DNA Barcoding of the Genus Magnisudis (Aulopiformes: Paralepididae) with a Coastal Record and Biological Features of Magnisudis atlantica
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
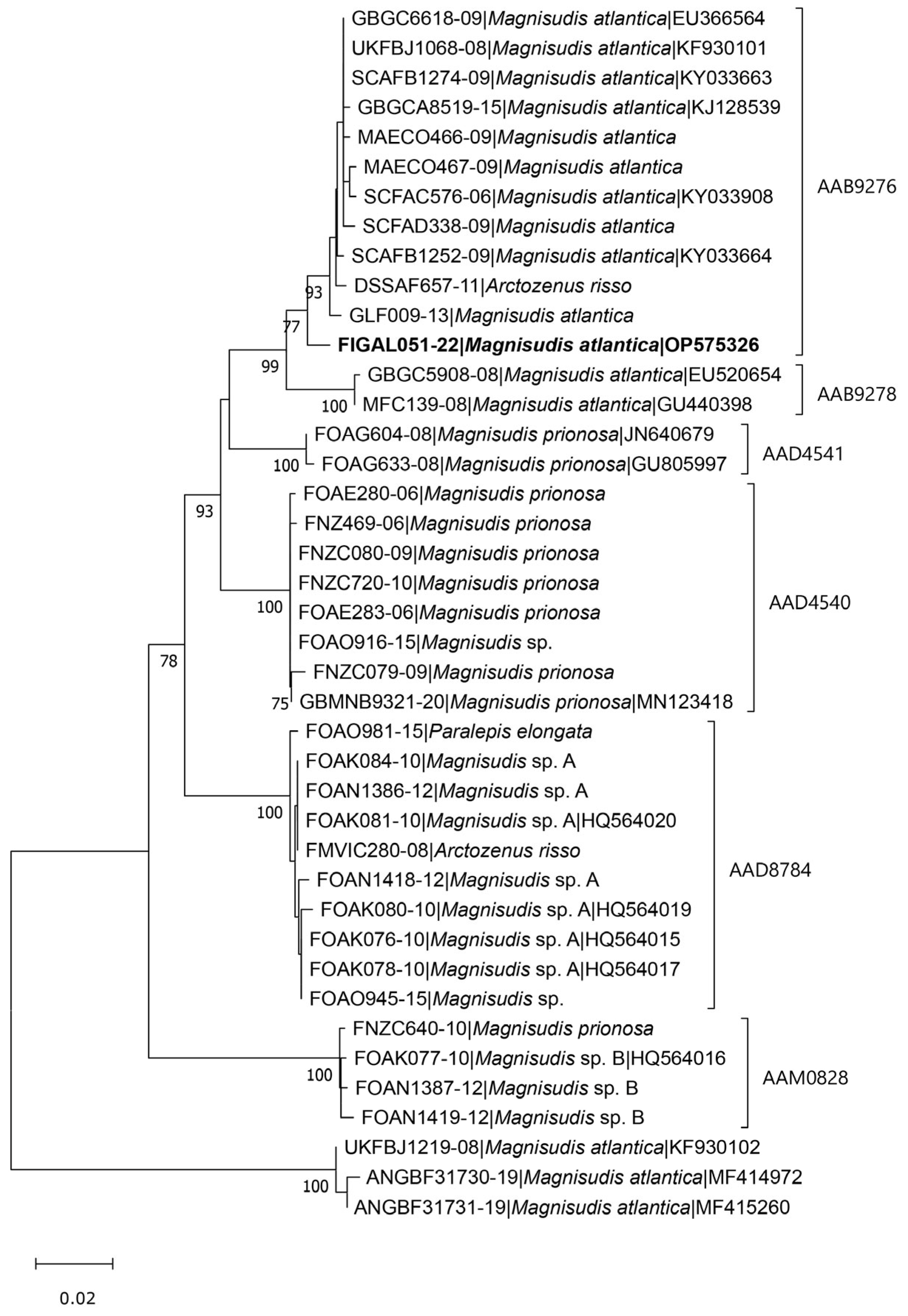
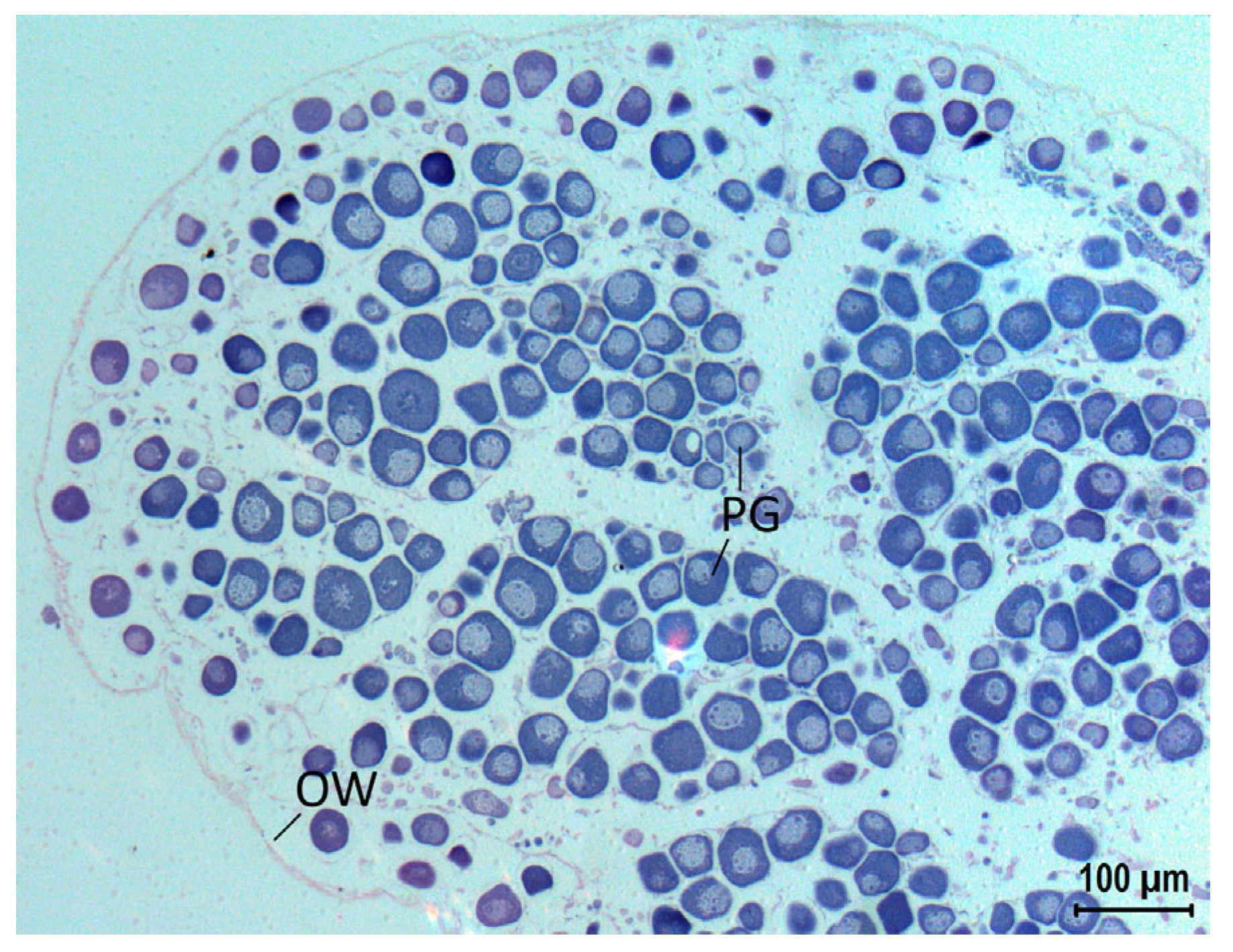
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identification through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Savolainen, V.; Cowan, R.S.; Vogler, A.P.; Roderick, G.K.; Lane, R. Towards writing the encyclopaedia of life: An introduction to DNA barcoding. Phil. Trans. R. Soc. 2005, 360, 1805–1811. [Google Scholar] [CrossRef]
- Kress, J.W.; García-Robledo, C.; Uriarte, M.; Erickson, D.L. DNA barcodes for ecology, evolution and conservation. Trends Ecol. Evol. 2015, 30, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Dayrat, B. Towards integrative taxonomy. Biol. J. Linn. Soc. 2005, 85, 407–415. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Gregory, T.R. The promise of DNA barcoding for taxonomy. Syst. Biol. 2005, 54, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The barcoding of life datasystem (www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.D.; Hanner, R.; Hebert, P.D.N. The campaign to DNA barcode all fishes, FISH-BOL. J. Fish Biol. 2009, 74, 329–356. [Google Scholar] [CrossRef]
- Priede, I.G. Deep-Sea Fishes: Biology, Diversity, Ecology and Fisheries; Cambridge University Press: Cambridge, UK, 2017; pp. 1–504. [Google Scholar]
- Teramura, A.; Koeda, K.; Matsuo, A.; Sato, M.P.; Senou, H.; Ho, H.C.; Suyama, Y.; Kikuchi, K.; Hirase, S. Assessing the effectiveness of DNA barcoding for exploring hidden genetic diversity in deep-sea fishes. Mar. Ecol. Prog. Ser. 2022, 701, 83–98. [Google Scholar] [CrossRef]
- Harry, R.R. Studies on the bathypelagic fishes of the family Paralepididae. 1. Survey of the genera. Pac. Sci. 1953, 7, 219–249. [Google Scholar]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V.H. Fishes of the World, 5th ed.; Wiley: Hoboken, NJ, USA, 2016; pp. 1–752. [Google Scholar]
- Russell, B.C. Paralepididae. In The Living Marine Resources of the Eastern Central Atlantic. Bony Fishes. Part 1. Elopiformes to Scorpaeniformes; Carpenter, K.E., De Angelis, N., Eds.; FAO: Rome, Italy, 2016; Volume 3, pp. 1844–1848. [Google Scholar]
