Identification of Putative Molecules for Adiponectin and Adiponectin Receptor and Their Roles in Learning and Memory in Lymnaea stagnalis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Snails
2.2. Definition of Nutritional Status
2.3. Identification of LymAdipo and LymAdipoR
2.4. In Situ Hybridization
2.5. Real-Time PCR
2.6. Measurement of the Hemolymph Glucose Concentration
2.7. Operant Conditioning of Escape Behavior
2.8. Statistics
3. Results
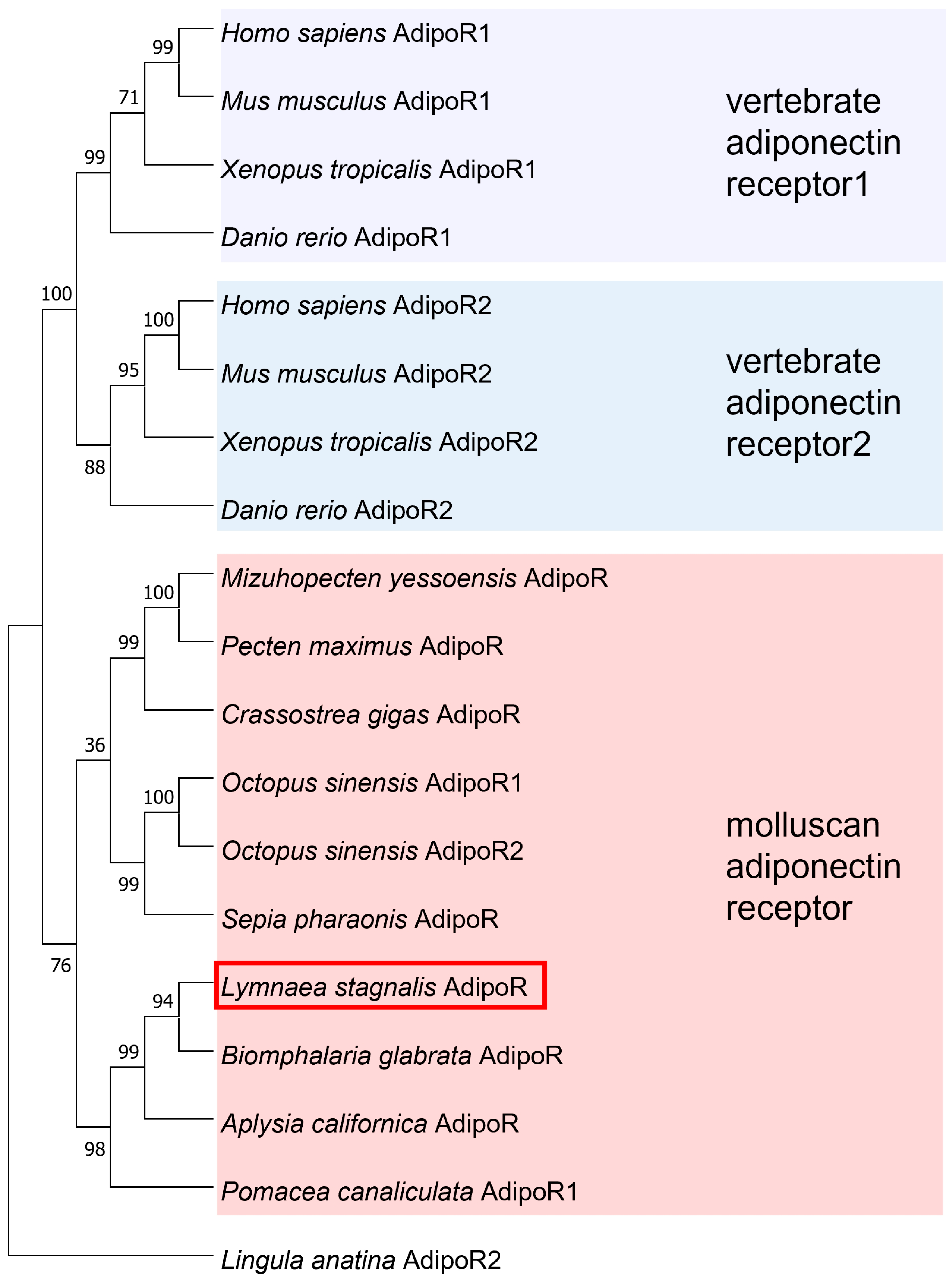
3.1. Identification of Putative Molecules of Adiponectin and Its Receptor in Lymnaea
3.2. Localization of LymAdipo and LymAdipoR in the Lymnaea CNS
3.3. Glucose Concentrations in the Hemolymph and Changes in the LymAdopo and LymAdipoR mRNA Expression Levels under Different Nutritional Conditions
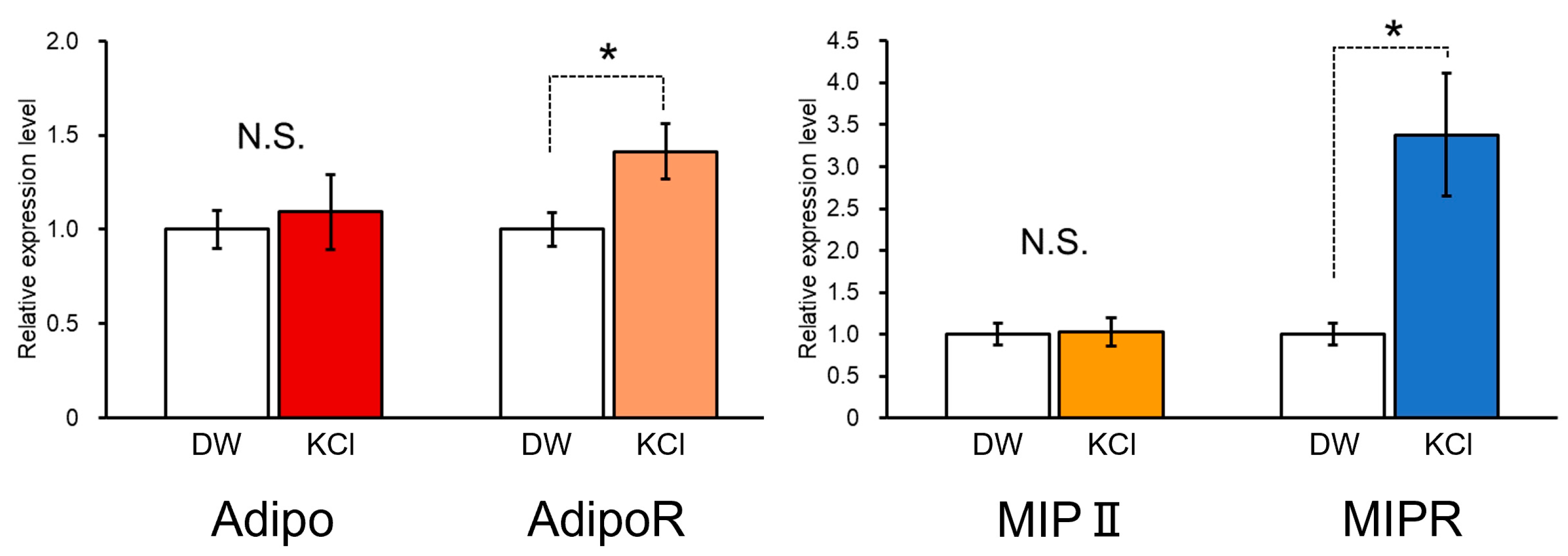
3.4. Establishment of Escape Behavior by Operant Conditioning and Change in the Expression Levels of LymAdipo and LymAdipoR during Memory Formation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Álvarez, B.; Koene, J.M.; Hollis, K.L.; Loy, I. Learning to anticipate mate presence shapes individual sex roles in the hermaphroditic pond snail, Lymnaea stagnalis. Anim. Cogn. 2022, 25, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, D.; Chikamoto, N.; Fujimoto, K.; Kitahashi, T.; Ito, E. Comparison between relative and absolute quantitative real-time PCR applied to single-cell analyses: Transcriptional levels in a key neuron for long-term memory in the pond snail. PLoS ONE 2022, 17, e0279017. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, D.; Sunada, H.; Totani, Y.; Watanabe, T.; Felletár, I.; Fitchett, A.; Eravci, M.; Anagnostopoulou, A.; Miki, R.; Okada, A.; et al. Molecular and functional characterization of an evolutionarily conserved CREB-binding protein in the Lymnaea CNS. FASEB J. 2022, 36, e22593. [Google Scholar] [CrossRef] [PubMed]
- Kemenes, G.; Benjamin, P.R.; Kemenes, I. The role of non-coding RNAs in the formation of long-term associative memory after single-trial learning in Lymnaea. Front. Behav. Neurosci. 2022, 16, 1005867. [Google Scholar] [CrossRef] [PubMed]
- Wingrove, J.; de Hoog, E.; Spencer, G.E. Disruptions in network plasticity precede deficits in memory following inhibition of retinoid signaling. J. Neurophysiol. 2023, 129, 41–55. [Google Scholar] [CrossRef]
