulin receptor substrate (IRS-1), glucokinase regulatory protein (GCKR), insulin-like growth factor I (IGF1), retinoic acid receptor responder 2 (RARRES2), and transcription factor 7 like 2 (TCF7L2) have all been linked with IR. The involvement of these SNPs in the pathogenesis of IR has been extensively reviewed elsewhere [35,36,37].
However, SNPs implicated in the pathogenesis of IR span beyond genes involved in the regulation of insulin signal transduction pathway and metabolic fuel metabolism. In line with this, SNPs in genes involved in regulating redox balance, as detailed below, have also been implicated in the pathogenesis of IR [10].
3. The Role of Oxidative Stress in Promoting Insulin Resistance
The insulin receptor is located in plasma membranes of insulin target cells and is characterized by an intermembrane glycoprotein consisting of four subunits: two alpha- and two beta-subunits forming two alpha-beta (α-β) dimers, bound by disulphide bridges [38,39]. Insulin, upon binding to its cognate receptor, induces a conformational change of the β-subunit that includes an extracellular-, a transmembrane-, and an intercellular-domain. Insulin receptor β-subunit has intrinsic tyrosine kinase activity which is activated by insulin-induced conformational change leading to trans autophosphorylation of three tyrosine residues (Tyr 1158, 1162, 1163) in the cytosolic domain of the receptor.
In order to activate the signal transduction pathway, the insulin receptor requires the interaction with the scaffolding proteins IRS1 and IRS2, two members of the family of insulin receptor substrate (IRS), and an adaptor protein called SHC. IRS1 and IRS2 promote propagation and amplification of insulin metabolic signals, whereas the SHC proteins promote and amplify insulin-activated mitogenic signals [40,41].
As a consequence of IRS binding to the insulin receptor, the phosphorylation of multiple tyrosine residues in the COOH-terminal tail of IRS proteins occurs, permitting the recruitment of Src homology 2 domain (SH2) located in the p85 regulatory subunit of phosphoinositide-3-kinase (PI3K) heterodimers. One of the actions of PI3K is to catalyse the synthesis of phosphatidylinositol-3,4,5-triphosphate (PIP3) from phosphatidylinositol-4,5-bisphosphate [42].
The synthesis of PIP3 leads to the amplification of downstream signals through the binding of mainly two effectors: phosphoinositide-dependent kinase 1 (PDK1) and protein-kinase B (Akt). Akt phosphorylation occurs on Thr308 and Ser473 residues by phosphoinositide-dependent kinase 1 (PDK1) and mechanistic target of rapamycin (mTOR), respectively; its phosphorylation acts as a key node in the transduction of insulin signal which occurs again through phosphorylation of downstream targets involved in cellular growth, survival, and metabolism such as glycogen synthase kinase 3, forkhead box O (FOXO) family of transcription factors, BH3-only protein BAD, mTORC1, endothelial nitric oxide synthase [43,44,45,46], and Akt substrate of 160 kDa protein through which vesicle rich in glucose transporter protein 4 is trafficked to plasma membrane [47].
Oxidative stress can promote IR by affecting the insulin pathway at different nodes via the activation of pathways that interfere with insulin signaling [48]. For example, oxidative stress, due to increased ROS production or defects in their detoxification processes, leads to the activation of different serine kinases such as inhibitor kB kinase β (IKK-β), protein kinase C (PKC), mitogen activated protein kinases (MAPK) as p38, extracellular signal-regulated kinase (ERK), c-Jun n-terminal kinases (JNK) which, in turn, can target key actors governing insulin signaling [49].
These kinases can target both the insulin receptor and IRS, depriving them of full signal transduction capacity and consequently leading to an attenuation of insulin response [50]. The phosphorylation of IRS1 on serine residues by JNK and MAPK [47,51,52] also hampers its phosphorylation on tyrosine residues making it more susceptible to proteasome-mediated degradation [49,53].
The activation of JNK by ROS can also occur through the oxidation and inactivation of the JNK-inhibiting MAPK phosphatase and subsequent apoptosis signal-regulating kinase 1 (ASK1) dissociation [54]. Similarly to JNK, as a result of its activation by pro-inflammatory mediators, IKK-β can phosphorylate multiple serine residues on IRS-1 and IRS-2 impairing the metabolic insulin signaling pathway via the inhibition of the tyrosine phosphorylation of IRS-1 mediated by the insulin receptor [51]. IKK-β is also central in the nuclear factor-κB (NF-kB) signaling pathway which also fuels oxidative stress by generating a vicious cycle that bridges together oxidative stress and inflammation [55].
Under basal conditions, NF-kB is sequestered as heterodimer in the cytoplasm by the NF-kB inhibitory protein (IkB) preventing its translocation to the nucleus and the induction of pro-inflammatory genes. However, in response to oxidative stress and pro-inflammatory stimuli, IKK-β phosphorylates IkB, which is ubiquitinated and undergoes proteasomal degradation, releases NF-kB. The NF-kB translocation to the nucleus results in the activation of the expression of genes encoding pro-inflammatory proteins and pro-oxidant enzymes such as inducible nitric oxide synthase (iNOS) and cyclooxygenase 2 (COX-2), promoting a vicious positive loop generated by inflammation and oxidative stress, termed oxinflammation [56,57,58,59].
In conclusion, oxinflammation and the consequent activation of MAPK and IKK-β can, as it is already known, lead to the development of IR by hampering the insulin signaling pathway [60,61] (Figure 1).
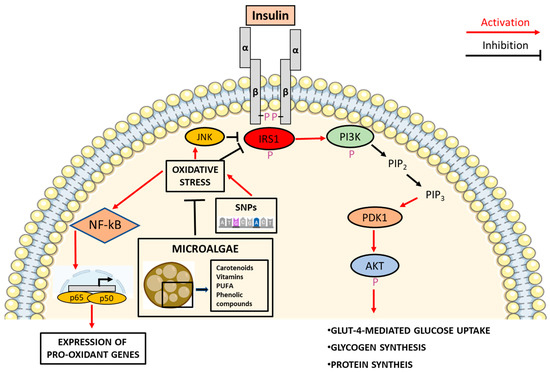
Figure 1.
Microalgae mitigate the ability of SNPs in inducing oxidative stress and deregulating insulin signaling.
4. SNPs and Their Involvement in Oxidative Stress
Oxidative stress refers to an imbalance between production of ROS and their removal by cell detoxification systems [62]. Some functional SNPs are present in genes involved in the regulation of redox status, impairing their expression, or altering the functionality of the proteins they encode [63] causing deleterious repercussions on redox balance, with the accumulation of oxygen free radicals and therefore oxidative stress.
Inside the cell, the major oxygen radical producers are the mitochondria, the endoplasmic reticulum, and the peroxisomes [64]. However, mitochondria represent the site where the greatest production of ROS occurs [65].
In the above-cited organelles, given the high production of ROS, enzymes with neutralizing action against free radicals are present, most of which are encoded by genes where numerous SNPs has been found. A list of the main antioxidant genes and the main functional SNPs found in them is reported in the following paragraphs and in Table 1.

Table 1.
List of the most representative SNPs involved in promoting IR and modulating oxidative stress.
4.1. Catalase (CAT)
Catalase is an enzyme normally found in all organisms exposed to oxygen and is encoded by the homonymous gene found in humans on chromosome 11p13 [97]. Catalase, one of the most efficient enzymes in the cell, carries out the dismutation reactions of several ROS, in particular H2O2, which is converted to H2O and O2. The presence of SNP rs1001179 (C262T) at the promoter region of the Cat gene negatively influences the binding of transcription factors and, consequently, the basal transcription and expression of the enzyme [66]. The presence of the T allele, when compared to the C allele, has been associated with reduced enzyme activity, which may contribute to increased levels of ROS and, consequently, to oxidative stress [67]. This SNPs has not only been associated with oxidative stress but also with an increased risk of developing IR [68] and related complications [69].
4.2. Superoxide Dismutase (SOD)
Superoxide dismutase (SOD) identifies a class of enzymes containing metal ions in the active site and capable of carrying out redox reactions to dismutate the superoxide anion (O−2) into molecular oxygen and hydrogen peroxide. In humans, three forms of SOD are detected: SOD-1, -2, and -3, with location in the cytoplasm, mitochondria, and extracellular space, respectively.
The presence of the functional SNP rs4880 (C47T) in the gene encoding MnSod (SOD2) leads to a missense mutation in exon 2 of nuclear chromosome 6q25 [70]. During (cytoplasmic) translation of the sixteenth codon of messenger RNA, the C47T SNP leads to the insertion of a valine instead of an alanine into the forming peptide chain, altering the conformation of mitochondrial targeting sequence and thus impairing the correct delivery of MnSOD to mitochondria [71]. The loss of efficiency in post-translational trafficking of this enzyme results in a reduction of the amount of MnSOD imported into mitochondria and in the diminished neutralizing potential of the superoxide ion [72] leading to IR [73] and poor cardiometabolic health [74].
4.3. Glutathione Peroxidase 1 (GPX1)
Glutathione peroxidase (GPx) is the nomenclature assigned to a family of enzymes with peroxidase activity. Eight genes, each encoding a different isoform of glutathione peroxidase (GPX1-8) with different cellular localisation and substrate, have been identified so far. The GPX1 gene located on the nuclear chromosome 3p21.3 encodes for the most representative and abundant enzyme of the family: GPX1 [75]. The presence of the functional SNP rs1050450 on Gpx1 gene, induces the substitution of a cytosine with a thymine resulting in the replacement of a leucine with a proline at position 198 [76].
This SNP causes the reduction of GPX1 enzyme activity [77], which has also been reported to correlate with the development of IR [78].
4.4. Glutathione-S-Transferases (GSTs)
Glutathione s-transferases (GST) identify a family of genes encoding proteins essential for the detoxification of endogenous and exogenous metabolite (carcinogens, antitumor drugs, environmental pollutants), including ROS [98]. The glutathione-s-transferases family comprises 16 genes further divided into six subfamilies: Gst-alpha (GstA), -mu (GstM), -omega (GstO), -pi (GstP), -theta (GstT), and -zeta (GstZ) [99].
Several SNPs in the GstM1, T1, and P1 genes have been associated with the increase in oxidative stress [100,101,102] and the development of IR as well as T2DM [103,104].
The SNP rs1695 located at the fifth exon of the GstP1 gene arouses the substitution of a guanine at position 313 with an adenine, causing the substitution of an isoleucine translated from codon 105 with a valine and a decrease in enzymatic activity [79,80,81].
The SNP rs1056806, located at the level of the GstM1 gene, generates the substitution of a cytosine with a guanine that has been found to predispose to an increased risk of obesity [63,82].
The SNP rs17856199 at the level of the GstT1 gene modifies the presence of an adenine with a cytosine, generating the substitution of a phenylalanine with a cysteine during the translation of codon 45 [82]. The reported amino acid substitution causes a decrease in hydrophobic interactions in the encoded protein, therefore affecting the functionality of the protein itself and diminishing its detoxifying capacity [83].
4.5. Paraoxonase 1 (PON1)
The family of genes encoding paraoxonase proteins encompasses at least three members and includes Pon1, Pon2, and Pon3, which are located in the long arm of chromosome 7 between positions q21.3 and q22.1 [105]. Most of PON1 proteins circulate in the bloodstream bound to high-density lipoproteins (HDL) where they exert antioxidant actions by preventing lipid oxidation and protecting low-density lipoproteins (LDL) from oxidation, thereby contributing to the prevention and cardiovascular disease [105,106,107,108].