- Fricke, R.; Eschmeyer, W.N.; Fong, J.D. Eschmeyer’s Catalog of Fishes: Species by Family/Subfamily. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp. (accessed on 25 November 2022).
- Ho, H.-C.; Oktaviyani, S.; Peristiwady, T.; Hui, T.-H. A rare species of scaled barracudina (Paralepididae) newly collected from the eastern Indian Ocean off Indonesia. Raffles Bull. Zool. 2021, 36, 486–490. [Google Scholar] [CrossRef]
- Krøyer, H.N. To nye Fiske for den danske Favna. Tidsskr. Fisk. 1868, 2, 70–71. [Google Scholar]
- Carl, H.; Nielsen, J.G. Magnisudis atlantica (Krøyer, 1868), Kort lakstobis, Atlas over danske saltvandsfisk. Available online: https://fiskeatlas.ku.dk/artstekster/Kort_lakstobis_Fiskeatlas.pdf (accessed on 26 November 2022).
- Porteiro, F.M.; Sutton, T.; Byrkjedal, I.; Orlov, A.M.; Heino, M.; Menezes, G.; Bergstad, O.A. Fishes of the Northern Mid-Atlantic Ridge Collected During the MAR-ECO Cruise in June-July 2004: An Annotated Checklist. Arquipelago 2017, 10, 1–126. Available online: https://nsuworks.nova.edu/occ_facreports/102 (accessed on 24 November 2022).
- Bañón, R.; Arronte, J.C.; Rodríguez-Cabello, C.; Piñeiro, C.G.; Punzón, A.; Serrano, A. Commented checklist of marine fishes from the Galicia Bank seamount (NW Spain). Zootaxa 2016, 4067, 293–333. [Google Scholar] [CrossRef] [PubMed]
- McEachran, J.D.; Fechhelm, J.D. Fishes of the Gulf of Mexico. Volume 1: Scorpaeniformes to Tetraodontiformes; University of Texas Press: Austin, TX, USA, 1998; pp. 1–1112. [Google Scholar]
- Rofen, R.R. Family Paralepididae. In Fishes of the Western North Atlantic; Mead, G.W., Bigelow, H.B., Olsen, Y.H., Breder, C.M., Schroeder, W.C., Cohen, D.M., Schultz, L.P., Merriman, D., Tee-Van, J., Eds.; Memoirs, Sears Foundation of Marine Research, Yale University: New Haven, CT, USA, 1966; No. I, Part 5; pp. 205–461. [Google Scholar]
- Devine, B.; Van Guelpen, L. Loss of gill rakers and teeth in adult specimens of barracudina Arctozenus risso (Aulopiformes: Paralepididae) from the western North Atlantic. J. Fish Biol. 2021, 98, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.-C.; Duhamel, G. A new species of the fish genus Arctozenus from the Kerguelen Islands, with comments on the lost teeth in adults (Aulopiformes: Paralepididae). Zootaxa 2019, 4651, 497–512. [Google Scholar] [CrossRef] [PubMed]