- Komatsuzaki, Y.; Lukowiak, K. Epicatechin alters the activity of a neuron necessary for long-term memory of aerial respiratory behavior in Lymnaea stagnalis. Zoo. Sci. 2022, 39, 365–373. [Google Scholar] [CrossRef]
- Pirger, Z.; Naskar, S.; László, Z.; Kemenes, G.; Reglődi, D.; Kemenes, I. Reversal of age-related learning deficiency by the vertebrate PACAP and IGF-1 in a novel invertebrate model of aging: The pond snail (Lymnaea stagnalis). J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69, 1331–1338. [Google Scholar] [CrossRef]
- Hussein, A.A.A.; Baz, E.S.; Mariën, J.; Tadros, M.M.; El-Shenawy, N.S.; Koene, J.M. Effect of photoperiod and light intensity on learning ability and memory formation of the pond snail Lymnaea stagnalis. Invert. Neurosci. 2020, 20, 18. [Google Scholar] [CrossRef]
- Nakai, J.; Chikamoto, N.; Fujimoto, K.; Totani, Y.; Hatakeyama, D.; Dyakonova, V.E.; Ito, E. Insulin and memory in invertebrates. Front. Behav. Neurosci. 2022, 16, 882932. [Google Scholar] [CrossRef]
- Rivi, V.; Benatti, C.; Actis, P.; Tascedda, F.; Blom, J.M.C. Behavioral and transcriptional effects of short or prolonged fasting on the memory performances of Lymnaea stagnalis. Neuroendocrinology, 2023, in press. [CrossRef]
- Azami, S.; Wagatsuma, A.; Sadamoto, H.; Hatakeyama, D.; Usami, T.; Fujie, M.; Koyanagi, R.; Azumi, K.; Fujito, Y.; Lukowiak, K.; et al. Altered gene activity correlated with long-term memory formation of conditioned taste aversion in Lymnaea. J. Neurosci. Res. 2006, 84, 1610–1620. [Google Scholar] [CrossRef] [PubMed]
- Murakami, J.; Okada, R.; Sadamoto, H.; Kobayashi, S.; Mita, K.; Sakamoto, Y.; Yamagishi, M.; Hatakeyama, D.; Otsuka, E.; Okuta, A.; et al. Involvement of insulin-like peptide in long-term synaptic plasticity and long-term memory of the pond snail Lymnaea stagnalis. J. Neurosci. 2013, 33, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Mita, K.; Yamagishi, M.; Fujito, Y.; Lukowiak, K.; Ito, E. An increase in insulin is important for the acquisition conditioned taste aversion in Lymnaea. Neurobiol. Learn. Mem. 2014, 116, 132–138. [Google Scholar] [CrossRef]
- Fruebis, J.; Tsaom, T.S.; Javorschi, S.; Ebbets-Reed, D.; Erickson, M.R.; Yen, F.T.; Bihain, B.E.; Lodish, H.F. Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc. Natl. Acad. Sci. USA 2001, 98, 2005–2010. [Google Scholar] [CrossRef]
- Yamauchi, T.; Kamon, J.; Waki, H.; Terauchi, Y.; Kubota, N.; Hara, K.; Mori, Y.; Ide, T.; Murakami, K.; Tsuboyama-Kasaoka, N.; et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 2001, 7, 941–946. [Google Scholar] [CrossRef]
