Introduction of Plant Transposon Annotation for Beginners
Abstract
:Simple Summary
Abstract
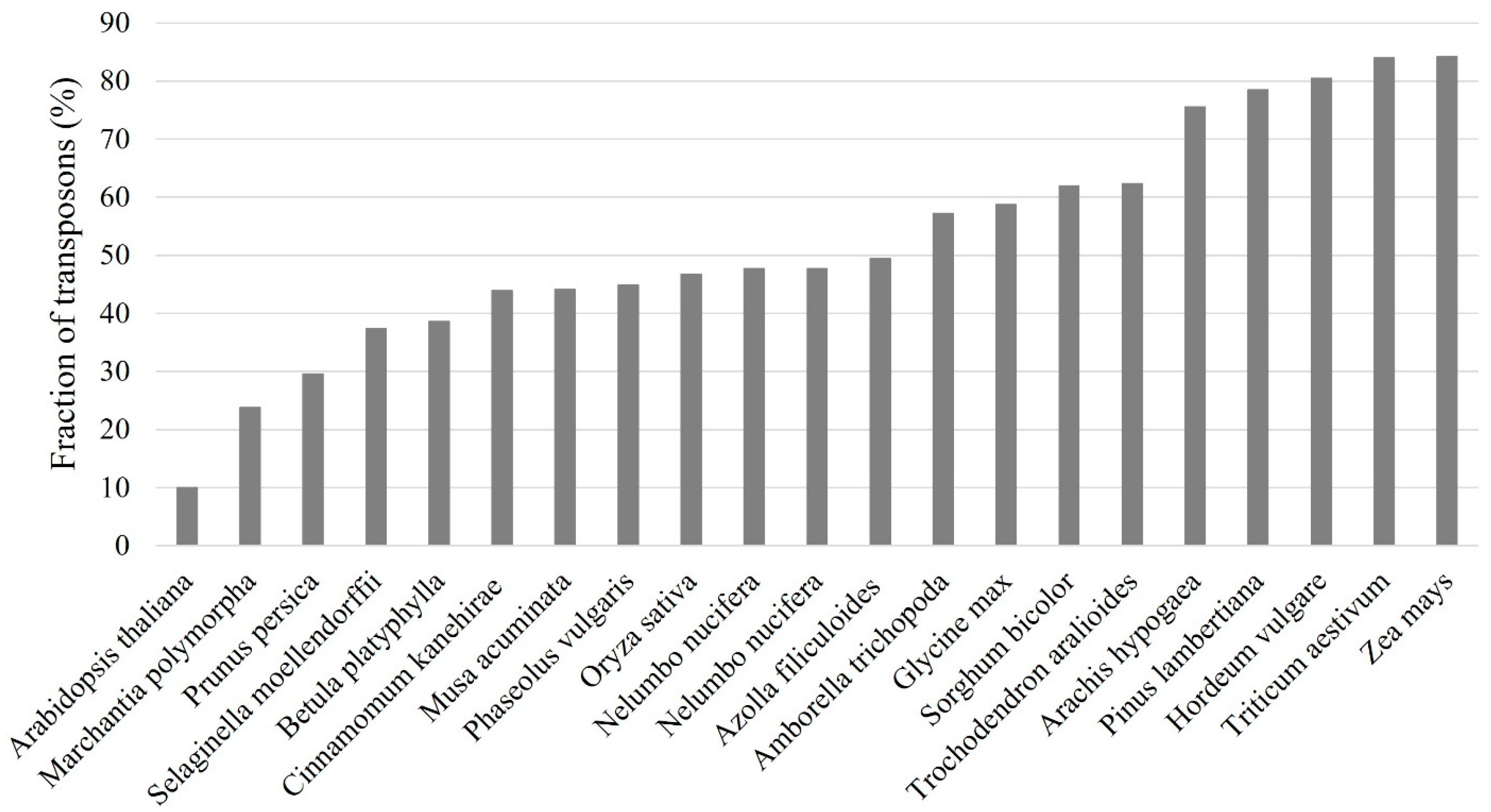
1. Introduction of Plant Transposons
2. Strategies of Transposon Discovery
2.1. Repeat-Based Annotation
2.2. Structure-Based Annotation
2.3. Homology-Based Annotation
3. Steps for Transposon Annotation
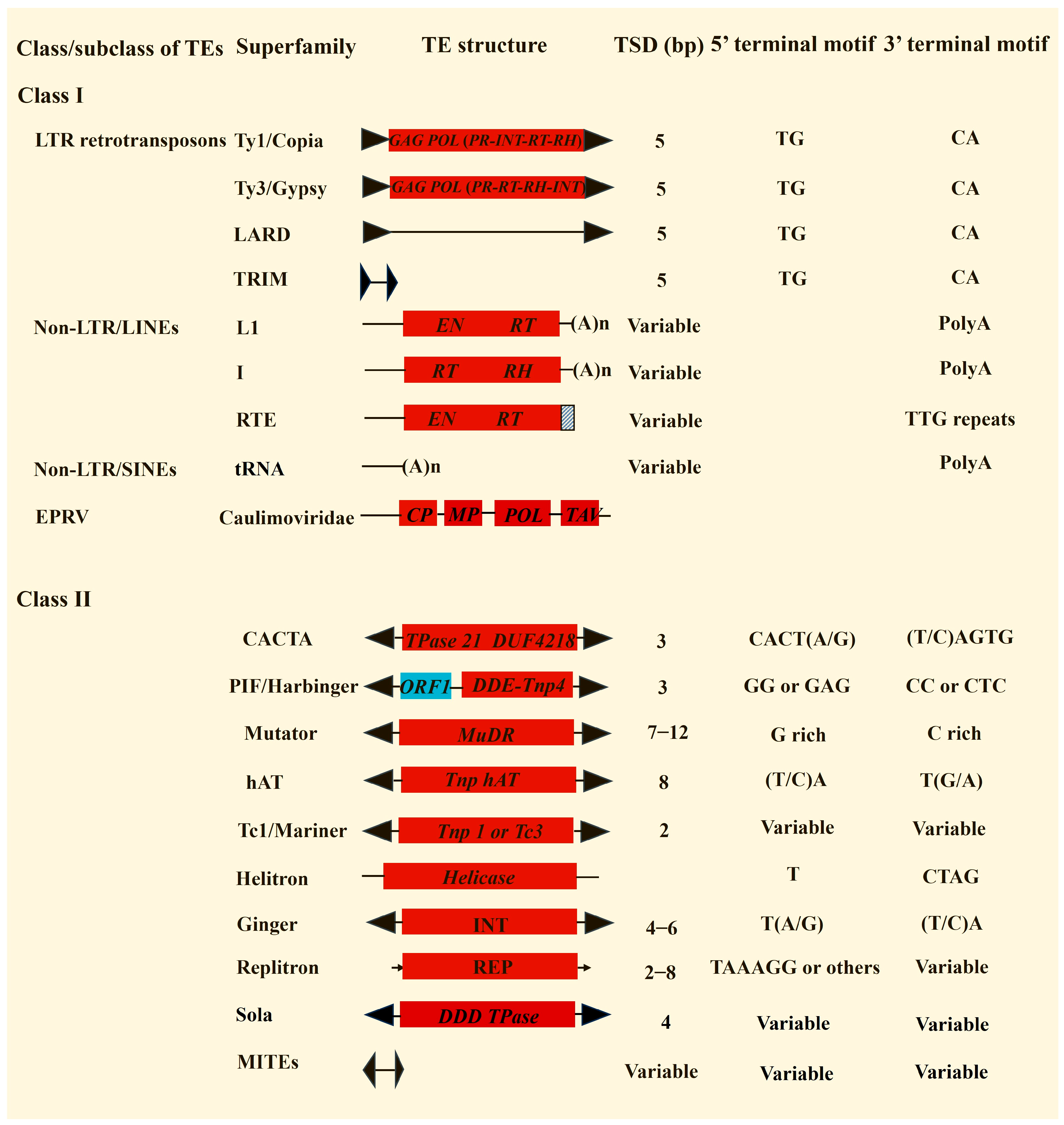
3.1. Brief Introduction of Plant Transposon Superfamilies and Their Annotations
3.1.1. LTR Retrotransposons
3.1.2. LINEs
3.1.3. SINEs
3.1.4. Endogenous Plant Pararetrovirus (EPRVs)
3.1.5. DNA Transposons with TIRs
Mutator Transposons
CACTA Transposons
hAT Transposons
PIF/Harbinger Transposons
Tc1/Mariner Transposons
Sola Transposons
Ginger Transposons
MITEs
TIR DNA Transposon Annotation
3.1.6. Helitron Transposons
3.1.7. Replitron Transposons
3.1.8. Others TE Annotation Resources
3.2. Classification of Transposons
3.2.1. Definition of Transposon Superfamilies
3.2.2. Definition of Transposon Families
3.2.3. Autonomous and Non-Autonomous Transposons
3.3. Quality Control and Improvement of Transposon Annotation
3.3.1. Identification and Exclusion of Misannotated Sequences
3.3.2. Elimination of Nested Transposons
3.3.3. Definition of Transposon Boundaries
3.3.4. Identification of Unannotated Transposons
3.4. Criteria for Good Transposon Database
4. Conclusions and Future Directions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kapitonov, V.V.; Jurka, J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 2008, 9, 411–412. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar]
- Bao, W.; Jurka, M.G.; Kapitonov, V.V.; Jurka, J. New superfamilies of eukaryotic DNA transposons and their internal divisions. Mol. Biol. Evol. 2009, 26, 983–993. [Google Scholar] [CrossRef]
- Bao, W.; Kapitonov, V.V.; Jurka, J. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob. DNA 2010, 1, 3. [Google Scholar] [CrossRef]
- Cerbin, S.; Wai, C.M.; VanBuren, R.; Jiang, N. GingerRoot: A novel DNA transposon encoding integrase-related transposase in plants and animals. Genome Biol. Evol. 2019, 11, 3181–3193. [Google Scholar] [CrossRef]