- Kenchington, E.L.; Baillie, S.M.; Kenchington, T.J.; Bentzen, P. Barcoding Atlantic Canada’s mesopelagic and upper bathypelagic marine fishes. PLoS ONE 2017, 12, e0185173. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.P.; Fielitz, C. Estimating divergence times of lizardfishes and their allies (Euteleostei: Aulopiformes) and the timing of deep-sea adaptations. Mol. Phylogenet. Evol. 2010, 57, 1194–1208. [Google Scholar] [CrossRef]
- Post, A. Results of the research cruises of FRV “Walther Herwig” to South America. LXVII. Revision of the subfamily Paralepidinae (Pisces, Aulopiformes, Alepisauroidei, Paralepididae). I. Taxonomy, morphology and geographical distribution. Arch. Fischereiwiss. 1987, 38, 75–131. [Google Scholar]
- Rees, D.J.; Byrkjedal, I.; Sutton, T.T. Pruning the Pearlsides: Reconciling morphology and molecules in mesopelagic fishes (Maurolicus: Sternoptychidae). Deep-Sea Research II 2017, 137, 246–257. [Google Scholar] [CrossRef]
- Brown-Peterson, N.J.; Wyanski, D.M.; Saborido-Rey, F.; Macewicz, B.J.; Lowerre-Barbieri, S.K. A standardized terminology for describing reproductive development in fishes. Mar. Coast. Fish. 2011, 3, 52–70. [Google Scholar] [CrossRef]
- Bañón, R.; Roura, A.; García-Fernández, C.; Alonso-Fernández, A.; de Carlos, A. Coastal habitat evidences and biological data of Alepisaurus ferox (Aulopiform; Alepisauridae) from northwestern Iberian Peninsula. Mar. Biodiv. 2022, 52, 22. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbour-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000; pp. 1–333. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Moura, T.; Silva, M.C.; Figueiredo, I. Barcoding deep-water chondrichthyans from mainland Portugal. Mar. Freshw. Res. 2015, 66, 508–517. [Google Scholar] [CrossRef]
- Post, A. Paralepididae. In Fishes of the North-Eastern Atlantic and the Mediterranean; Whitehead, P.J.P., Bauchot, M.-L., Hureau, J.-C., Neilsen, J., Tortonese, E., Eds.; UNESCO: Paris, France, 1984; pp. 498–508. [Google Scholar]