- Kadowaki, T.; Yamauchi, T.; Kubota, N.; Hara, K.; Ueki, K.; Tobe, K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J. Clin. Invest. 2006, 116, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Scherer, P.E. Adipokines as novel biomarkers and regulators of the metabolic syndrome. Ann. N. Y. Acad. Sci. 2010, 1212, E1–E19, Erratum in: Ann. N. Y. Acad. Sci. 2011, 1226, 50. [Google Scholar] [CrossRef]
- Yamakado, S.; Cho, H.; Inada, M.; Morikawa, M.; Jiang, Y.H.; Saito, K.; Nakaishi, K.; Watabe, S.; Takagi, H.; Kaneda, M.; et al. Urinary adiponectin as a new diagnostic index for chronic kidney disease due to diabetic nephropathy. BMJ Open Diabetes Res. Care. 2019, 7, e000661. [Google Scholar] [CrossRef]
- Kishore, U.; Gaboriaud, C.; Waters, P.; Shrive, A.K.; Greenhough, T.J.; Reid, K.B.; Sim, R.B.; Arlaud, G.J. C1q and tumor necrosis factor superfamily: Modularity and versatility. Trends. Immunol. 2004, 25, 551–561. [Google Scholar] [CrossRef]
- Yamauchi, T.; Kamon, J.; Ito, Y.; Tsuchida, A.; Yokomizo, T.; Kita, S.; Sugiyama, T.; Miyagishi, M.; Hara, K.; Tsunoda, M.; et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 2003, 423, 762–769, Erratum in: Nature 2004, 431, 1123. [Google Scholar] [CrossRef]
- Hug, C.; Wang, J.; Ahmad, N.S.; Bogan, J.S.; Tsao, T.S.; Lodish, H.F. T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc. Natl. Acad. Sci. USA 2004, 101, 10308–10313. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, H.; Fujii, Y.; Okada-Iwabu, M.; Iwabu, M.; Nakamura, Y.; Hosaka, T.; Motoyama, K.; Ikeda, M.; Wakiyama, M.; Terada, T.; et al. Crystal structures of the human adiponectin receptors. Nature 2015, 520, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Huang, S.; Liu, S.; Sun, J.; Liu, Z.; Yang, W.; Wang, L.; Song, L. A novel Adiponectin receptor (AdipoR) involved in regulating cytokines production and apoptosis of haemocytes in oyster Crassostrea gigas. Dev. Comp. Immunol. 2020, 110, 103727. [Google Scholar] [CrossRef]
- Gerdol, M.; Greco, S.; Pallavicini, A. Extensive tandem duplication events drive the expansion of the C1q-domain-containing gene family in bivalves. Mar. Drugs 2019, 17, 583. [Google Scholar] [CrossRef] [PubMed]
- Grinchenko, A.V.; von Kriegsheim, A.; Shved, N.A.; Egorova, A.E.; Ilyaskina, D.V.; Karp, T.D.; Goncharov, N.V.; Petrova, I.Y.; Kumeiko, V.V. A novel C1q domain-containing protein isolated from the mollusk Modiolus kurilensis recognizing glycans enriched with acidic galactans and mannans. Mar. Drugs 2021, 19, 668. [Google Scholar] [CrossRef]