- Craig, R.J. Replitrons: A major group of eukaryotic transposons encoding HUH endonuclease. Proc. Natl. Acad. Sci. USA 2023, 120, e2301424120. [Google Scholar] [CrossRef] [PubMed]
- Vassilieff, H.; Haddad, S.; Jamilloux, V.; Choisne, N.; Sharma, V.; Giraud, D.; Wan, M.; Serfraz, S.; Geering, A.D.W.; Teycheney, P.Y.; et al. CAULIFINDER: A pipeline for the automated detection and annotation of caulimovirid endogenous viral elements in plant genomes. Mob. DNA 2022, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Richert-Pöggeler, K.R.; Noreen, F.; Schwarzacher, T.; Harper, G.; Hohn, T. Induction of infectious petunia vein clearing (pararetro) virus from endogenous provirus in petunia. EMBO J. 2003, 22, 4836–4845. [Google Scholar] [CrossRef] [PubMed]
- Havecker, E.R.; Gao, X.; Voytas, D.F. The diversity of LTR retrotransposons. Genome Biol. 2004, 5, 225. [Google Scholar] [CrossRef] [PubMed]
- Witte, C.P.; Le, Q.H.; Bureau, T.; Kumar, A. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc. Natl. Acad. Sci. USA 2001, 98, 13778–13783. [Google Scholar] [CrossRef]
- Gao, D.; Li, Y.; Do Kim, K.; Abernathy, B.; Jackson, S.A. Landscape and evolutionary dynamics of terminal repeat retrotransposons in miniature in plant genomes. Genome Biol. 2016, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Bureau, T.E.; Wessler, S.R. Tourist: A large family of small inverted repeat elements frequently associated with maize genes. Plant Cell. 1992, 4, 1283–1294. [Google Scholar]
- Bennetzen, J.L.; Wang, H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef]
- Gao, D.; Jiang, N.; Wing, R.A.; Jiang, J.; Jackson, S.A. Transposons play an important role in the evolution and diversification of centromeres among closely related species. Front. Plant Sci. 2015, 6, 216. [Google Scholar] [CrossRef]
- Serrato-Capuchina, A.; Matute, D.R. The role of transposable elements in speciation. Genes 2018, 9, 254. [Google Scholar] [CrossRef]