- Jones, R. Ecology of the Barracudinas (Aulopiformes: Paralepididae), a Ubiquitous but Understudied Mesopelagic Predatory Fish Family, in the Gulf of Mexico. Ph.D. Thesis, Florida Atlantic University, Boca Raton, FL, USA, August 2018. [Google Scholar]
- Franz, G.P.; Warth, P.; Grunow, B.; Konstantinidis, P. Osteology of the white barracudina, Arctozenus risso (Bonaparte) (Aulopiformes: Paralepididae), Ichthyol. Herpetol. 2022, 110, 115–130. [Google Scholar] [CrossRef]
- Thompson, B.A. Order Aulopiformes. In The Living Marine Resources of the Western Central Atlantic; Carpenter, K.E., Ed.; FAO: Rome, Italy, 2002; Volume 2, Part 1, pp. 933–934. [Google Scholar]
- Bañón, R.; Alonso-Fernández, A.; Quigley, D.T.G.; Miranda, A.; Arronte, J.C. Unusual shallow inshore records of Cornish blackfish Schedophilus medusophagus (Stromateoidei: Centrolophidae) from Galician waters (NW Spain). Cah. Biol. Mar. 2012, 53, 271–277. [Google Scholar]

| L (mm) | %SL | %HL | in SL | in HL | |
|---|---|---|---|---|---|
| Total length | 402 | ||||
| Standard length | 361 | ||||
| Pre-dorsal fin length | 212 | 58.7 | 1.7 | ||
| Pre-pelvic fin length | 220 | 60.9 | 1.6 | ||
| Pre-anal fin length | 280 | 77.6 | 1.3 | ||
| Pre-adipose fin length | 314 | 87.0 | 1.1 | ||
| Interdorsal adipose length | 81 | 22.4 | 4.5 | ||
| Pre-anus length | 232 | 64.3 | 1.6 | ||
| Head length | 89 | 24.7 | 4.1 | ||
| Head depth | 37 | 10.2 | 9.8 | ||
| Snout length | 35 | 9.7 | 39.3 | 10.3 | 2.5 |
| Post-orbital length | 35 | 9.7 | 39.3 | 10.3 | 2.5 |
| Pre-nostril length | 24 | 6.6 | 27 | 15 | 3.7 |
| Upper jaw length | 38 | 10.5 | 42.7 | 9.5 | 2.3 |
| Lower jaw length | 40 | 11.1 | 44.9 | 9.0 | 2.2 |
| Eye diameter | 22 | 6.1 | 24.7 | 16.4 | 4.0 |
| Interorbital width | 14 | 3.9 | 15.7 | 25.8 | 6.4 |
| Mouth gape | 40 | 11.1 | 44.9 | 9.0 | 2.2 |
| Body depth at pectoral fin origin | 42 | 11.6 | 47.2 | 8.6 | 2.1 |
| Body depth (maximum) | 50 | 13.9 | 56.2 | 7.2 | 1.8 |
| Anal fin base | 51 | 14.1 | 57.3 | 7.1 | 1.7 |
| Dorsal fin base | 19 | 5.3 | 21.3 | 19.0 | 4.7 |
| Adipose fin base | 9 | 2.5 | 10.1 | 40.1 | 9.9 |
| Pectoral fin length | 45 | 12.5 | 50.6 | 8.0 | 2.0 |
