IS481EU Shows a New Connection between Eukaryotic and Prokaryotic DNA Transposons
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
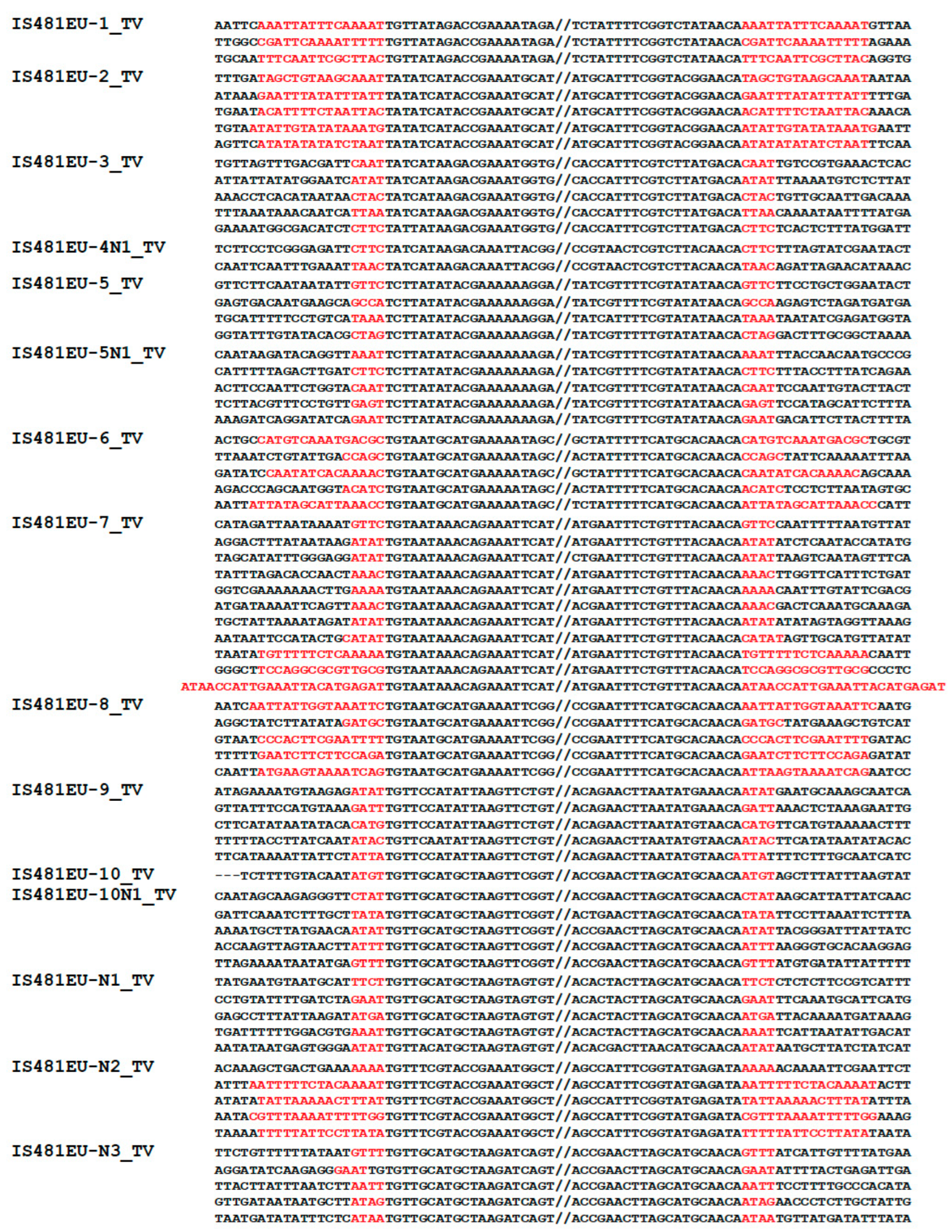
2.1. Characterization of IS481EU Families
2.2. Protein Dataset and Phylogenetic Analysis
3. Results
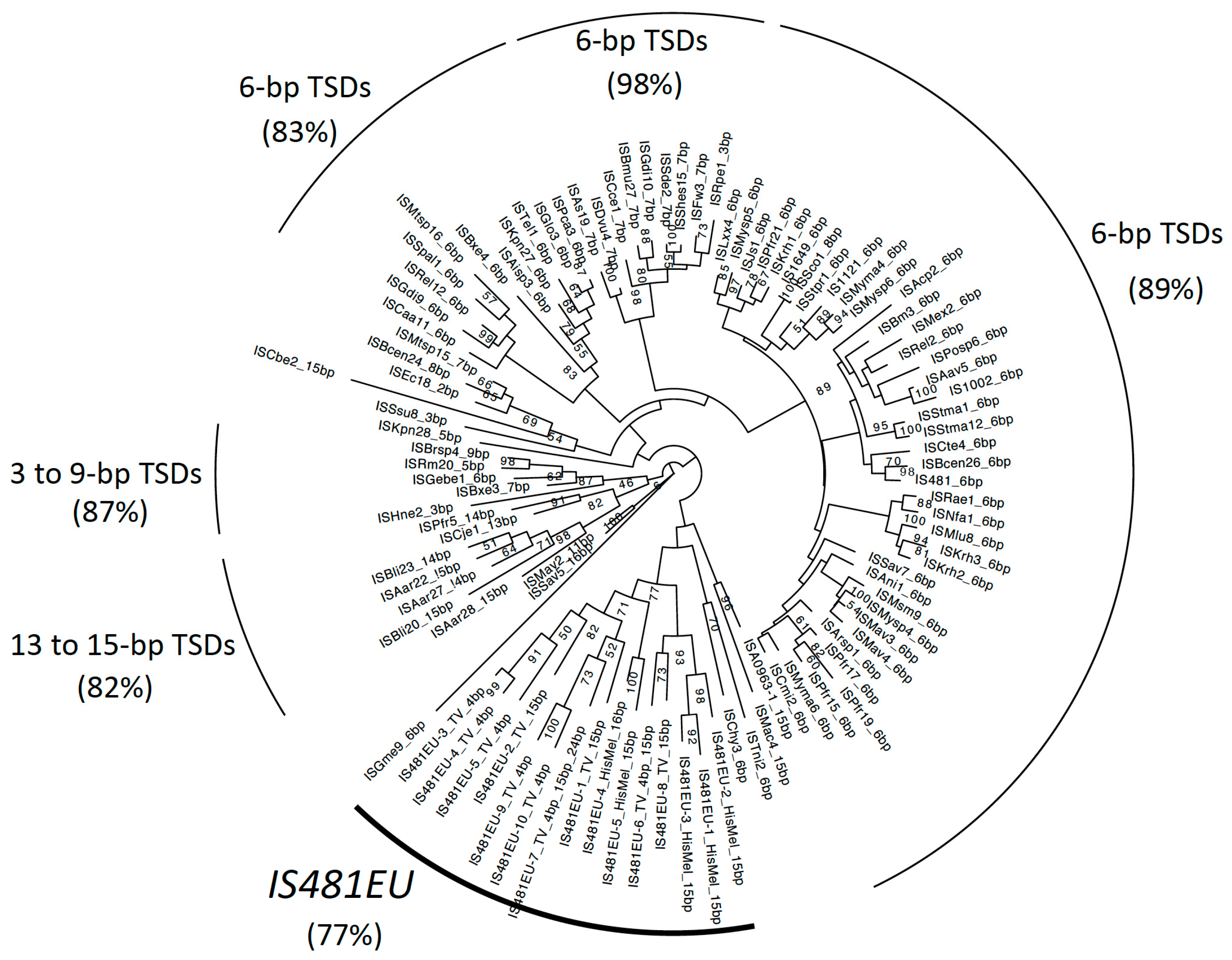
3.1. Eukaryotic TEs Related to the Prokaryotic IS481 Family
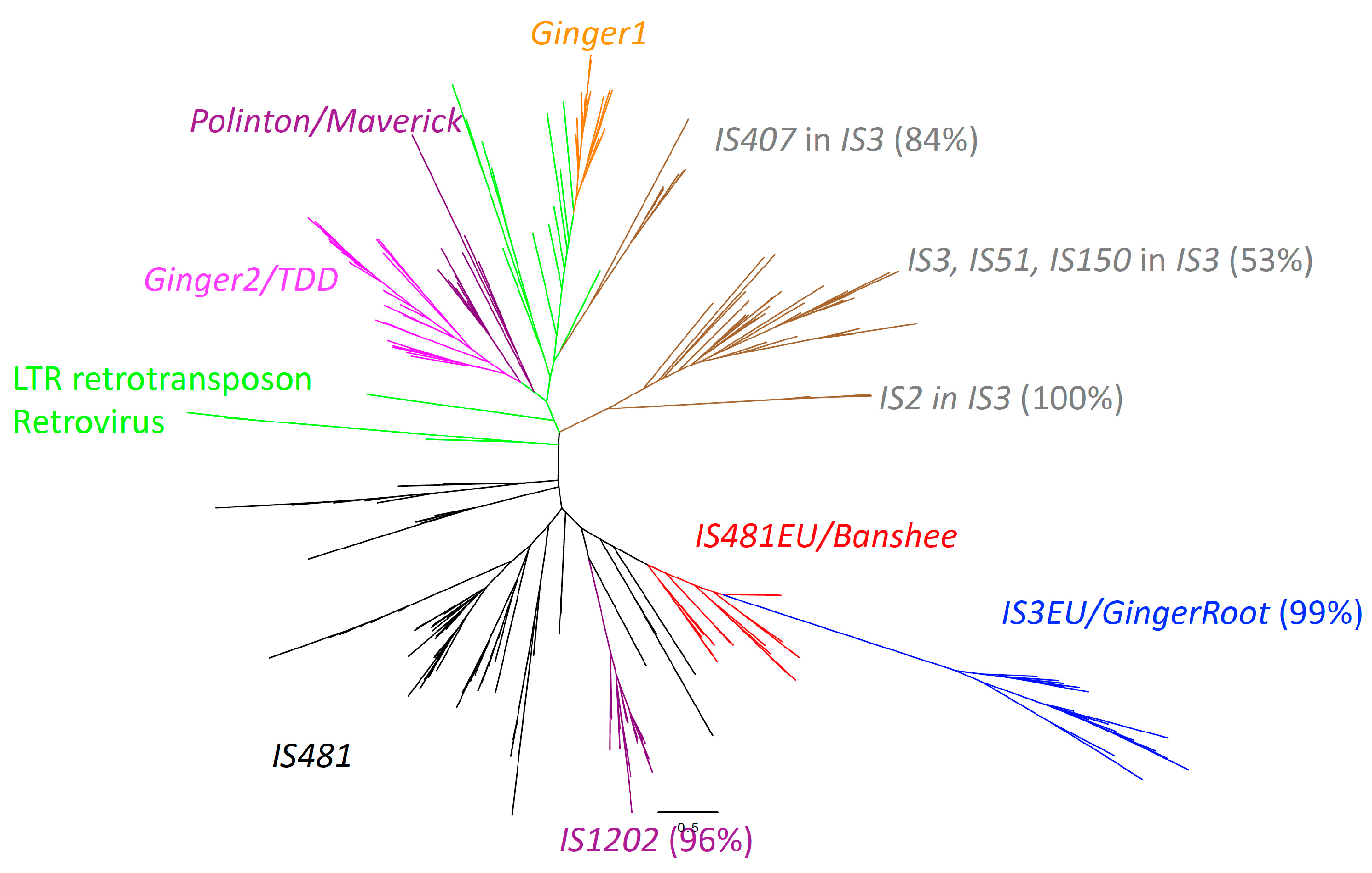
3.2. The Phylogenetic Relationship between IS481EU and IS481
3.3. The Origins of IS3EU/GingerRoot and IS481EU in Eukaryotes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Curcio, M.J.; Derbyshire, K.M. The outs and ins of transposition: From mu to kangaroo. Nat. Rev. Mol. Cell Biol. 2003, 4, 865–877. [Google Scholar] [CrossRef]
- Finnegan, D.J. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989, 5, 103–107. [Google Scholar] [CrossRef]
- Aziz, R.K.; Breitbart, M.; Edwards, R.A. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010, 38, 4207–4217. [Google Scholar] [CrossRef]
- Hickman, A.B.; Chandler, M.; Dyda, F. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 50–69. [Google Scholar] [CrossRef] [PubMed]
- Majorek, K.A.; Dunin-Horkawicz, S.; Steczkiewicz, K.; Muszewska, A.; Nowotny, M.; Ginalski, K.; Bujnicki, J.M. The RNase H-like superfamily: New members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 2014, 42, 4160–4179. [Google Scholar] [CrossRef] [PubMed]
- Dyda, F.; Hickman, A.B.; Jenkins, T.M.; Engelman, A.; Craigie, R.; Davies, D.R. Crystal structure of the catalytic domain of HIV-1 integrase: Similarity to other polynucleotidyl transferases. Science 1994, 266, 1981–1986. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005, 3, e181. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Tao, X.; Yuan, S.; Zhang, Y.; Li, P.; Beilinson, H.A.; Zhang, Y.; Yu, W.; Pontarotti, P.; Escriva, H.; et al. Discovery of an Active RAG Transposon Illuminates the Origins of V (D) J Recombination. Cell 2016, 166, 102–114. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Varani, A.; Ton-Hoang, B.; Chandler, M. Everyman’s Guide to Bacterial Insertion Sequences. Microbiol. Spectr. 2015, 3, MDNA3-0030-2014. [Google Scholar] [CrossRef] [PubMed]
- Ross, K.; Varani, A.M.; Snesrud, E.; Huang, H.; Alvarenga, D.O.; Zhang, J.; Wu, C.; McGann, P.; Chandler, M. TnCentral: A Prokaryotic Transposable Element Database and Web Portal for Transposon Analysis. mBio 2021, 12, e0206021. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.K. Structural and sequence diversity of eukaryotic transposable elements. Genes Genet. Syst. 2020, 94, 233–252. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.K.; Jurka, J. A superfamily of DNA transposons targeting multicopy small RNA genes. PLoS ONE 2013, 8, e68260. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.W.; Wessler, S.R. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. USA 2011, 108, 7884–7889. [Google Scholar] [CrossRef]
- Tellier, M.; Bouuaert, C.C.; Chalmers, R. Mariner and the ITm Superfamily of Transposons. Microbiol. Spectr. 2015, 3, MDNA3-0033-2014. [Google Scholar] [CrossRef]
- Shi, S.; Puzakov, M.; Guan, Z.; Xiang, K.; Diaby, M.; Wang, Y.; Wang, S.; Song, C.; Gao, B. Prokaryotic and Eukaryotic Horizontal Transfer of Sailor (DD82E), a New Superfamily of IS630-Tc1-Mariner DNA Transposons. Biology 2021, 10, 1005. [Google Scholar] [CrossRef]
- Yusa, K. piggyBac Transposon. Microbiol. Spectr. 2015, 3, MDNA3-0028-2014. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Sim, C.; Hong, Y.S.; Hogan, J.R.; Fraser, M.J.; Robertson, H.M.; Collins, F.H. Molecular evolutionary analysis of the widespread piggyBac transposon family and related "domesticated" sequences. Mol. Genet. Genom. 2003, 270, 173–180. [Google Scholar] [CrossRef]