Two SNPs in the Pon1 gene were found to be particularly involved in the promotion of IR: the first one is the functional SNP rs854560 that causes the substitution of an adenine for a thymine in the 55 codon of the mRNA, leading to the substitution of a leucine with a methionine [84]. The reported amino acid substitution was found to reduce Pon1 transcript levels and, consequently, protein expression, with a reduction in its antioxidant capacity [85,86,87].
Second, the functional SNP rs662 is underpinned by the substitution of an adenine for a guanine at codon 192 with a consequence replacement of a glutamine with an arginine in the resulting protein. This substitution induces a downstream decrease in the activity of the antioxidant enzyme PON1 [84,88]. In line with the effects of other SNPs involved in redox homeostasis, SNPs rs854560 and rs662 have both been associated with IR and T2DM [89,90,91] by a decrease in the antioxidant activity of PON1.
4.6. Nuclear Factor-Erythroid 2-Related Factor 2 (Nrf2)
Nuclear factor-erythroid 2-related factor 2 (Nrf2) is an ubiquitous transcription factor encoded by the Nrf2 gene located on chromosome 2 at position q31 [92]. The functional SNP rs35652124 causes the substitution of a cytosine for a thymine in the promoter region of the Nrf2 gene at position -653, which impairs NRF2 transcription and, therefore, its protective effects against oxidative stress [92,93].
The functional SNP rs6721961 causes the substitution of a cytosine for an adenine nucleotide upstream of the promoter region of the Nrf2 gene, precisely 617 bp upstream of the start of the coding region, causing a decreased gene expression of Nrf2 [96].
Not surprisingly, the SNPs rs35652124 and rs6721961 have both been associated with increased risk for the development of IR and T2DM [13,94,95].
5. Microalgae as a Source of Nutraceuticals
Microalgae are unicellular eukaryotic and prokaryotic microorganisms, including cyanobacteria, found in fresh and salt water and able to perform the photosynthesis reactions [30]. Microalgae have been extensively studied in recent years as they are characterized by a high growth rate, high adaptability to different growth conditions, and the ability to accumulate biologically active molecules depending on the metabolism triggered by the conditions in which they are grown. As photosynthetic organisms, autotrophic microalgae do not require organic compounds to support their growth, instead they need water, light as source of energy, CO2 as carbon source, and nitrogen and phosphorous as nutrients. However heterotrophic microalgae can use organic compounds, mainly glucose, as a source of energy [109,110]. In light of these specific characteristics, autotrophic microalgae cultivation is environmentally sustainable as they do not require large amounts of water for their growth and are also capable of using atmospheric CO2 during photosynthesis, fixing carbon in a plethora of organic molecules while generating oxygen [111]. The ability of microalgae to fix carbon has been quantified as 1.83 kg of CO2 per kg of microalgal (bio)mass [112].
Microalgae have pleiotropic applications and as such are being explored for biofuel production in solid, liquid, and gaseous states; biomass production; as well as a functional and nutritional additive [113]. In the nutraceutical field, microalgae are attracting increasing attention as they possess a complete nutritional profile and, most importantly, represent an inducible source of bioactive molecules. In support of their nutritional value as well as safety profile, several microalgae are approved for human consumption by the food and drug administration (FDA) and the European food safety authority (EFSA) also as an ingredient for functional foods such as pasta, yogurt, and biscuits [114].
In terms of their nutritional composition, microalgae are generally rich in proteins, lipids, polysaccharides, pigments, vitamins, and more. Microalgae protein content ranges from 40% to 70% of the dry weight of the cell [115] and remarkably, the nutritional value of microalgae protein has been reported not to be inferior to that of animal proteins [30]. The lipid content of microalgae varies between 20% and 50% of cell dry weight and encompasses triacylglycerols, glycolipids, phytosterols, phospholipids, and some microalgae also contain esterified PUFAs (poly-unsaturated fatty acids) such as arachidonic acid (ARA) and eicosapentaenoic acid (EPA). Microalgae also contain carotenoids, chlorophyll A and B [116], as well as water and lipid soluble vitamins including vitamin A, B1, B2, B6, B12, C, E, biotin, and folic acid [117,118].
Despite their wide nutritional composition, microalgae are united by their antioxidant potential ascribed to a wide array of bioactive molecules able to dampen oxidative stress, with plausible repercussions on insulin sensitivity.
In this regard, a study conducted in a rat model of diet-induced MetS, showed that the inclusion of the microalgae Tetraselmis chuii in the diet provided benefits by promoting the hepatic production of antioxidant enzymes such as GPX, GSH, SOD, and decreasing the expression of the pro-inflammatory genes Tgfβ1, Il-1β, Tnf-α, and Nf-kb1 [119]. In a further study conducted on equines suffering from MetS, in which there is both a strong accumulation of oxidative stress-related molecules and a decrease in SOD enzyme activity, it was observed that the supplementation with blue-green algae Arthrospira platensis resulted in the increase in SOD enzyme activity compared to the control cohort and in a decrease in body weight and insulin resistance [26,120].
To date no clinical trial has investigated the implications of microalgae antioxidant molecules on human metabolic health. Future research is also required to define the digestibility of microalgae and the bioavailability of the antioxidant molecules that they contain, in order to counteract oxidative stress and insulin resistance [119,121].
Thus, in the following paragraphs, the role of microalgae-derived nutraceuticals as putative tools to tackle oxidative stress are described.
6. Microalgal Molecules with Antioxidant Action
As described above, microalgae represent natural bioreactors able to upregulate the production of specific molecules in response to exogenous stressors. These molecules include bioactives that can directly or indirectly scavenge free oxygen radicals, thereby countering oxidative stress [122]. In this regard, carotenoids, vitamins, phenolic compounds, and omega-3 fatty acids are the key classes of molecules responsible for the antioxidant properties ascribed to microalgae.
6.1. Carotenoids
The health promoting effects attributed to carotenoids are dependent upon their antioxidants and anti-inflammatory properties, in concert with the role of some carotenoids to act as vitamin A precursors [123]. The major dietary sources of carotenoids are represented by fruits, vegetables, legumes, and cereals. However, it must not be overlooked that microalgae also represent a source of carotenoids. Remarkably, compared with the other plant-derived carotenoids, their production in the microalgae is more efficient, cost effective, and not limited by regions and seasons [124].
The carotenoids produced by microalgae, most widely known for their antioxidant properties, are β-carotene, Lutein, astaxanthin, zeaxanthin, fucoxanthin, β-cryptoxanthin, violaxanthin, canthaxanthin, among others [125] (Table 2).

Table 2.
List of the main microalgae synthesizing carotenoids, vitamins, PUFA, and phenolic compounds.
Β-carotene is particularly abundant in Dunaliella salina which is considered to be the largest producer of this carotenoid (up to 13% of its biomass), but it is produced also in Chlorella sorokiniana, Nannochloropsis gaditana, among others [126]. Lutein is a xanthophyll member of the carotenoid family and occurs in both animal as well as plant products like in egg yolk, spinach, corn, and kale [131]. Lutein can be produced by microalgae as was already reported in Chlorella protothecoides, Chlorella sorokiniana, and Dunaliella salina [132]. Astaxanthin can be found in nature mainly in salmon, crustaceans, or krill, but it is also produced by chemical synthesis and in microalgae, mainly Haematococcus pluvialis [134] and Chlorella zofingiensis [184]. Zeaxanthin is also present in plant and animal products [185] but can be produced by microalgae, particularly in Nannochloropsis oceanica as well as in Chlorella saccharophilia and Synechococcus sp. [137]. Fucoxanthin is the main carotenoid pigment, component of photosynthetic light-harvesting complexes, in marine ecosystems and a member of the xanthophyll family, it can be produced by microalgae and, in particular, Phaeodactilum tricornutum [139], but it can also be produced in Isochrysis galbana, Odontella sinensis, and Chaetoceros calcitrans [140]. β-cryptoxanthin is a retinol precursor carotenoid with a chemical structure similar to β-carotene but more polar. Unlike β-carotene, however, β-cryptoxanthin is not present in many foods, and the richest are squash, persimmons, hot peppers, tangerines, and papaya [127]. In addition to the foods listed, this carotenoid is also produced in certain microalgae including Pandorina morum and the cyanobacteria Arthrospira platensis [142]. Violaxanthin (VX) is a carotenoid found mainly in orange-colored fruit, but it also produced in microalgae, for example, Chlorella vulgaris, Nannochloropsis oceanica, and Dunaliella salina [144,145]. Canthaxanthin (CX) similar to astaxanthin is contained in bacteria, algae, and some fungi [186], but it can also be produced in microalgae, and examples are Chlorella vulgaris [146] and Dactylococcus dissociatus [147].
All aforementioned carotenoids exert antioxidant effects via different mechanisms. In particular, they can directly scavenge reactive molecules, particularly singlet molecular oxygen (1O2) and peroxyl radicals as well as modulate molecular pathways and transcription factors which regulate the expression of genes involved in the ROS detoxification systems. In support of this, β-carotene is able to quench 1O2 by acquiring the singlet excitation energy, generating triplet-state β-carotene and ground-state oxygen, and subsequently dissipating the energy in the form of heat thereby reconstituting the carotenoid normal energy state [128]. To further support to the antioxidant potential of carotenoids, astaxanthin and β-cryptoxanthin have also been proven to act as ROS scavengers, as demonstrated by different antioxidant activity assay such as ferric reducing antioxidant power (FRAP), trolox equivalent antioxidant capacity (TEAC), oxygen radical absorbance capacity (ORAC), 2,2-diphenyl-1-picrylhydrazyl (DPPH), electron spin resonance spectroscopy (ESR) [187], 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) cation radical (ABTS), and total antioxidant capacity (TAC) [143]. However, as mentioned above, the ability of carotenoids to exploit antioxidant effects, span beyond their capacity to directly scavenge ROS. Indeed, some carotenoids are able to prevent oxidative stress by activating molecular pathways that culminate in the upregulation of antioxidant genes, with these effects being dependent upon NRF2 activation. In the absence of stimuli which perturbate the cell redox balance, the Nrf2 transcription factor is sequestered in the cytoplasm by the Kelch-like ECH associated protein 1 (Keap-1) repressor, which also plays a key role in promoting NRF2 proteasomal degradation [188]. Instead, in response to oxidative stress, the oxidation of specific cysteine residues in the Keap-1 repressor leads to its dissociation from NRF2, which is free to translocate into the nucleus, bind antioxidant responsive element (ARE) regions, and therefore induce the expression of antioxidant genes [189]. Carotenoids have been found to be capable of increasing NRF2-mediated expression of antioxidant genes through the by-products of carotenoids oxidation, which is the result of direct scavenging action and induction of NRF2 translocation to the nucleus. Thus, carotenoids not only directly scavenge ROS, but they also indirectly counteract oxidative stress by inducing genes involved in the cellular machinery responsible for detoxifying ROS [190]. In support of this, supplementation of astaxanthin in subjects affected by T2DM resulted in the upregulation of NRF2 [135]. The induction of this transcription factor in response to carotenoids also holds true for β-carotene [129], lutein [133], zeaxanthin [138], fucoxanthin [141], and β-cryptoxanthin [136] (Figure 2).
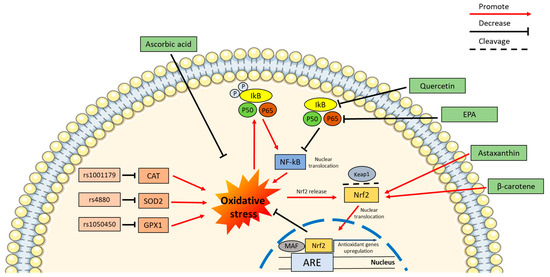
Figure 2.