| Pelvic fin length | 14 | 3.9 | 15.7 | 25.8 | 6.4 |
| Caudal peduncle length | 13 | 3.6 | 14.6 | 27.8 | 6.8 |
| Caudal peduncle depth | 13 | 3.6 | 14.6 | 27.8 | 6.8 |
| Meristic | |||||
| Dorsal fin rays | 10 | ||||
| Anal fin rays | 23 | ||||
| Pectoral fin rays | 17 | ||||
| Pelvic fin rays | 9 | ||||
| Branchiostegal rays | 6 | ||||
| Gill rakers | 7 + 29 | ||||
| Premaxillary teeth | 63 | ||||
| Dentary teeth | 40 | ||||
| Lateral line scales | 63 |
| BIN | N | Maximum Intraspecific | Minimum Interspecific | |||||
|---|---|---|---|---|---|---|---|---|
| AAB9276 | AAB9278 | AAD4541 | AAD4540 | AAD8784 | AAM0828 | |||
| AAB9276 | 12 | 0.0184 | ||||||
| AAB9278 | 2 | 0.000 | 0.0246 | |||||
| AAD4541 | 2 | 0.0017 | 0.0459 | 0.0522 | ||||
| AAD4540 | 8 | 0.0048 | 0.0476 | 0.0492 | 0.0380 | |||
| AAD8784 | 10 | 0.0077 | 0.0691 | 0.0737 | 0.0617 | 0.0523 | ||
| AAM0828 | 4 | 0.0046 | 0.0983 | 0.1045 | 0.0934 | 0.0906 | 0.0906 | |
| Unknown | 3 | 0.0016 | 0.1649 | 0.1690 | 0.1614 | 0.1633 | 0.1525 | 0.1695 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bañón, R.; Almón, B.; Rábade, S.; Ríos, M.B.; de Carlos, A. DNA Barcoding of the Genus Magnisudis (Aulopiformes: Paralepididae) with a Coastal Record and Biological Features of Magnisudis atlantica. Biology 2023, 12, 349. https://doi.org/10.3390/biology12030349
Bañón R, Almón B, Rábade S, Ríos MB, de Carlos A. DNA Barcoding of the Genus Magnisudis (Aulopiformes: Paralepididae) with a Coastal Record and Biological Features of Magnisudis atlantica. Biology. 2023; 12(3):349. https://doi.org/10.3390/biology12030349
Chicago/Turabian StyleBañón, Rafael, Bruno Almón, Sonia Rábade, María Berta Ríos, and Alejandro de Carlos. 2023. "DNA Barcoding of the Genus Magnisudis (Aulopiformes: Paralepididae) with a Coastal Record and Biological Features of Magnisudis atlantica" Biology 12, no. 3: 349. https://doi.org/10.3390/biology12030349
APA StyleBañón, R., Almón, B., Rábade, S., Ríos, M. B., & de Carlos, A. (2023). DNA Barcoding of the Genus Magnisudis (Aulopiformes: Paralepididae) with a Coastal Record and Biological Features of Magnisudis atlantica. Biology, 12(3), 349. https://doi.org/10.3390/biology12030349