- Gao, D.; Gill, N.; Kim, H.; Walling, J.G.; Zhang, W.; Fan, C.; Yu, Y.; Ma, J.; SanMiguel, P.; Jiang, N.; et al. A lineage-specific centromere retrotransposon in Oryza brachyantha. Plant J. 2009, 60, 820–831. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Jamilloux, V.; Daron, J.; Choulet, F.; Quesneville, H. De novo annotation of transposable elements: Tackling the fat genome issue. Proc. IEEE 2016, 105, 474–481. [Google Scholar] [CrossRef]
- Yandell, M.; Ence, D. A beginner’s guide to eukaryotic genome annotation. Nat. Rev. Genet. 2012, 13, 329. [Google Scholar] [CrossRef] [PubMed]
- Jangam, D.; Feschotte, C.; Betrán, E. Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet. 2017, 33, 817–831. [Google Scholar] [CrossRef] [PubMed]
- TE Hub Consortium; Elliott, T.A.; Heitkam, T.; Hubley, R.; Quesneville, H.; Suh, A.; Wheeler, T.J. TE Hub: A community-oriented space for sharing and connecting tools, data, resources, and methods for transposable element annotation. Mob. DNA 2021, 12, 16. [Google Scholar]
- Lerat, E. Identifying repeats and transposable elements in sequenced genomes: How to find your way through the dense forest of programs. Heredity 2010, 104, 520–533. [Google Scholar] [CrossRef]
- Storer, J.M.; Hubley, R.; Rosen, J.; Smit, A.F.A. Methodologies for the de novo discovery of transposable element families. Genes 2022, 13, 709. [Google Scholar] [CrossRef]
- Mokhtar, M.M.; Alsamman, A.M.; El Allali, A. PlantLTRdb: An interactive database for 195 plant species LTR-retrotransposons. Front. Plant Sci. 2023, 14, 1134627. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.R.; Eddy, S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002, 12, 1269–1276. [Google Scholar] [CrossRef]
- McCarthy, E.M.; McDonald, J.F. LTR STRUC: A novel search and identification program for LTR retrotransposons. Bioinformatics 2003, 19, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, H. LTR-FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007, 35, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Ellinghaus, D.; Kurtz, S.; Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 2008, 9, 18. [Google Scholar] [CrossRef]