SNPs induced oxidative stress, that could be counteracted by microalgae. Representation of the effect of some polymorphisms on the promotion of oxidative stress leading to a self-maintenance loop via the inflammation pathway. Oxidative stress promotes the phosphorylation of IkB, with the release of NF-kB and its translocation into the nucleus with a consequent expression promotion of pro-inflammatory genes. The molecules produced by microalgae, such as quercetin and EPA, can prevent phosphorylation of IkB and block the self-maintaining loop described above. Other microalgae metabolites such as the carotenoids astaxanthin and β-carotene can promote the translocation of the transcription factor NRF2 from the cytoplasm to the nucleus, as a consequence of the cleavage of its binding to the inhibitor Keap1, promoted by oxidative stress. The NRF2 translocation to the nucleus induced by carotenoids promotes the expression of antioxidant genes through its binding to ARE domains. In addition to the mechanisms mentioned, some microalgae metabolites such as ascorbic acid can reduce the oxidative stress by a direct scavenging activity.
The inhibition of the NF-kB signaling represents an additional mechanism underpinning the antioxidant potential of carotenoids. Indeed, carotenoids can hamper the nuclear translocation of the transcription factor NF-kB and the expression of pro-inflammatory molecules as well as genes involved in oxidative stress [130]. It has been hypothesized that carotenoids may exert indirect antioxidant activity by blocking IKK kinase activity and thereby allowing NF-kB sequestration in cytoplasm [191] (Figure 2). Furthermore, in addition to the NF-kB pathway itself, carotenoids can also regulate the action of the MAPKs ERK, JNK, and p38, which have been identified to be involved in the promotion of NF-kB activation [192,193].
Importantly, the ability of carotenoids to dampen oxidative stress and inflammation has also been associated with an improvement in insulin sensitivity, further supporting the role of microalgae as a source of nutraceuticals able to improve metabolic health [194,195,196].
6.2. Vitamins
Microalgae are also identified as a source of vitamins. In this regard, vitamins C and E contribute to the antioxidant potential of microalgae [152] (Table 2).
Vitamin C can be obtained from different microalgae such as Chlorella whose content can reach 266.67 μg/g Fwt [148], Nannochloris, and Dunaliella [152]. Vitamin C exerts its antioxidant effects by activating the NRF2 transcription factor as well as by acting as a direct ROS scavenger [149]. Additionally, this vitamin can counter ROS production by inhibiting the NF-kB signaling pathway, which also results in an anti-inflammatory effect [150] (Figure 2).
There is still a lack of consensus on the role of Vitamin C in improving insulin sensitivity. Nevertheless, Shi Lipeng and co-workers performed a meta-analysis of available data from randomized controlled trials on the effect of vitamin C supplementation in patients with T2DM. The result of the analysis, despite some controversies, showed a positive relationship between vitamin C intake, glycaemic control, and insulin sensitivity [151].
With regard to Vitamin E, it is produced, in particular, in Tetraselmis (6.32 mg/g DW), Chlamydomonas (4 mg/g DW), Chlorella (2 mg/g DW), and Dunaliella (1.90 mg/g DW) [152]. To the same extent as vitamin C, the antioxidant properties of Vitamin E are also well established and are dependent on its ability to directly scavenge ROS as well as indirectly by inducing Nrf2 and downregulating the NF-kB pathway [153].
In terms of its metabolic effects, in overweight subjects, vitamin E reduced oxidative stress and improve insulin sensitivity in a plasma concentration-dependent manner [154]. Previous studies have also postulated that the decrease in oxidative stress leads to an improvement in the chemical–physical state of plasma membranes [155] and in this sense, vitamin E has the ability to decrease the curvature of the plasma membrane influencing the activity of enzymes such as protein kinase C and diacylglycerol kinase [156], improving the cell’s response to insulin.
6.3. PUFA
Microalgae have also been shown to produce polyunsaturated fatty acids (PUFA) including ω-3 fatty acids. The ability of microalgae to produce ω-3 fatty acids varies depending on the algal species, for examples: C18:3 ω-3 α-linolenic acid (ALA) is mainly produced in Dunaliella primolecta [157], C. vulgaris, Chlorococcum amblystomatis, Scenedesmus obliquus, and Tetraselmis chuii [158]; C20:5 ω-3 EPA can be produced by Nannochloropsis oceanica [164] and Phaeodactylum tricornutum [165]; C22:6 ω-3 docosahexaenoic acid (DHA) is produced by Schizochytrium sp. [166] and Isochrysis galbana [167]; C16:3 ω-3 hexadecatrienoic acid is mainly produced in C. vulgaris and C18:4 ω-3 stearidonic acid mainly in Chlorococcum amblystomatis, Scenedesmus obliquus, and Tetraselmis chuii [158] (Table 2).
ω-3 fatty acids have been widely shown to exert anti-inflammatory effects [159,160,161], with the inhibition of inflammation also leading to a decrease in ROS generation. Beside the modulation of inflammation, the ability of ω-3 fatty acids to improve cellular redox status may also depend upon an improvement in mitochondrial function, mitochondrial-ER tethering, and a decrease in ER stress [197]. ω-3 fatty acids possess a modest direct free radical scavenging activity (DHA 28%, EPA 24% compared to the positive control quercetin with 100% scavenging activity) [162], but they mainly reduce oxidative stress indirectly by preserving the IkB inhibitor and thus preventing the translocation of NF-kB into the nucleus [168] (Figure 2).
In addition to the aforementioned mechanisms, ω-3 has also been found to dampen ROS levels by enhancing the expression of genes encoding enzymes with antioxidant activity, such as Gpx and heme oxygenase 1 (Ho-1), through Nrf2 gene expression activation [162,163].
In light of this, it is plausible that ω-3 fatty acids may improve IR. Nonetheless, the effects of ω-3 fatty acids on insulin resistance remain controversial with some reports indicating an improvement of insulin sensitivity upon their supplementation [198], whereas other studies fail to confirm this effect [199]. These controversies may be explained by the heterogeneity of the study population, particularly with regard to their ω-3 fatty acid status and the doses as well as the EPA/DHA ratio used in these studies.
6.4. Phenolic Compounds
Microalgae can also produce phenolic compounds which have a high ROS scavenging capacity in light of their chemical structure characterized by aromatic ring bearing one or more hydroxyl groups. In keeping with this, microalgae contain several classes of acidic phenols such as gallic, sinapic, and caffeic acids and also flavonoids, including isoflavones, flavanones, and flavonols [200]. Examples of microalgae in which phenolic compounds can be produced are Chlorella vulgaris, Haematococcus pluvial is, Diacronema lutheri, Phaeodactylum tricornutum, Tetraselmis suecica, and Porphyridium purpureum [169] (Table 2).
Phenolic compounds directly counter oxidative stress by acting as electron donors, being oxidized to quinones at the end of the subtraction of the stray electron from the reactive molecules [170,171]. Phenolic compounds may also reduce oxidative stress indirectly through NRF2 activation and subsequent expression of antioxidant genes such as Gst but mainly Ho-1 [172] and via the inhibition of NF-kB pathway [173].
One of the phenolic compounds with insulin sensitizing and antioxidant effects which could be synthetized in microalgae is represented by the flavonol quercetin. In particular, this phenolic compound can be produced by Tetraselmis suecica and Nannochloropsis gaditana [174]. It was reported that quercetin can directly scavenge free radicals due to the presence of two antioxidant pharmacophores in the molecule, induce the expression of NRF2, and also inhibit ROS-associated inflammation by blocking IKK-β and the nuclear translocation of NF-kB [175,176,177] (Figure 2). Quercetin can be integrated into the diet to stimulate insulin secretion, to protect beta cells from ROS, and to ameliorate the antioxidant defence and inflammation, therefore improving IR [178].
Another phenolic compound which can be synthesized in microalgae is represented by caffeic acid (CA). It can be produced in several microalgae including Phaeodactylum tricornutum, Tetraselmis suecica, Nannochloropsis gaditana, among others [179,180]. CA is capable of directly scavenging radicals such as the superoxide anion with both enzymatic and non-enzymatic reactions [181], to exert stimulatory effect on NRF2 presumably liberating NRF2 from the NRF2-keap1 complex [182], and it can also inhibit the NF-kB pathway, reducing inflammation as well oxidative stress, consequently improving insulin sensitivity [183].
7. Conclusions
In conclusion, microalgae represent a novel food rich in nutrients such as proteins, lipids, carbohydrates, vitamins, and minerals, but most importantly, it is an inducible source of bioactive molecules with antioxidant action. Specifically, unlike common seaweeds and plants, functionalized microalgae approved and under approval for human consumption can constitute a valuable nutritional tool for precision nutrition interventions in individuals carrying SNPs which affect their redox status. Thus, particularly in genetically predisposed individuals, the potential of microalgae to restore redox balance may also prevent the development of metabolic aberrations directly related to oxidative stress, such as insulin resistance. However, the fact that these effects may also be elicited by other organisms, such as seaweed, must not be overlooked [201,202]. Nevertheless, studies directly investigating the impact of microalgae intake in individuals with an increased susceptibility to oxidative stress are lacking, particularly those aimed at elucidating the relationship between antioxidant rich microalgae, oxidative stress, and insulin resistance. In addition, the bioavailability of the metabolites produced in green microalgae, the sustainable methods for their extraction and purification for a green economy, and the sensory quality of microalgae formulated food products represent some issues that have to be improved [118]. Despite this, microalgae, in light of their ability to act as bioreactors and accumulate a wide array of antioxidant molecules, remain a promising tool to improve insulin sensitivity by restoring redox balance.