- Garcia, A.; Estêvão, J.; Costas, B.; Cruz, A.; Fernández-Boo, S. Evaluation of the Ruditapes decussatus immune response after differential injected doses of Perkinsus olseni. J. Invertebr. Pathol. 2022, 195, 107849. [Google Scholar] [CrossRef]
- Buechler, C.; Wanninger, J.; Neumeier, M. Adiponectin receptor binding proteins—Recent advances in elucidating adiponectin signalling pathways. FEBS Lett. 2010, 584, 4280–4286. [Google Scholar] [CrossRef]
- Galvan, M.D.; Hulsebus, H.; Heitker, T.; Zeng, E.; Bohlson, S.S. Complement protein C1q and adiponectin stimulate Mer tyrosine kinase-dependent engulfment of apoptotic cells through a shared pathway. J. Innate. Immun. 2014, 6, 780–792. [Google Scholar] [CrossRef]
- Mazrooie, R.; Rohampour, K.; Zamani, M.; Hosseinmardi, N.; Zeraati, M. Intracerebroventricular administration of adiponectin attenuates streptozotocin-induced memory impairment in rats. Physiol. Int. 2017, 104, 150–157. [Google Scholar] [CrossRef]
- Pousti, F.; Ahmadi, R.; Mirahmadi, F.; Hosseinmardi, N.; Rohampour, K. Adiponectin modulates synaptic plasticity in hippocampal dentate gyrus. Neurosci. Lett. 2018, 662, 227–232. [Google Scholar] [CrossRef]
- Ng, R.C.; Cheng, O.Y.; Jian, M.; Kwan, J.S.; Ho, P.W.; Cheng, K.K.; Yeung, P.K.; Zhou, L.L.; Hoo, R.L.; Chung, S.K.; et al. Chronic adiponectin deficiency leads to Alzheimer’s disease-like cognitive impairments and pathologies through AMPK inactivation and cerebral insulin resistance in aged mice. Mol. Neurodegener. 2016, 11, 71. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.W.; Abid, N.B.; Jo, M.H.; Jo, M.G.; Yoon, G.H.; Kim, M.O. Suppression of adiponectin receptor 1 promotes memory dysfunction and Alzheimer’s disease-like pathologies. Sci. Rep. 2017, 7, 12435. [Google Scholar] [CrossRef]
- Ma, L.; Wang, R.; Dong, W.; Zhao, Z. Caloric restriction can improve learning and memory in C57/BL mice probably via regulation of the AMPK signaling pathway. Exp. Gerontol. 2018, 102, 28–35. [Google Scholar] [CrossRef]
- Kobayashi, S.; Kojima, S.; Yamanaka, M.; Sadamoto, H.; Nakamura, H.; Fujito, Y.; Kawai, R.; Sakakibara, M.; Ito, E. Operant conditioning of escape behavior in the pond snail, Lymnaea stagnalis. Zool. Sci. 1998, 15, 683–690. [Google Scholar] [CrossRef]