- Feschotte, C. Merlin, a new superfamily of DNA transposons identified in diverse animal genomes and related to bacterial IS1016 insertion sequences. Mol. Biol. Evol. 2004, 21, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Jurka, M.G.; Kapitonov, V.V.; Jurka, J. New superfamilies of eukaryotic DNA transposons and their internal divisions. Mol. Biol. Evol. 2009, 26, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R. Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories. Mob. DNA 2017, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Hua-Van, A.; Capy, P. Analysis of the DDE motif in the Mutator superfamily. J. Mol. Evol. 2008, 67, 670–681. [Google Scholar] [CrossRef] [PubMed]
- Eisen, J.A.; Benito, M.I.; Walbot, V. Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 1994, 22, 2634–2636. [Google Scholar] [CrossRef][Green Version]
- Han, M.J.; Xiong, C.L.; Zhang, H.B.; Zhang, M.Q.; Zhang, H.H.; Zhang, Z. The diversification of PHIS transposon superfamily in eukaryotes. Mob. DNA 2015, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, E.A.; Arkhipova, I.R. A single-copy IS5-like transposon in the genome of a bdelloid rotifer. Mol. Biol. Evol. 2009, 26, 1921–1929. [Google Scholar] [CrossRef]
- Han, M.J.; Xu, H.E.; Zhang, H.H.; Feschotte, C.; Zhang, Z. Spy: A new group of eukaryotic DNA transposons without target site duplications. Genome Biol. Evol. 2014, 6, 1748–1757. [Google Scholar] [CrossRef]
- Bao, W.; Kapitonov, V.V.; Jurka, J. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob. DNA 2010, 1, 3. [Google Scholar] [CrossRef]