Author Contributions
M.M. and D.S.: idealization, intellectual input; M.M.: literature search and writing initial version of the manuscript; D.S., C.S., A.P. and L.M.N.: manuscript editing and supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported “Local Research grants” from University of Ferrara, Italy (FAR 2022) to L.M.N. and to C.S. Moreover, this work was also supported with funding from the LEGA ITALIANA PER LA LOTTA CONTRO I TUMORI (LILT, Ferrara) (project number: C/A/0243/02/18).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful to Simonetta Pancaldi (Department of Environmental Sciences and Prevention, University of Ferrara, Italy) for helpful collaboration. Some figure details were created with BioRender.com, accessed on 8 March 2023 and smart.servier.com, accessed on 14 December 2022.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shatoff, E.; Bundschuh, R. Single nucleotide polymorphisms affect RNA-protein interactions at a distance through modulation of RNA secondary structures. PLoS Comput. Biol. 2020, 16, e1007852. [Google Scholar] [CrossRef]
- Alwi, Z.B. The Use of SNPs in Pharmacogenomics Studies. Malays. J. Med. Sci. 2005, 12, 4–12. [Google Scholar]
- Sun, W.; Yao, S.; Tang, J.; Liu, S.; Chen, J.; Deng, D.; Zeng, C. Integrative analysis of super enhancer SNPs for type 2 diabetes. PLoS ONE 2018, 13, e0192105. [Google Scholar] [CrossRef]
- Di Pino, A.; DeFronzo, R.A. Insulin Resistance and Atherosclerosis: Implications for Insulin-Sensitizing Agents. Endocr. Rev. 2019, 40, 1447–1467. [Google Scholar] [CrossRef]
- Sergi, D.; Boulestin, H.; Campbell, F.M.; Williams, L.M. The Role of Dietary Advanced Glycation End Products in Metabolic Dysfunction. Mol. Nutr. Food Res. 2021, 65, e1900934. [Google Scholar] [CrossRef]
- Li, M.; Chi, X.; Wang, Y.; Setrerrahmane, S.; Xie, W.; Xu, H. Trends in insulin resistance: Insights into mechanisms and therapeutic strategy. Signal Transduct. Target. Ther. 2022, 7, 216. [Google Scholar] [CrossRef]
- Sergi, D.; Naumovski, N.; Heilbronn, L.K.; Abeywardena, M.; O’Callaghan, N.; Lionetti, L.; Luscombe-Marsh, N. Mitochondrial (Dys)function and Insulin Resistance: From Pathophysiological Molecular Mechanisms to the Impact of Diet. Front. Physiol. 2019, 10, 532. [Google Scholar] [CrossRef]
- Chaurasia, B.; Summers, S.A. Ceramides in Metabolism: Key Lipotoxic Players. Annu. Rev. Physiol. 2021, 83, 303–330. [Google Scholar] [CrossRef]
- Patel, B.M.; Goyal, R.K. Liver and insulin resistance: New wine in old bottle!!! Eur. J. Pharmacol. 2019, 862, 172657. [Google Scholar] [CrossRef]
- Rupérez, A.I.; Gil, A.; Aguilera, C.M. Genetics of oxidative stress in obesity. Int. J. Mol. Sci. 2014, 15, 3118–3144. [Google Scholar] [CrossRef]
- Yucesoy, B.; Johnson, V.J.; Lummus, Z.L.; Kissling, G.E.; Fluharty, K.; Gautrin, D.; Malo, J.L.; Cartier, A.; Boulet, L.P.; Sastre, J.; et al. Genetic variants in antioxidant genes are associated with diisocyanate-induced asthma. Toxicol. Sci. 2012, 129, 166–173. [Google Scholar] [CrossRef]
- Fletcher, N.M.; Belotte, J.; Saed, M.G.; Memaj, I.; Diamond, M.P.; Morris, R.T.; Saed, G.M. Specific point mutations in key redox enzymes are associated with chemoresistance in epithelial ovarian cancer. Free Radic. Biol. Med. 2017, 102, 122–132. [Google Scholar] [CrossRef]
- Zazueta, C.; Jimenez-Uribe, A.P.; Pedraza-Chaverri, J.; Buelna-Chontal, M. Genetic Variations on Redox Control in Cardiometabolic Diseases: The Role of Nrf2. Antioxidants 2022, 11, 507. [Google Scholar] [CrossRef]
- Durani, L.W.; Jaafar, F.; Tan, J.K.; Tajul Arifin, K.; Mohd Yusof, Y.A.; Wan Ngah, W.Z.; Makpol, S. Targeting genes in insulin-associated signalling pathway, DNA damage, cell proliferation and cell differentiation pathways by tocotrienol-rich fraction in preventing cellular senescence of human diploid fibroblasts. Clin. Ter. 2015, 166, e365–e373. [Google Scholar] [CrossRef]
- Gerber, P.A.; Rutter, G.A. The Role of Oxidative Stress and Hypoxia in Pancreatic Beta-Cell Dysfunction in Diabetes Mellitus. Antioxid. Redox Signal. 2017, 26, 501–518. [Google Scholar] [CrossRef]
- Jakubczyk, K.; Dec, K.; Kałduńska, J.; Kawczuga, D.; Kochman, J.; Janda, K. Reactive oxygen species—Sources, functions, oxidative damage. Pol. Merkur. Lek. 2020, 48, 124–127. [Google Scholar]
- Madreiter-Sokolowski, C.T.; Thomas, C.; Ristow, M. Interrelation between ROS and Ca. Redox Biol. 2020, 36, 101678. [Google Scholar] [CrossRef]
- Voeikov, V.L. Reactive oxygen species (ROS): Pathogens or sources of vital energy? Part 2. Bioenergetic and bioinformational functions of ROS. J. Altern. Complement. Med. 2006, 12, 265–270. [Google Scholar] [CrossRef]
- Li, R.; Jia, Z.; Trush, M.A. Defining ROS in Biology and Medicine. React. Oxyg. Species 2016, 1, 9–21. [Google Scholar] [CrossRef]
- Yang, S.; Lian, G. ROS and diseases: Role in metabolism and energy supply. Mol. Cell. Biochem. 2020, 467, 1–12. [Google Scholar] [CrossRef]
- Sergi, D.; Williams, L.M. Potential relationship between dietary long-chain saturated fatty acids and hypothalamic dysfunction in obesity. Nutr. Rev. 2020, 78, 261–277. [Google Scholar] [CrossRef] [PubMed]
- Unger, R.H. Lipotoxic diseases. Annu. Rev. Med. 2002, 53, 319–336. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Liu, H.; Li, C. Dietary Regulation of Oxidative Stress in Chronic Metabolic Diseases. Foods 2021, 10, 1854. [Google Scholar] [CrossRef] [PubMed]
- Gantenbein, K.V.; Kanaka-Gantenbein, C. Mediterranean Diet as an Antioxidant: The Impact on Metabolic Health and Overall Wellbeing. Nutrients 2021, 13, 1951. [Google Scholar] [CrossRef]
- Sanz, J.M.; Sergi, D.; Colombari, S.; Capatti, E.; Situlin, R.; Biolo, G.; Di Girolamo, F.G.; Lazzer, S.; Šimunič, B.; Pišot, R.; et al. Dietary Acid Load but Not Mediterranean Diet Adherence Score is Associated With Metabolic and Cardiovascular Health State: A Population Observational Study From Northern Italy. Front. Nutr. 2022, 9, 828587. [Google Scholar] [CrossRef]
- Zanella, L.; Vianello, F. Potential of Microalgae as Functional Foods Applied to Mitochondria Protection and Healthy Aging Promotion. Nutraceuticals 2023, 3, 119–152. [Google Scholar] [CrossRef]
- Pagels, F.; Vasconcelos, V.; Guedes, A.C. Carotenoids from Cyanobacteria: Biotechnological Potential and Optimization Strategies. Biomolecules 2021, 11, 735. [Google Scholar] [CrossRef]
- Radyukina, N.L.; Mikheeva, L.E.; Karbysheva, E.A. Low Molecular Weight Antioxidants in Cyanobacteria and Plant Cells. Biol. Bull. Rev. 2019, 9, 520–531. [Google Scholar] [CrossRef]
- Walker, T.L.; Purton, S.; Becker, D.K.; Collet, C. Microalgae as bioreactors. Plant Cell Rep. 2005, 24, 629–641. [Google Scholar] [CrossRef]
- Dolganyuk, V.; Belova, D.; Babich, O.; Prosekov, A.; Ivanova, S.; Katserov, D.; Patyukov, N.; Sukhikh, S. Microalgae: A Promising Source of Valuable Bioproducts. Biomolecules 2020, 10, 1153. [Google Scholar] [CrossRef]
- McCarthy, M.I. Genomics, type 2 diabetes, and obesity. N. Engl. J. Med. 2010, 363, 2339–2350. [Google Scholar] [CrossRef]
- Morris, A.P.; Voight, B.F.; Teslovich, T.M.; Ferreira, T.; Segrè, A.V.; Steinthorsdottir, V.; Strawbridge, R.J.; Khan, H.; Grallert, H.; Mahajan, A.; et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012, 44, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.F.; Yeo, G.S.H. The genetics of obesity: From discovery to biology. Nat. Rev. Genet. 2022, 23, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Bays, H.E.; Bindlish, S.; Clayton, T.L. Obesity, diabetes mellitus, and cardiometabolic risk: An Obesity Medicine Association (OMA) Clinical Practice Statement (CPS) 2023. Obes. Pillars 2023, 5, 100056. [Google Scholar] [CrossRef]
- Povel, C.M.; Boer, J.M.; Onland-Moret, N.C.; Dollé, M.E.; Feskens, E.J.; van der Schouw, Y.T. Single nucleotide polymorphisms (SNPs) involved in insulin resistance, weight regulation, lipid metabolism and inflammation in relation to metabolic syndrome: An epidemiological study. Cardiovasc. Diabetol. 2012, 11, 133. [Google Scholar] [CrossRef]
- Liu, P.H.; Chang, Y.C.; Jiang, Y.D.; Chen, W.J.; Chang, T.J.; Kuo, S.S.; Lee, K.C.; Hsiao, P.C.; Chiu, K.C.; Chuang, L.M. Genetic variants of TCF7L2 are associated with insulin resistance and related metabolic phenotypes in Taiwanese adolescents and Caucasian young adults. J. Clin. Endocrinol. Metab. 2009, 94, 3575–3582. [Google Scholar] [CrossRef]
- Perumalsamy, S.; Wan Ahmad, W.A.; Zaman Huri, H. Single Nucleotide Polymorphism rs17173608 in the Chemerin Encoding Gene: Is It a Predictor of Insulin Resistance and Severity of Coronary Artery Disease in Non-Obese Type 2 Diabetes? Healthcare 2021, 9, 623. [Google Scholar] [CrossRef]
- Hancock, M.L.; Meyer, R.C.; Mistry, M.; Khetani, R.S.; Wagschal, A.; Shin, T.; Ho Sui, S.J.; Näär, A.M.; Flanagan, J.G. Insulin Receptor Associates with Promoters Genome-wide and Regulates Gene Expression. Cell 2019, 177, 722–736. [Google Scholar] [CrossRef]
- Nielsen, J.; Brandt, J.; Boesen, T.; Hummelshøj, T.; Slaaby, R.; Schluckebier, G.; Nissen, P. Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling. J. Mol. Biol. 2022, 434, 167458. [Google Scholar] [CrossRef]
- Copps, K.D.; White, M.F. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia 2012, 55, 2565–2582. [Google Scholar] [CrossRef]
- Guo, S. Insulin signaling, resistance, and the metabolic syndrome: Insights from mouse models into disease mechanisms. J. Endocrinol. 2014, 220, T1–T23. [Google Scholar] [CrossRef] [PubMed]