- Benatti, C.; Rivi, V.; Colliva, C.; Radighieri, G.; Tascedda, F.; Blom, J.M.C. Redefining operant conditioning of escape behaviour in Lymnaea stagnalis. ISJ 2020, 17, 129–137. [Google Scholar] [CrossRef]
- Totani, Y.; Nakai, J.; Hatakeyama, D.; Dyakonova, V.E.; Lukowiak, K.; Ito, E. CNS serotonin content mediating food deprivation-enhanced learning is regulated by hemolymph tryptophan concentration and autophagic flux in the pond snail. Nutr. Neurosci. 2023, 26, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Sadamoto, H.; Takahashi, H.; Okada, T.; Kenmoku, H.; Toyota, M.; Asakawa, Y. De novo sequencing and transcriptome analysis of the central nervous system of mollusc Lymnaea stagnalis by deep RNA sequencing. PLoS ONE 2012, 7, e42546. [Google Scholar] [CrossRef]
- Hatakeyama, D.; Fujito, Y.; Sakakibara, M.; Ito, E. Expression and distribution of transcription factor CCAAT/enhancer-binding protein in the central nervous system of Lymnaea stagnalis. Cell. Tissue Res. 2004, 318, 631–641. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Sadamoto, H.; Sato, H.; Kobayashi, S.; Murakami, J.; Aonuma, H.; Ando, H.; Fujito, Y.; Hamano, K.; Awaji, M.; Lukowiak, K.; et al. CREB in the pond snail Lymnaea stagnalis: Cloning, gene expression, and function in identifiable neurons of the central nervous system. J. Neurobiol. 2004, 58, 455–466. [Google Scholar] [CrossRef]
- Kubota, N.; Yano, W.; Kubota, T.; Yamauchi, T.; Itoh, S.; Kumagai, H.; Kozono, H.; Takamoto, I.; Okamoto, S.; Shiuchi, T.; et al. Adiponectin stimulates AMP-activated protein kinase in the hypothalamus and increases food intake. Cell. Metab. 2007, 6, 55–68. [Google Scholar] [CrossRef]
- Cowley, M.A.; Smart, J.L.; Rubinstein, M.; Cerdán, M.G.; Diano, S.; Horvath, T.L.; Cone, R.D.; Low, M.J. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 2001, 411, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Arquier, N.; Bjordal, M.; Hammann, P.; Kuhn, L.; Léopold, P. Brain adiponectin signaling controls peripheral insulin response in Drosophila. Nat. Commun. 2021, 12, 5633. [Google Scholar] [CrossRef]
- Hu, E.; Liang, P.; Spiegelman, B.M. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J. Biol. Chem. 1996, 271, 10697–10703. [Google Scholar] [CrossRef]