- Steinbiss, S.; Willhoeft, U.; Gremme, G.; Kurtz, S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 2009, 37, 7002–7013. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.; Jiang, N. LTR_retriever: A highly accurate and sensitive program for identification of long terminal-repeat retrotransposons. Plant Physiol. 2017, 176, 1410–1422. [Google Scholar] [CrossRef]
- Orozco-arias, S.; Liu, J.; Id, R.T.; Ceballos, D.; Silva, D.; Id, D.; Ming, R.; Guyot, R. Inpactor, integrated and parallel analyzer and classifier of LTR retrotransposons and its application for pineapple LTR retrotransposons diversity and dynamics. Biology 2018, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Wenke, T.; Döbel, T.; Sörensen, T.R.; Junghans, H.; Weisshaar, B.; Schmidt, T. Targeted identification of short interspersed nuclear element families shows their widespread existence and extreme heterogeneity in plant genomes. Plant Cell 2011, 23, 3117–3128. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, N.; Sun, Y. AnnoSINE: A short interspersed nuclear elements annotation tool for plant genomes. Plant Physiol. 2022, 188, 955–970. [Google Scholar] [CrossRef] [PubMed]
- Gremme, G.; Steinbiss, S.; Kurtz, S. GenomeTools: A comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013, 10, 645–656. [Google Scholar] [CrossRef]
- Su, W.; Gu, X.; Peterson, T. TIR-Learner, a New Ensemble Method for TIR Transposable Element Annotation, Provides Evidence for Abundant New Transposable Elements in the Maize Genome. Mol. Plant. 2019, 12, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Liang, C. Generic Repeat Finder: A High-Sensitivity Tool for Genome-Wide De Novo Repeat Detection. Plant Physiol. 2019, 180, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wessler, S.R. MITE-Hunter: A program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010, 38, e199. [Google Scholar] [CrossRef]
- Ye, C.; Ji, G.; Liang, C. detectMITE: A novel approach to detect miniature inverted repeat transposable elements in genomes. Sci. Rep. 2016, 6, 19688. [Google Scholar] [CrossRef]
- Hu, J.; Zheng, Y.; Shang, X. MiteFinderII: A novel tool to identify miniature inverted-repeat transposable elements hidden in eukaryotic genomes. BMC Med. Genom. 2018, 11, 101. [Google Scholar] [CrossRef]