- Kearney, A.L.; Norris, D.M.; Ghomlaghi, M.; Kin Lok Wong, M.; Humphrey, S.J.; Carroll, L.; Yang, G.; Cooke, K.C.; Yang, P.; Geddes, T.A.; et al. Akt phosphorylates insulin receptor substrate to limit PI3K-mediated PIP3 synthesis. Elife 2021, 10, e66942. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Shi, X.; Sheng, K.; Han, G.; Li, W.; Zhao, Q.; Jiang, B.; Feng, J.; Li, J.; Gu, Y. PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (Review). Mol. Med. Rep. 2019, 19, 783–791. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef]
- Junaid, M.; Akter, Y.; Afrose, S.S.; Tania, M.; Khan, M.A. Biological Role of AKT and Regulation of AKT Signaling Pathway by Thymoquinone: Perspectives in Cancer Therapeutics. Mini Rev. Med. Chem. 2021, 21, 288–301. [Google Scholar] [CrossRef]
- Khalid, M.; Alkaabi, J.; Khan, M.A.B.; Adem, A. Insulin Signal Transduction Perturbations in Insulin Resistance. Int. J. Mol. Sci. 2021, 22, 8590. [Google Scholar] [CrossRef]
- McKeegan, K.; Mason, S.A.; Trewin, A.J.; Keske, M.A.; Wadley, G.D.; Della Gatta, P.A.; Nikolaidis, M.G.; Parker, L. Reactive oxygen species in exercise and insulin resistance: Working towards personalized antioxidant treatment. Redox Biol. 2021, 44, 102005. [Google Scholar] [CrossRef]
- Evans, J.L.; Maddux, B.A.; Goldfine, I.D. The molecular basis for oxidative stress-induced insulin resistance. Antioxid. Redox Signal. 2005, 7, 1040–1052. [Google Scholar] [CrossRef]
- Evans, J.L.; Goldfine, I.D.; Maddux, B.A.; Grodsky, G.M. Are oxidative stress-activated signaling pathways mediators of insulin resistance and beta-cell dysfunction? Diabetes 2003, 52, 1–8. [Google Scholar] [CrossRef]
- Gao, Z.; Hwang, D.; Bataille, F.; Lefevre, M.; York, D.; Quon, M.J.; Ye, J. Serine phosphorylation of insulin receptor substrate 1 by inhibitor kappa B kinase complex. J. Biol. Chem. 2002, 277, 48115–48121. [Google Scholar] [CrossRef] [PubMed]
- Gual, P.; Le Marchand-Brustel, Y.; Tanti, J.F. Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie 2005, 87, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, V.; Werner, E.D.; Giraud, J.; Lee, Y.H.; Shoelson, S.E.; White, M.F. Phosphorylation of Ser307 in insulin receptor substrate-1 blocks interactions with the insulin receptor and inhibits insulin action. J. Biol. Chem. 2002, 277, 1531–1537. [Google Scholar] [CrossRef] [PubMed]
- Gehi, B.R.; Gadhave, K.; Uversky, V.N.; Giri, R. Intrinsic disorder in proteins associated with oxidative stress-induced JNK signaling. Cell. Mol. Life Sci. 2022, 79, 202. [Google Scholar] [CrossRef] [PubMed]
- Lingappan, K. NF-κB in Oxidative Stress. Curr. Opin. Toxicol. 2018, 7, 81–86. [Google Scholar] [CrossRef]
- Kim, K.N.; Heo, S.J.; Yoon, W.J.; Kang, S.M.; Ahn, G.; Yi, T.H.; Jeon, Y.J. Fucoxanthin inhibits the inflammatory response by suppressing the activation of NF-κB and MAPKs in lipopolysaccharide-induced RAW 264.7 macrophages. Eur. J. Pharmacol. 2010, 649, 369–375. [Google Scholar] [CrossRef]
- Linnewiel-Hermoni, K.; Motro, Y.; Miller, Y.; Levy, J.; Sharoni, Y. Carotenoid derivatives inhibit nuclear factor kappa B activity in bone and cancer cells by targeting key thiol groups. Free. Radic. Biol. Med. 2014, 75, 105–120. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Valacchi, G.; Virgili, F.; Cervellati, C.; Pecorelli, A. OxInflammation: From Subclinical Condition to Pathological Biomarker. Front. Physiol. 2018, 9, 858. [Google Scholar] [CrossRef]
- Bloomgarden, Z.T. Inflammation and insulin resistance. Diabetes Care 2003, 26, 1619–1623. [Google Scholar] [CrossRef]
- Tong, Y.; Xu, S.; Huang, L.; Chen, C. Obesity and insulin resistance: Pathophysiology and treatment. Drug Discov. Today 2022, 27, 822–830. [Google Scholar] [CrossRef]
- Fakhri, S.; Abbaszadeh, F.; Moradi, S.Z.; Cao, H.; Khan, H.; Xiao, J. Effects of Polyphenols on Oxidative Stress, Inflammation, and Interconnected Pathways during Spinal Cord Injury. Oxidative Med. Cell. Longev. 2022, 2022, 8100195. [Google Scholar] [CrossRef] [PubMed]
- Gusti, A.M.T.; Qusti, S.Y.; Alshammari, E.M.; Toraih, E.A.; Fawzy, M.S. Antioxidants-Related Superoxide Dismutase (SOD), catalase (CAT), glutathione peroxidase (GPX), glutathione-S-transferase (GST), and nitric oxide synthase (NOS) gene variants analysis in an obese population: A preliminary case-control study. Antioxidants 2021, 10, 595. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wong, H.S. Are mitochondria the main contributor of reactive oxygen species in cells? J. Exp. Biol. 2021, 224, jeb221606. [Google Scholar] [CrossRef] [PubMed]
- Tirichen, H.; Yaigoub, H.; Xu, W.; Wu, C.; Li, R.; Li, Y. Mitochondrial Reactive Oxygen Species and Their Contribution in Chronic Kidney Disease Progression Through Oxidative Stress. Front. Physiol. 2021, 12, 627837. [Google Scholar] [CrossRef] [PubMed]
- Khodayari, S.; Salehi, Z.; Fakhrieh Asl, S.; Aminian, K.; Mirzaei Gisomi, N.; Torabi Dalivandan, S. Catalase gene C-262T polymorphism: Importance in ulcerative colitis. J. Gastroenterol. Hepatol. 2013, 28, 819–822. [Google Scholar] [CrossRef]
- Ahn, J.; Nowell, S.; McCann, S.E.; Yu, J.; Carter, L.; Lang, N.P.; Kadlubar, F.F.; Ratnasinghe, L.D.; Ambrosone, C.B. Associations between catalase phenotype and genotype: Modification by epidemiologic factors. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1217–1222. [Google Scholar] [CrossRef]
- Hebert-Schuster, M.; Fabre, E.E.; Nivet-Antoine, V. Catalase polymorphisms and metabolic diseases. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 397–402. [Google Scholar] [CrossRef]
- Dos Santos, K.G.; Canani, L.H.; Gross, J.L.; Tschiedel, B.; Souto, K.E.; Roisenberg, I. The catalase -262C/T promoter polymorphism and diabetic complications in Caucasians with type 2 diabetes. Dis. Markers 2006, 22, 355–359. [Google Scholar] [CrossRef]
- Church, S.L.; Grant, J.W.; Meese, E.U.; Trent, J.M. Sublocalization of the gene encoding manganese superoxide dismutase (MnSOD/SOD2) to 6q25 by fluorescence in situ hybridization and somatic cell hybrid mapping. Genomics 1992, 14, 823–825. [Google Scholar] [CrossRef]
- Siokas, V.; Stamati, P.; Pateraki, G.; Liampas, I.; Aloizou, A.M.; Tsirelis, D.; Nousia, A.; Sgantzos, M.; Nasios, G.; Bogdanos, D.P.; et al. Analysis of SOD2 rs4880 Genetic Variant in Patients with Alzheimer’s Disease. Curr. Issues Mol. Biol. 2022, 44, 4406–4414. [Google Scholar] [CrossRef] [PubMed]
- Sutton, A.; Imbert, A.; Igoudjil, A.; Descatoire, V.; Cazanave, S.; Pessayre, D.; Degoul, F. The manganese superoxide dismutase Ala16Val dimorphism modulates both mitochondrial import and mRNA stability. Pharmacogenet. Genom. 2005, 15, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Pourvali, K.; Abbasi, M.; Mottaghi, A. Role of Superoxide Dismutase 2 Gene Ala16Val Polymorphism and Total Antioxidant Capacity in Diabetes and its Complications. Avicenna J. Med. Biotechnol. 2016, 8, 48–56. [Google Scholar] [PubMed]
- Decharatchakul, N.; Settasatian, C.; Settasatian, N.; Komanasin, N.; Kukongviriyapan, U.; Intharaphet, P.; Senthong, V. Association of genetic polymorphisms in SOD2, SOD3, GPX3, and GSTT1 with hypertriglyceridemia and low HDL-C level in subjects with high risk of coronary artery disease. PeerJ 2019, 7, e7407. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, M.; Vats, P. Reactive metabolites and antioxidant gene polymorphisms in Type 2 diabetes mellitus. Redox Biol. 2014, 2, 170–177. [Google Scholar] [CrossRef]
- Nikic, P.; Dragicevic, D.; Jerotic, D.; Savic, S.; Djukic, T.; Stankovic, B.; Kovacevic, L.; Simic, T.; Matic, M. Polymorphisms of Antioxidant Enzymes SOD2 (rs4880) and GPX1 (rs1050450) Are Associated with Bladder Cancer Risk or its Aggressiveness. Medicina 2023, 59, 131. [Google Scholar] [CrossRef]
- Jerotic, D.; Ranin, J.; Bukumiric, Z.; Djukic, T.; Coric, V.; Savic-Radojevic, A.; Todorovic, N.; Asanin, M.; Ercegovac, M.; Milosevic, I.; et al. SOD2 rs4880 and GPX1 rs1050450 polymorphisms do not confer risk of COVID-19, but influence inflammation or coagulation parameters in Serbian cohort. Redox Rep. 2022, 27, 85–91. [Google Scholar] [CrossRef]
- Huang, J.Q.; Zhou, J.C.; Wu, Y.Y.; Ren, F.Z.; Lei, X.G. Role of glutathione peroxidase 1 in glucose and lipid metabolism-related diseases. Free Radic. Biol. Med. 2018, 127, 108–115. [Google Scholar] [CrossRef]
- Hu, X.; Xia, H.; Srivastava, S.K.; Pal, A.; Awasthi, Y.C.; Zimniak, P.; Singh, S.V. Catalytic efficiencies of allelic variants of human glutathione S-transferase P1-1 toward carcinogenic anti-diol epoxides of benzo[c]phenanthrene and benzo[g]chrysene. Cancer Res. 1998, 58, 5340–5343. [Google Scholar]
- Mandal, R.K.; Mittal, R.D. Glutathione S-Transferase P1 313 (A > G) Ile105Val Polymorphism Contributes to Cancer Susceptibility in Indian Population: A Meta-analysis of 39 Case-Control Studies. Indian J. Clin. Biochem. 2020, 35, 8–19. [Google Scholar] [CrossRef]
- Gong, J.Y.; Peng, S.Y.; Xing, K.; Fan, L.; Tan, S.L.; Luo, Z.Y.; Yuan, H.Y.; Xu, P.; Luo, J.Q. Evaluating the role of GSTP1 genetic polymorphism (rs1695, 313A>G) as a predictor in cyclophosphamide-induced toxicities. Medicine 2021, 100, e24423. [Google Scholar] [CrossRef] [PubMed]
- Gusti, A.M.T.; Qusti, S.Y.; Bahijri, S.M.; Toraih, E.A.; Bokhari, S.; Attallah, S.M.; Alzahrani, A.; Alshehri, W.M.A.; Alotaibi, H.; Fawzy, M.S. Glutathione S-Transferase (GSTT1 rs17856199) and Nitric Oxide Synthase (NOS2 rs2297518) Genotype Combination as Potential Oxidative Stress-Related Molecular Markers for Type 2 Diabetes Mellitus. Diabetes Metab. Syndr. Obes. 2021, 14, 1385–1403. [Google Scholar] [CrossRef] [PubMed]