- Stephenson, R.; Lewis, V. Behavioural evidence for a sleep-like quiescent state in a pulmonate mollusc, Lymnaea stagnalis (Linnaeus). J. Exp. Biol. 2011, 214, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, G.P.; Benjamin, P.R. The whole-body withdrawal response of Lymnaea stagnalis. I. Identification of central motoneurones and muscles. J. Exp. Biol. 1991, 158, 63–95. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.I.; Winlow, W. Coordination of locomotor and cardiorespiratory networks of Lymnaea stagnalis by a pair of identified interneurones. J. Exp. Biol. 1991, 158, 37–62. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Takasaki, M.; Lukowiak, K.; Syed, N. Inhibition of the respiratory pattern-generating neurons by an identified whole-body withdrawal interneuron of Lymnaea stagnalis. J. Exp. Biol. 1996, 199, 1887–1898. [Google Scholar] [CrossRef] [PubMed]







| Protein | Accession Number |
|---|---|
| Homo sapiens Adipo | ABZ10942.1 |
| Mus musculus Adipo | NP_033735.3 |
| Xenopus tropicalis Adipo | XP_002938627.3 |
| Danio rerio Adipo | NP_001373470.1 |
| Biomphalaria glabrata C1qC | XP_013083362.1 |
| Homo sapiens C1qC | AAH09016.1 |
| Mus musculus C1qC | AAH43945.1 |
| Xenopus tropicalis C1qC | XP_031750749.1 |
| Danio rerio C1qC | AJP77512.1 |
| Homo sapiens C1qTNF7 | EAW92731.1 |
| Mus musculus C1qTNF7 | AAY21932.1 |
| Xenopus tropicalis C1qTNF7 | XP_017946491.1 |
| Danio rerio C1qTNF7 | XP_693031.2 |
| Homo sapiens C1qTNF1 | AAQ88790.1 |
| Mus musculus C1qTNF1 | AAY21926.1 |
| Xenopus laevis C1qTNF1 | XP_018094785.1 |
| Danio rerio C1qTNF1 | NP_001017875.1 |
| Pomacea canaliculata C1qB | XP_025098395.1 |
| Pomacea canaliculata C1qC | XP_025086549.1 |
| Mizuhopecten yessoensis C1qTNF2 | XP_021344498.1 |
| Pecten maximus C1qC | XP_033753106.1 |
| Pecten maximus C1qTNF2 | XP_033744519.1 |
| Octopus sinensis C1qTNF7 | XP_029636373.1 |
| Crassostrea gigas C1qC | XP_034310292.1 |
| Crassostrea gigas C1qTNF7 | XP_011449385.2 |
| Biomphalaria glabrata C1qTNF7 | KAI8775437.1 |
| Lingula anatine Adipolin | XP_013384435.1 |
| Homo sapiens AdipoR1 | KAI4084514.1 |
| Homo sapiens AdipoR2 | KAI4064009.1 |
| Mus musculus AdipoR1 | AAH14875.1 |
| Mus musculus AdipoR2 | AAH24094.2 |
| Crassostrea gigas AdipoR | XP_019926543.1 |
| Danio rerio AdipoR1 | NP_001314683.1 |
| Danio rerio AdipoR2 | NP_001020677.1 |
| Xenopus tropicalis AdipoR1 | NP_001007928.1 |
| Xenopus tropicalis AdipoR2 | XP_031754470.1 |
| Drosophila melanogaster AdipoR | NP_651061.1 |
| Octopus sinensis AdipoR1 | XP_036366622.1 |
| Octopus sinensis AdipoR2 | XP_036366621.1 |
| Aplysia californica AdipoR | XP_005097206.1 |
| Pomacea canaliculata AdipoR1 | XP_025108208.1 |
| Mizuhopecten yessoensis AdipoR | XP_021370275.1 |