- Feschotte, C.; Pritham, E.J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007, 41, 331–368. [Google Scholar] [CrossRef]
- Kohany, O.; Gentles, A.J.; Hankus, L.; Jurka, J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 2006, 7, 474. [Google Scholar] [CrossRef]
- Cerbin, S.; Wai, C.M.; VanBuren, R.; Jiang, N. GingerRoot: A Novel DNA Transposon Encoding Integrase-Related Transposase in Plants and Animals. Genome Biol. Evol. 2019, 11, 3181–3193. [Google Scholar] [CrossRef] [PubMed]
- Llorens, C.; Futami, R.; Covelli, L.; Dominguez-Escriba, L.; Viu, J.M.; Tamarit, D.; Aguilar-Rodríguez, J.; Vicente-Ripolles, M.; Fuster, G.; Bernet, G.P.; et al. The Gypsy Database (GyDB) of mobile genetic elements: Release 2.0. Nucleic Acids Res. 2011, 39, D70–D74. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Do, C.B.; Mahabhashyam, M.S.; Brudno, M.; Batzoglou, S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005, 15, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Engelman, A.; Cherepanov, P. Retroviral Integrase Structure and DNA Recombination Mechanism. Microbiol. Spectr. 2014, 2, 1–22. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 2008, 9, 411–412. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. Self-synthesizing DNA transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 2006, 103, 4540–4545. [Google Scholar] [CrossRef]
- Lesbats, P.; Engelman, A.N.; Cherepanov, P. Retroviral DNA Integration. Chem. Rev. 2016, 116, 12730–12757. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.R.; Grey, H.; McKenzie, G.; Jones, A.C.; Richardson, J.M. A bend, flip and trap mechanism for transposon integration. Elife 2016, 5, e15537. [Google Scholar] [CrossRef] [PubMed]