- Masoodi, T.A.; Rao Talluri, V.; Shaik, N.A.; Al-Aama, J.Y.; Hasan, Q. Functional genomics based prioritization of potential nsSNPs in EPHX1, GSTT1, GSTM1 and GSTP1 genes for breast cancer susceptibility studies. Genomics 2012, 99, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Collazo, A.A.; Pérez-Méndez, O.; López-Olmos, V.; Delgado-Rizo, V.; Muñoz-Valle, J.F.; Martínez-López, E.; Villanueva-Quintero, D.G.; Domínguez-Díaz, C.; Fafutis-Morris, M.; Alvarado-Navarro, A. Association between rs662 (A > G) and rs854560 (A > T) polymorphisms in PON1 gene and the susceptibility for psoriasis in mestizo population of Western Mexico. Mol. Biol. Rep. 2021, 48, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Leviev, I.; Negro, F.; James, R.W. Two alleles of the human paraoxonase gene produce different amounts of mRNA. An explanation for differences in serum concentrations of paraoxonase associated with the (Leu-Met54) polymorphism. Arterioscler. Thromb. Vasc. Biol. 1997, 17, 2935–2939. [Google Scholar] [CrossRef]
- Brophy, V.H.; Jampsa, R.L.; Clendenning, J.B.; McKinstry, L.A.; Jarvik, G.P.; Furlong, C.E. Effects of 5′ regulatory-region polymorphisms on paraoxonase-gene (PON1) expression. Am. J. Hum. Genet. 2001, 68, 1428–1436. [Google Scholar] [CrossRef]
- Grzegorzewska, A.E.; Adamska, P.; Iwańczyk-Skalska, E.; Ostromecka, K.; Niepolski, L.; Marcinkowski, W.; Mostowska, A.; Warchoł, W.; Żaba, C.; Jagodziński, P.P. Paraoxonase 1 concerning dyslipidaemia, cardiovascular diseases, and mortality in haemodialysis patients. Sci. Rep. 2021, 11, 6773. [Google Scholar] [CrossRef]
- Li, W.F.; Pan, M.H.; Chung, M.C.; Ho, C.K.; Chuang, H.Y. Lead exposure is associated with decreased serum paraoxonase 1 (PON1) activity and genotypes. Environ. Health Perspect. 2006, 114, 1233–1236. [Google Scholar] [CrossRef]
- Gupta, N.; Singh, S.; Maturu, V.N.; Sharma, Y.P.; Gill, K.D. Paraoxonase 1 (PON1) polymorphisms, haplotypes and activity in predicting cad risk in North-West Indian Punjabis. PLoS ONE 2011, 6, e17805. [Google Scholar] [CrossRef]
- Alegría-Torres, J.A.; García-Domínguez, M.L.; Cruz, M.; Aradillas-García, C. Q192R polymorphism of paraoxonase 1 gene associated with insulin resistance in Mexican children. Arch. Med. Res. 2015, 46, 78–83. [Google Scholar] [CrossRef]
- Gomathi, P.; Iyer, A.C.; Murugan, P.S.; Sasikumar, S.; Raj, N.B.A.J.; Ganesan, D.; Nallaperumal, S.; Murugan, M.; Selvam, G.S. Association of paraoxonase-1 gene polymorphisms with insulin resistance in South Indian population. Gene 2018, 650, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Marzec, J.M.; Christie, J.D.; Reddy, S.P.; Jedlicka, A.E.; Vuong, H.; Lanken, P.N.; Aplenc, R.; Yamamoto, T.; Yamamoto, M.; Cho, H.Y.; et al. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J. 2007, 21, 2237–2246. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.M.; Maltagliati, A.J. Nrf2 at the heart of oxidative stress and cardiac protection. Physiol. Genom. 2018, 50, 77–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, H.; Liu, J.; Ouyang, Y.; Wang, D.; Bao, W.; Liu, L. Association between the NF-E2 Related Factor 2 Gene Polymorphism and Oxidative Stress, Anti-Oxidative Status, and Newly-Diagnosed Type 2 Diabetes Mellitus in a Chinese Population. Int. J. Mol. Sci. 2015, 16, 16483–16496. [Google Scholar] [CrossRef] [PubMed]
- Tao, T.T.; Lin, X.H.; Tang, S.J.; Gui, W.W.; Zhu, W.F.; Li, H. Association of genetic variants in the Sirt1 and Nrf2 genes with the risk of metabolic syndrome in a Chinese Han population. BMC Endocr. Disord. 2022, 22, 84. [Google Scholar] [CrossRef]
- Suzuki, T.; Shibata, T.; Takaya, K.; Shiraishi, K.; Kohno, T.; Kunitoh, H.; Tsuta, K.; Furuta, K.; Goto, K.; Hosoda, F.; et al. Regulatory nexus of synthesis and degradation deciphers cellular Nrf2 expression levels. Mol. Cell. Biol. 2013, 33, 2402–2412. [Google Scholar] [CrossRef]
- Shen, Y.; Li, D.; Tian, P.; Shen, K.; Zhu, J.; Feng, M.; Wan, C.; Yang, T.; Chen, L.; Wen, F. The catalase C-262T gene polymorphism and cancer risk: A systematic review and meta-analysis. Medicine 2015, 94, e679. [Google Scholar] [CrossRef]
- Hayes, J.D.; Pulford, D.J. The glutathione S-transferase supergene family: Regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 445–600. [Google Scholar] [CrossRef]
- Nebert, D.W.; Vasiliou, V. Analysis of the glutathione S-transferase (GST) gene family. Hum. Genom. 2004, 1, 460–464. [Google Scholar] [CrossRef]
- Yan, C.; Duan, L.; Fu, C.; Tian, C.; Zhang, B.; Shao, X.; Zhu, G. Association Between Glutathione S-Transferase (GST) Polymorphisms and Schizophrenia in a Chinese Han Population. Neuropsychiatr. Dis. Treat. 2020, 16, 479–487. [Google Scholar] [CrossRef]
- Elofey, S.G.H.; Shafik, N.F.; Radwan, N.H.; Mansour, O.M.; Allam, R.M.; Shouman, S.; AbdelGawad, I.A. Relation between GSTP1 polymorphism and oxidative stress in patients with hepatocellular carcinoma. J. Egypt. Natl. Canc. Inst. 2020, 32, 38. [Google Scholar] [CrossRef]
- Mbah Ntepe, L.J.; Habib, R.; Judith Laure, N.; Raza, S.; Nepovimova, E.; Kuca, K.; Batool, S.; Muhammad Nurulain, S. Oxidative Stress and Analysis of Selected SNPs of ACHE (rs 2571598), BCHE (rs 3495), CAT (rs 7943316), SIRT1 (rs 10823108), GSTP1 (rs 1695), and Gene GSTM1, GSTT1 in Chronic Organophosphates Exposed Groups from Cameroon and Pakistan. Int. J. Mol. Sci. 2020, 21, 6432. [Google Scholar] [CrossRef] [PubMed]
- Bid, H.K.; Konwar, R.; Saxena, M.; Chaudhari, P.; Agrawal, C.G.; Banerjee, M. Association of glutathione S-transferase (GSTM1, T1 and P1) gene polymorphisms with type 2 diabetes mellitus in north Indian population. J. Postgrad. Med. 2010, 56, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Amer, M.A.; Ghattas, M.H.; Abo-Elmatty, D.M.; Abou-El-Ela, S.H. Influence of glutathione S-transferase polymorphisms on type-2 diabetes mellitus risk. Genet. Mol. Res. 2011, 10, 3722–3730. [Google Scholar] [CrossRef] [PubMed]
- Hegele, R.A. Paraoxonase genes and disease. Ann. Med. 1999, 31, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Mackness, M.I.; Arrol, S.; Abbott, C.; Durrington, P.N. Protection of low-density lipoprotein against oxidative modification by high-density lipoprotein associated paraoxonase. Atherosclerosis 1993, 104, 129–135. [Google Scholar] [CrossRef]
- Shin, M.J.; Park, E.; Lee, J.H.; Chung, N. Relationship between insulin resistance and lipid peroxidation and antioxidant vitamins in hypercholesterolemic patients. Ann. Nutr. Metab. 2006, 50, 115–120. [Google Scholar] [CrossRef]
- Soran, H.; Schofield, J.D.; Durrington, P.N. Antioxidant properties of HDL. Front. Pharmacol. 2015, 6, 222. [Google Scholar] [CrossRef]
- Blair, M.F.; Kokabian, B.; Gude, V.G. Light and growth medium effect on Chlorella vulgaris biomass production. J. Environ. Chem. Eng. 2014, 2, 665–674. [Google Scholar] [CrossRef]
- Nzayisenga, J.C.; Eriksson, K.; Sellstedt, A. Mixotrophic and heterotrophic production of lipids and carbohydrates by a locally isolated microalga using wastewater as a growth medium. Bioresour. Technol. 2018, 257, 260–265. [Google Scholar] [CrossRef]
- Ahmad, A.; Hassan, S.W.; Banat, F. An overview of microalgae biomass as a sustainable aquaculture feed ingredient: Food security and circular economy. Bioengineered 2022, 13, 9521–9547. [Google Scholar] [CrossRef] [PubMed]
- Gendy, T.S.; El-Temtamy, S.A. Commercialization potential aspects of microalgae for biofuel production: An overview. Egypt. J. Pet. 2013, 22, 43–51. [Google Scholar] [CrossRef]
- Khan, M.I.; Shin, J.H.; Kim, J.D. The promising future of microalgae: Current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Fact. 2018, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Ferreira de Oliveira, A.P.; Bragotto, A.P.A. Microalgae-based products: Food and public health. Future Foods 2022, 6, 100157. [Google Scholar] [CrossRef]
- Ejike, C.E.C.C.; Collins, S.A.; Balasuriya, N.; Swanson, A.K.; Mason, B.; Udenigwe, C.C. Prospects of microalgae proteins in producing peptide-based functional foods for promoting cardiovascular health. Trends Food Sci. Technol. 2017, 59, 30–36. [Google Scholar] [CrossRef]
- Silva, S.C.; Ferreira, I.C.F.R.; Dias, M.M.; Barreiro, M.F. Microalgae-Derived Pigments: A 10-Year Bibliometric Review and Industry and Market Trend Analysis. Molecules 2020, 25, 3406. [Google Scholar] [CrossRef]
- Ramos-Romero, S.; Torrella, J.R.; Pagès, T.; Viscor, G.; Torres, J.L. Edible Microalgae and Their Bioactive Compounds in the Prevention and Treatment of Metabolic Alterations. Nutrients 2021, 13, 563. [Google Scholar] [CrossRef]
- Ampofo, J.; Abbey, L. Microalgae: Bioactive Composition, Health Benefits, Safety and Prospects as Potential High-Value Ingredients for the Functional Food Industry. Foods 2022, 11, 1744. [Google Scholar] [CrossRef]
- Tamel Selvan, K.; Goon, J.A.; Makpol, S.; Tan, J.K. Effects of Microalgae on Metabolic Syndrome. Antioxidants 2023, 12, 449. [Google Scholar] [CrossRef]
- Nawrocka, D.; Kornicka, K.; Śmieszek, A.; Marycz, K. Spirulina platensis Improves Mitochondrial Function Impaired by Elevated Oxidative Stress in Adipose-Derived Mesenchymal Stromal Cells (ASCs) and Intestinal Epithelial Cells (IECs), and Enhances Insulin Sensitivity in Equine Metabolic Syndrome (EMS) Horses. Mar. Drugs 2017, 15, 237. [Google Scholar] [CrossRef]
- Paterson, S.; Gómez-Cortés, P.; de la Fuente, M.A.; Hernández-Ledesma, B. Bioactivity and Digestibility of Microalgae. Nutrients 2023, 15, 477. [Google Scholar] [CrossRef] [PubMed]
- Simioni, C.; Zauli, G.; Martelli, A.M.; Vitale, M.; Sacchetti, G.; Gonelli, A.; Neri, L.M. Oxidative stress: Role of physical exercise and antioxidant nutraceuticals in adulthood and aging. Oncotarget 2018, 9, 17181–17198. [Google Scholar] [CrossRef] [PubMed]