| Lingula anatina AdipoR2 | XP_013397290.1 |
| Biomphalaria glabrata AdipoR | KAI8771842.1 |
| Sepia pharaonis AdipoR | CAE1287042.1 |
| Pecten maximus AdipoR | XP_033744837.1 |
| Primer | Sequence (5′-3′) | Product Size (bp) | |
|---|---|---|---|
| 18S_PCR | Forward | CTC CTT CGT GCT AGG GAT TG | 106 |
| Reverse | GTA CAA AGG GCA GGG ACG TA | ||
| β-tubulin_PCR | Forward | CAA GCG CAT CTC TGA GCA GTT | 108 |
| Reverse | TTG GAT TCC GCC TCT GTG AA | ||
| LymAdipo_PCR | Forward | TGC TGA GCA TGG AGA ACC AG | 111 |
| Reverse | CCG TGT TAC TTC CGG TTC CA | ||
| LymAdipoR_PCR | Forward | TCC AGT GGC AAG AAA AGG CA | 108 |
| Reverse | CAA CAC GTT CAC TGT GGC AG | ||
| MIPII_PCR | Forward | AGA GGG CCA ATC ATC TTG CAG | 77 |
| Reverse | GGA AGC CAG CCA AAT TCG AG | ||
| MIPR_PCR | Forward | AGA CAG ACT ACT ATA GAA AAG GAG GTA AAG GAA | 118 |
| Reverse | ACA ACT CCA TAT GAC CAA ACA TCT GA | ||
| LymAdipo_in situ | Forward | TGC TGC CCG TAG TTC TAC AC | 471 |
| Reverse | AGC TGT CTC CCA GGT TGA GA | ||
| LymAdipoR_in situ | Forward | TTC TAT TGT CGC CTG GAG CC | 421 |
| Reverse | AGT CCC CCA GGG TAA GTC TG | ||
| M13 | Forward | GTA AAA CGA CGG CCA GT | - |
| Reverse | CAG GAA ACA GCT ATG AC | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujimoto, K.; Totani, Y.; Nakai, J.; Chikamoto, N.; Namiki, K.; Hatakeyama, D.; Ito, E. Identification of Putative Molecules for Adiponectin and Adiponectin Receptor and Their Roles in Learning and Memory in Lymnaea stagnalis. Biology 2023, 12, 375. https://doi.org/10.3390/biology12030375
Fujimoto K, Totani Y, Nakai J, Chikamoto N, Namiki K, Hatakeyama D, Ito E. Identification of Putative Molecules for Adiponectin and Adiponectin Receptor and Their Roles in Learning and Memory in Lymnaea stagnalis. Biology. 2023; 12(3):375. https://doi.org/10.3390/biology12030375
Chicago/Turabian StyleFujimoto, Kanta, Yuki Totani, Junko Nakai, Nozomi Chikamoto, Kengo Namiki, Dai Hatakeyama, and Etsuro Ito. 2023. "Identification of Putative Molecules for Adiponectin and Adiponectin Receptor and Their Roles in Learning and Memory in Lymnaea stagnalis" Biology 12, no. 3: 375. https://doi.org/10.3390/biology12030375
APA StyleFujimoto, K., Totani, Y., Nakai, J., Chikamoto, N., Namiki, K., Hatakeyama, D., & Ito, E. (2023). Identification of Putative Molecules for Adiponectin and Adiponectin Receptor and Their Roles in Learning and Memory in Lymnaea stagnalis. Biology, 12(3), 375. https://doi.org/10.3390/biology12030375