- Arias-Palomo, E.; Berger, J.M. An Atypical AAA+ ATPase Assembly Controls Efficient Transposition through DNA Remodeling and Transposase Recruitment. Cell 2015, 162, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Montano, S.P.; Pigli, Y.Z.; Rice, P.A. The mu transpososome structure sheds light on DDE recombinase evolution. Nature 2012, 491, 413–417. [Google Scholar] [CrossRef]
- Kojima, K.K. AcademH, a lineage of Academ DNA transposons encoding helicase found in animals and fungi. Mob. DNA 2020, 11, 15. [Google Scholar] [CrossRef] [PubMed]

| Phylum | Order | Family | Species | Genome Assembly | IS481EU Family |
|---|---|---|---|---|---|
| Parabasalia | Trichomonadida | Trichomonadidae | Trichomonas vaginalis | ASM289133v1 | IS481EU-1_TV to 10_TV, IS481EU-N1_TV to N3_TV, 4N1_TV, 5N1_TV, 8N1_TV, and 10N1_TV. |
| Trichomonas tenax | PRJEB22701 | IS481EU-1_TrTenax | |||
| Trichomonas gallinae | MiGF1c1.0 | IS481EU-1_TrGallinae to IS481EU-5_TrGallinae | |||
| Tritrichomonadida | Tritrichomonadidae | Tritrichomonas foetus | TF_PacBio | IS481EU-1_TrFoetus | |
| Dientamoebidae | Histomonas meleagridis | ASM2018611v1 | IS481EU-1_HisMel to 5_HisMel |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kojima, K.K.; Bao, W. IS481EU Shows a New Connection between Eukaryotic and Prokaryotic DNA Transposons. Biology 2023, 12, 365. https://doi.org/10.3390/biology12030365
Kojima KK, Bao W. IS481EU Shows a New Connection between Eukaryotic and Prokaryotic DNA Transposons. Biology. 2023; 12(3):365. https://doi.org/10.3390/biology12030365
Chicago/Turabian StyleKojima, Kenji K., and Weidong Bao. 2023. "IS481EU Shows a New Connection between Eukaryotic and Prokaryotic DNA Transposons" Biology 12, no. 3: 365. https://doi.org/10.3390/biology12030365
APA StyleKojima, K. K., & Bao, W. (2023). IS481EU Shows a New Connection between Eukaryotic and Prokaryotic DNA Transposons. Biology, 12(3), 365. https://doi.org/10.3390/biology12030365