- Böhm, V.; Lietz, G.; Olmedilla-Alonso, B.; Phelan, D.; Reboul, E.; Bánati, D.; Borel, P.; Corte-Real, J.; de Lera, A.R.; Desmarchelier, C.; et al. From carotenoid intake to carotenoid blood and tissue concentrations—Implications for dietary intake recommendations. Nutr. Rev. 2021, 79, 544–573. [Google Scholar] [CrossRef]
- Ren, Y.; Sun, H.; Deng, J.; Huang, J.; Chen, F. Carotenoid Production from Microalgae: Biosynthesis, Salinity Responses and Novel Biotechnologies. Mar. Drugs 2021, 19, 713. [Google Scholar] [CrossRef]
- Raposo, M.F.; de Morais, A.M.; de Morais, R.M. Carotenoids from Marine Microalgae: A Valuable Natural Source for the Prevention of Chronic Diseases. Mar. Drugs 2015, 13, 5128–5155. [Google Scholar] [CrossRef]
- Wang, J.; Hu, X.; Chen, J.; Wang, T.; Huang, X.; Chen, G. The Extraction of β-Carotene from Microalgae for Testing Their Health Benefits. Foods 2022, 11, 502. [Google Scholar] [CrossRef] [PubMed]
- Burri, B.J.; La Frano, M.R.; Zhu, C. Absorption, metabolism, and functions of β-cryptoxanthin. Nutr. Rev. 2016, 74, 69–82. [Google Scholar] [CrossRef]
- Miazek, K.; Beton, K.; Śliwińska, A.; Brożek-Płuska, B. The Effect of β-Carotene, Tocopherols and Ascorbic Acid as Anti-Oxidant Molecules on Human and Animal In Vitro/In Vivo Studies: A Review of Research Design and Analytical Techniques Used. Biomolecules 2022, 12, 1087. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, L.; Gao, Y.; Xie, Z.; Zhang, Y.; Pan, Z.; Tu, Y.; Wang, H.; Han, Q.; Hu, X.; et al. β-carotene provides neuro protection after experimental traumatic brain injury via the Nrf2-ARE pathway. J. Integr. Neurosci. 2019, 18, 153–161. [Google Scholar] [CrossRef]
- Metibemu, D.S.; Ogungbe, I.V. Carotenoids in Drug Discovery and Medicine: Pathways and Molecular Targets Implicated in Human Diseases. Molecules 2022, 27, 6005. [Google Scholar] [CrossRef]
- Ochoa Becerra, M.; Mojica Contreras, L.; Hsieh Lo, M.; Mateos Díaz, J.; Castillo Herrera, G. Lutein as a functional food ingredient: Stability and bioavailability. J. Funct. Foods 2020, 66, 103771. [Google Scholar] [CrossRef]
- Zheng, H.; Wang, Y.; Li, S.; Nagarajan, D.; Varjani, S.; Lee, D.-J.; Chang, J.-S. Recent advances in lutein production from microalgae. Renew. Sustain. Energy Rev. 2022, 153, 111795. [Google Scholar] [CrossRef]
- Frede, K.; Ebert, F.; Kipp, A.P.; Schwerdtle, T.; Baldermann, S. Lutein Activates the Transcription Factor Nrf2 in Human Retinal Pigment Epithelial Cells. J. Agric. Food Chem. 2017, 65, 5944–5952. [Google Scholar] [CrossRef] [PubMed]
- Régnier, P.; Bastias, J.; Rodriguez-Ruiz, V.; Caballero-Casero, N.; Caballo, C.; Sicilia, D.; Fuentes, A.; Maire, M.; Crepin, M.; Letourneur, D.; et al. Astaxanthin from Haematococcus pluvialis Prevents Oxidative Stress on Human Endothelial Cells without Toxicity. Mar. Drugs 2015, 13, 2857–2874. [Google Scholar] [CrossRef] [PubMed]
- Rad, N.R.; Movahedian, A.; Feizi, A.; Aminorroaya, A.; Aarabi, M.H. Antioxidant effects of astaxanthin and metformin combined therapy in type 2 diabetes mellitus patients: A randomized double-blind controlled clinical trial. Res. Pharm. Sci. 2022, 17, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Sahin, K.; Orhan, C.; Akdemir, F.; Tuzcu, M.; Sahin, N.; Yılmaz, I.; Juturu, V. β-Cryptoxanthin ameliorates metabolic risk factors by regulating NF-κB and Nrf2 pathways in insulin resistance induced by high-fat diet in rodents. Food Chem. Toxicol. 2017, 107, 270–279. [Google Scholar] [CrossRef]
- Bourdon, L.; Jensen, A.A.; Kavanagh, J.M.; McClure, D.D. Microalgal production of zeaxanthin. Algal Res. 2021, 55, 102266. [Google Scholar] [CrossRef]
- Zou, X.; Gao, J.; Zheng, Y.; Wang, X.; Chen, C.; Cao, K.; Xu, J.; Li, Y.; Lu, W.; Liu, J.; et al. Zeaxanthin induces Nrf2-mediated phase II enzymes in protection of cell death. Cell Death Dis. 2014, 5, e1218. [Google Scholar] [CrossRef]
- Pajot, A.; Hao Huynh, G.; Picot, L.; Marchal, L.; Nicolau, E. Fucoxanthin from Algae to Human, an Extraordinary Bioresource: Insights and Advances in up and Downstream Processes. Mar. Drugs 2022, 20, 222. [Google Scholar] [CrossRef]
- Khaw, Y.S.; Yusoff, F.M.; Tan, H.T.; Noor Mazli, N.A.I.; Nazarudin, M.F.; Shaharuddin, N.A.; Omar, A.R.; Takahashi, K. Fucoxanthin Production of Microalgae under Different Culture Factors: A Systematic Review. Mar. Drugs 2022, 20, 592. [Google Scholar] [CrossRef]
- Gammone, M.A.; D’Orazio, N. Anti-obesity activity of the marine carotenoid fucoxanthin. Mar. Drugs 2015, 13, 2196–2214. [Google Scholar] [CrossRef] [PubMed]
- Smaoui, S.; Barkallah, M.; Ben Hlima, H.; Fendri, I.; Mousavi Khaneghah, A.; Michaud, P.; Abdelkafi, S. Microalgae Xanthophylls: From Biosynthesis Pathway and Production Techniques to Encapsulation Development. Foods 2021, 10, 2835. [Google Scholar] [CrossRef] [PubMed]
- Brahma, D.; Dutta, D. Antioxidant property of beta-cryptoxanthin produced by Kocuria marina DAGII. Mater. Today Proc. 2022, 57, 1833–1837. [Google Scholar] [CrossRef]
- Ahmed, F.; Fanning, K.; Netzel, M.; Turner, W.; Li, Y.; Schenk, P.M. Profiling of carotenoids and antioxidant capacity of microalgae from subtropical coastal and brackish waters. Food Chem. 2014, 165, 300–306. [Google Scholar] [CrossRef]
- Kim, J.; Kim, M.; Lee, S.; Jin, E. Development of a Chlorella vulgaris mutant by chemical mutagenesis as a producer for natural violaxanthin. Algal Res. 2020, 46, 101790. [Google Scholar] [CrossRef]
- Ahmad, M.T.; Shariff, M.; Yusoff, F.M.; Goh, Y.M.; Banerjee, S. Applications of microalga Chlorella vulgaris in aquaculture. Rev. Aquac. 2020, 12, 328–346. [Google Scholar] [CrossRef]
- Grama, B.S.; Chader, S.; Khelifi, D.; Agathos, S.N.; Jeffryes, C. Induction of canthaxanthin production in a Dactylococcus microalga isolated from the Algerian Sahara. Bioresour. Technol. 2014, 151, 297–305. [Google Scholar] [CrossRef]
- Yusuf, N.; Athirah, N.M.; Suhaila, A. Antioxidative Responses of Chlorella vulgaris Under Different Growth Phases. Squalen Bull. Mar. Fish. Postharvest Biotechnol. 2022, 17, 111–120. [Google Scholar] [CrossRef]
- Gęgotek, A.; Jarocka-Karpowicz, I.; Skrzydlewska, E. Cytoprotective Effect of Ascorbic Acid and Rutin against Oxidative Changes in the Proteome of Skin Fibroblasts Cultured in a Three-Dimensional System. Nutrients 2020, 12, 1074. [Google Scholar] [CrossRef]
- Gęgotek, A.; Skrzydlewska, E. Antioxidative and Anti-Inflammatory Activity of Ascorbic Acid. Antioxidants 2022, 11, 1993. [Google Scholar] [CrossRef]
- Shi, L.; Du, X.; Guo, P.; Huang, L.; Qi, P.; Gong, Q. Ascorbic acid supplementation in type 2 diabetes mellitus: A protocol for systematic review and meta-analysis. Medicine 2020, 99, e23125. [Google Scholar] [CrossRef] [PubMed]
- Del Mondo, A.; Smerilli, A.; Sané, E.; Sansone, C.; Brunet, C. Challenging microalgal vitamins for human health. Microb. Cell Fact. 2020, 19, 201. [Google Scholar] [CrossRef] [PubMed]
- Quoc, Q.L.; Bich, T.C.T.; Kim, S.H.; Park, H.S.; Shin, Y.S. Administration of vitamin E attenuates airway inflammation through restoration of Nrf2 in a mouse model of asthma. J. Cell. Mol. Med. 2021, 25, 6721–6732. [Google Scholar] [CrossRef] [PubMed]
- Manning, P.J.; Sutherland, W.H.; Walker, R.J.; Williams, S.M.; De Jong, S.A.; Ryalls, A.R.; Berry, E.A. Effect of high-dose vitamin E on insulin resistance and associated parameters in overweight subjects. Diabetes Care 2004, 27, 2166–2171. [Google Scholar] [CrossRef] [PubMed]
- Paolisso, G.; Giugliano, D. Oxidative stress and insulin action: Is there a relationship? Diabetologia 1996, 39, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Bradford, A.; Atkinson, J.; Fuller, N.; Rand, R.P. The effect of vitamin E on the structure of membrane lipid assemblies. J. Lipid Res. 2003, 44, 1940–1945. [Google Scholar] [CrossRef]
- Lang, I.; Hodac, L.; Friedl, T.; Feussner, I. Fatty acid profiles and their distribution patterns in microalgae: A comprehensive analysis of more than 2000 strains from the SAG culture collection. BMC Plant Biol. 2011, 11, 124. [Google Scholar] [CrossRef]
- Conde, T.A.; Neves, B.F.; Couto, D.; Melo, T.; Neves, B.; Costa, M.; Silva, J.; Domingues, P.; Domingues, M.R. Microalgae as Sustainable Bio-Factories of Healthy Lipids: Evaluating Fatty Acid Content and Antioxidant Activity. Mar. Drugs 2021, 19, 357. [Google Scholar] [CrossRef]
- Djuric, Z.; Turgeon, D.K.; Sen, A.; Ren, J.; Herman, K.; Ramaswamy, D.; Zhao, L.; Ruffin, M.T.; Normolle, D.P.; Smith, W.L.; et al. The Anti-inflammatory Effect of Personalized Omega-3 Fatty Acid Dosing for Reducing Prostaglandin E. Cancer Prev. Res. 2017, 10, 729–737. [Google Scholar] [CrossRef]
- Rogero, M.M.; Calder, P.C. Obesity, Inflammation, Toll-Like Receptor 4 and Fatty Acids. Nutrients 2018, 10, 432. [Google Scholar] [CrossRef]
- Simonetto, M.; Infante, M.; Sacco, R.L.; Rundek, T.; Della-Morte, D. A Novel Anti-Inflammatory Role of Omega-3 PUFAs in Prevention and Treatment of Atherosclerosis and Vascular Cognitive Impairment and Dementia. Nutrients 2019, 11, 2279. [Google Scholar] [CrossRef] [PubMed]
- Meital, L.T.; Windsor, M.T.; Perissiou, M.; Schulze, K.; Magee, R.; Kuballa, A.; Golledge, J.; Bailey, T.G.; Askew, C.D.; Russell, F.D. Omega-3 fatty acids decrease oxidative stress and inflammation in macrophages from patients with small abdominal aortic aneurysm. Sci. Rep. 2019, 9, 12978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, S.; Mao, L.; Leak, R.K.; Shi, Y.; Zhang, W.; Hu, X.; Sun, B.; Cao, G.; Gao, Y.; et al. Omega-3 fatty acids protect the brain against ischemic injury by activating Nrf2 and upregulating heme oxygenase 1. J. Neurosci. 2014, 34, 1903–1915. [Google Scholar] [CrossRef] [PubMed]
- Sá, M.; Ferrer-Ledo, N.; Wijffels, R.; Crespo, J.G.; Barbosa, M.; Galinha, C.F. Monitoring of eicosapentaenoic acid (EPA) production in the microalgae Nannochloropsis oceanica. Algal Res. 2020, 45, 101766. [Google Scholar] [CrossRef]
- Sivakumar, R.; Sachin, S.; Priyadarshini, R.; Ghosh, S. Sustainable production of eicosapentaenoic acid-rich oil from microalgae: Towards an algal biorefinery. J. Appl. Microbiol. 2022, 132, 4170–4185. [Google Scholar] [CrossRef]
- Sahin, D.; Tas, E.; Altindag, U.H. Enhancement of docosahexaenoic acid (DHA) production from Schizochytrium sp. S31 using different growth medium conditions. AMB Express 2018, 8, 7. [Google Scholar] [CrossRef]
- Blasio, M.; Balzano, S. Fatty Acids Derivatives from Eukaryotic Microalgae, Pathways and Potential Applications. Front. Microbiol. 2021, 12, 718933. [Google Scholar] [CrossRef]
- Quispe, R.; Alfaddagh, A.; Kazzi, B.; Zghyer, F.; Marvel, F.A.; Blumenthal, R.S.; Sharma, G.; Martin, S.S. Controversies in the Use of Omega-3 Fatty Acids to Prevent Atherosclerosis. Curr. Atheroscler. Rep. 2022, 24, 571–581. [Google Scholar] [CrossRef]
- Goiris, K.; Muylaert, K.; Voorspoels, S.; Noten, B.; De Paepe, D.; Baart, G.J.E.; De Cooman, L. Detection of flavonoids in microalgae from different evolutionary lineages. J. Phycol. 2014, 50, 483–492. [Google Scholar] [CrossRef]
- Fukumoto, L.R.; Mazza, G. Assessing antioxidant and prooxidant activities of phenolic compounds. J. Agric. Food Chem. 2000, 48, 3597–3604. [Google Scholar] [CrossRef]
- Martinez-Gomez, A.; Caballero, I.; Blanco, C.A. Phenols and Melanoidins as Natural Antioxidants in Beer. Structure, Reactivity and Antioxidant Activity. Biomolecules 2020, 10, 400. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Liu, M.; Ding, Y.; Yao, M.; Zhu, Y.; Zhao, J.; Cheng, L.; Bai, J.; Wang, F.; Cao, J.; et al. A phenolic amide (LyA) isolated from the fruits of. Aging 2019, 11, 12361–12374. [Google Scholar] [CrossRef] [PubMed]
- Karunaweera, N.; Raju, R.; Gyengesi, E.; Münch, G. Plant polyphenols as inhibitors of NF-κB induced cytokine production-a potential anti-inflammatory treatment for Alzheimer’s disease? Front. Mol. Neurosci. 2015, 8, 24. [Google Scholar] [CrossRef]
- Zhou, L.; Li, K.; Duan, X.; Hill, D.; Barrow, C.; Dunshea, F.; Martin, G.; Suleria, H. Bioactive compounds in microalgae and their potential health benefits. Food Biosci. 2022, 49, 101932. [Google Scholar] [CrossRef]
- Guo, X.D.; Zhang, D.Y.; Gao, X.J.; Parry, J.; Liu, K.; Liu, B.L.; Wang, M. Quercetin and quercetin-3-O-glucuronide are equally effective in ameliorating endothelial insulin resistance through inhibition of reactive oxygen species-associated inflammation. Mol. Nutr. Food Res. 2013, 57, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Tsao, J.P.; Bernard, J.R.; Hsu, H.C.; Hsu, C.L.; Liao, S.F.; Cheng, I.S. Short-Term Oral Quercetin Supplementation Improves Post-Exercise Insulin Sensitivity, Antioxidant Capacity and Enhances Subsequent Cycling Time to Exhaustion in Healthy Adults: A Pilot Study. Front. Nutr. 2022, 9, 875319. [Google Scholar] [CrossRef] [PubMed]
- Moratilla-Rivera, I.; Sánchez, M.; Valdés-González, J.A.; Gómez-Serranillos, M.P. Natural Products as Modulators of Nrf2 Signaling Pathway in Neuroprotection. Int. J. Mol. Sci. 2023, 24, 3748. [Google Scholar] [CrossRef]
- Dhanya, R. Quercetin for managing type 2 diabetes and its complications, an insight into multitarget therapy. Biomed. Pharmacother. 2022, 146, 112560. [Google Scholar] [CrossRef]
- Haoujar, I.; Cacciola, F.; Abrini, J.; Mangraviti, D.; Giuffrida, D.; Oulad El Majdoub, Y.; Kounnoun, A.; Miceli, N.; Fernanda Taviano, M.; Mondello, L.; et al. The Contribution of Carotenoids, Phenolic Compounds, and Flavonoids to the Antioxidative Properties of Marine Microalgae Isolated from Mediterranean Morocco. Molecules 2019, 24, 4037. [Google Scholar] [CrossRef]
- Pyne, S.; Paria, K. Optimization of extraction process parameters of caffeic acid from microalgae by supercritical carbon dioxide green technology. BMC Chem. 2022, 16, 31. [Google Scholar] [CrossRef]
- Khan, F.A.; Maalik, A.; Murtaza, G. Inhibitory mechanism against oxidative stress of caffeic acid. J. Food Drug Anal. 2016, 24, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Shahin, N.N.; Shamma, R.N.; Ahmed, I.S. A Nano-Liposomal Formulation of Caffeic Acid Phenethyl Ester Modulates Nrf2 and NF-κβ Signaling and Alleviates Experimentally Induced Acute Pancreatitis in a Rat Model. Antioxidants 2022, 11, 1536. [Google Scholar] [CrossRef]
- Choudhary, S.; Mourya, A.; Ahuja, S.; Sah, S.P.; Kumar, A. Plausible anti-inflammatory mechanism of resveratrol and caffeic acid against chronic stress-induced insulin resistance in mice. Inflammopharmacology 2016, 24, 347–361. [Google Scholar] [CrossRef]
- Liu, J.; Sun, Z.; Gerken, H.; Liu, Z.; Jiang, Y.; Chen, F. Chlorella zofingiensis as an alternative microalgal producer of astaxanthin: Biology and industrial potential. Mar. Drugs 2014, 12, 3487–3515. [Google Scholar] [CrossRef] [PubMed]
- Murillo, A.G.; Hu, S.; Fernandez, M.L. Zeaxanthin: Metabolism, Properties, and Antioxidant Protection of Eyes, Heart, Liver, and Skin. Antioxidants 2019, 8, 390. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, B.A.; Farrona, S.; Ventura, M.R.; Abranches, R. Canthaxanthin, a Red-Hot Carotenoid: Applications, Synthesis, and Biosynthetic Evolution. Plants 2020, 9, 1039. [Google Scholar] [CrossRef]
- Dose, J.; Matsugo, S.; Yokokawa, H.; Koshida, Y.; Okazaki, S.; Seidel, U.; Eggersdorfer, M.; Rimbach, G.; Esatbeyoglu, T. Free Radical Scavenging and Cellular Antioxidant Properties of Astaxanthin. Int. J. Mol. Sci. 2016, 17, 103. [Google Scholar] [CrossRef]
- Deshmukh, P.; Unni, S.; Krishnappa, G.; Padmanabhan, B. The Keap1-Nrf2 pathway: Promising therapeutic target to counteract ROS-mediated damage in cancers and neurodegenerative diseases. Biophys. Rev. 2017, 9, 41–56. [Google Scholar] [CrossRef]
- Taira, J.; Sonamoto, M.; Uehara, M. Dual Biological Functions of a Cytoprotective Effect and Apoptosis Induction by Bioavailable Marine Carotenoid Fucoxanthinol through Modulation of the Nrf2 Activation in RAW264.7 Macrophage Cells. Mar. Drugs 2017, 15, 305. [Google Scholar] [CrossRef]
- Bohn, T.; Bonet, M.L.; Borel, P.; Keijer, J.; Landrier, J.F.; Milisav, I.; Ribot, J.; Riso, P.; Winklhofer-Roob, B.; Sharoni, Y.; et al. Mechanistic aspects of carotenoid health benefits—Where are we now? Nutr. Res. Rev. 2021, 34, 276–302. [Google Scholar] [CrossRef]
- Chauhan, A.; Islam, A.U.; Prakash, H.; Singh, S. Phytochemicals targeting NF-κB signaling: Potential anti-cancer interventions. J. Pharm. Anal. 2022, 12, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Hong, P.; Zheng, X. β-carotene attenuates lipopolysaccharide-induced inflammation via inhibition of the NF-κB, JAK2/STAT3 and JNK/p38 MAPK signaling pathways in macrophages. Anim. Sci. J. 2019, 90, 140–148. [Google Scholar] [CrossRef]
- Starska-Kowarska, K. Dietary Carotenoids in Head and Neck Cancer-Molecular and Clinical Implications. Nutrients 2022, 14, 531. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, M.; Nakamura, M.; Ogawa, K.; Ikoma, Y.; Yano, M. High-serum carotenoids associated with lower risk for developing type 2 diabetes among Japanese subjects: Mikkabi cohort study. BMJ Open Diabetes Res. Care 2015, 3, e000147. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Nagashimada, M.; Zhuge, F.; Zhan, L.; Nagata, N.; Tsutsui, A.; Nakanuma, Y.; Kaneko, S.; Ota, T. Astaxanthin prevents and reverses diet-induced insulin resistance and steatohepatitis in mice: A comparison with vitamin E. Sci. Rep. 2015, 5, 17192. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, Y.; Obana, A.; Muto, S.; Asaoka, R.; Tanito, M.; Ermakov, I.V.; Bernstein, P.S.; Gellermann, W. Relationships between Skin Carotenoid Levels and Metabolic Syndrome. Antioxidants 2021, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Lepretti, M.; Martucciello, S.; Burgos Aceves, M.A.; Putti, R.; Lionetti, L. Omega-3 Fatty Acids and Insulin Resistance: Focus on the Regulation of Mitochondria and Endoplasmic Reticulum Stress. Nutrients 2018, 10, 350. [Google Scholar] [CrossRef]
- Abbott, K.A.; Burrows, T.L.; Acharya, S.; Thota, R.N.; Garg, M.L. DHA-enriched fish oil reduces insulin resistance in overweight and obese adults. Prostaglandins Leukot. Essent. Fat. Acids 2020, 159, 102154. [Google Scholar] [CrossRef]
- Brown, T.J.; Brainard, J.; Song, F.; Wang, X.; Abdelhamid, A.; Hooper, L.; Group, P. Omega-3, omega-6, and total dietary polyunsaturated fat for prevention and treatment of type 2 diabetes mellitus: Systematic review and meta-analysis of randomised controlled trials. BMJ 2019, 366, l4697. [Google Scholar] [CrossRef]
- Safafar, H.; van Wagenen, J.; Møller, P.; Jacobsen, C. Carotenoids, Phenolic Compounds and Tocopherols Contribute to the Antioxidative Properties of Some Microalgae Species Grown on Industrial Wastewater. Mar. Drugs 2015, 13, 7339–7356. [Google Scholar] [CrossRef]
- Murakami, S.; Hirazawa, C.; Mizutani, T.; Yoshikawa, R.; Ohya, T.; Ma, N.; Owaki, Y.; Owaki, T.; Ito, T.; Matsuzaki, C. The anti-obesity and anti-diabetic effects of the edible seaweed. Food Sci. Nutr. 2023, 11, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Ayora, H.; Pérez-Jiménez, J. 10—Antioxidant capacity of seaweeds: In vitro and in vivo assessment. In Marine Phenolic Compounds; Pérez-Correa, J.R., Mateos, R., Domínguez, H., Eds.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 299–341. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).