- Crescente, J.M.; Zavallo, D.; Helguera, M.; Vanzetti, L.S. MITE Tracker: An accurate approach to identify miniature inverted-repeat transposable elements in large genomes. BMC Bioinform. 2018, 19, 348. [Google Scholar] [CrossRef]
- Xiong, W.; He, L.; Lai, J.; Dooner, H.K.; Du, C. HelitronScanner uncovers a large overlooked cache of Helitron transposons in many plant genomes. Proc. Natl. Acad. Sci. USA 2014, 111, 10263–10268. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef]
- Garcia, T.; Duitama, J.; Zullo, S.S.; Gil, J.; Ariani, A.; Dohle, S.; Palkovic, A.; Skeen, P.; Bermudez-Santana, C.I.; Debouck, D.G.; et al. Comprehensive genomic resources related to domestication and crop improvement traits in Lima bean. Nat. Commun. 2021, 12, 702. [Google Scholar] [CrossRef]
- El Baidouri, M.; Kim, K.D.; Abernathy, B.; Arikit, S.; Maumus, F.; Panaud, O.; Meyers, B.C.; Jackson, S.A. A new approach for annotation of transposable elements using small RNA mapping. Nucleic Acids Res. 2015, 43, e84. [Google Scholar] [CrossRef]
- Rho, M.; Tang, H. MGEScan-Non-LTR: Computational Identification and Classification of Autonomous Non-LTR Retrotransposons in Eukaryotic Genomes. Nucleic Acids Res. 2009, 37, e143. [Google Scholar] [CrossRef]
- Malik, H.S.; Eickbush, T.H. The RTE class of non-LTR retrotransposons is widely distributed in animals and is the origin of many SINEs. Mol. Biol. Evol. 1998, 15, 1123–1134. [Google Scholar] [CrossRef]
- Gao, D.; Chu, Y.; Xia, H.; Xu, C.; Heyduk, K.; Abernathy, B.; Ozias-Akins, P.; Leebens-Mack, J.H.; Jackson, S.A. Horizontal Transfer of Non-LTR Retrotransposons from Arthropods to Flowering Plants. Mol. Biol. Evol. 2018, 35, 354–364. [Google Scholar] [CrossRef]
- Deragon, J.M.; Zhang, X. Short interspersed elements (SINEs) in plants: Origin, classification, and use as phylogenetic markers. Syst. Biol. 2006, 55, 949–956. [Google Scholar] [CrossRef]
- Umeda, M.; Ohtsubo, H.; Ohtsubo, E. Diversification of the rice Waxy gene by insertion of mobile DNA elements into introns. Jpn. J. Genet. 1991, 66, 569–586. [Google Scholar] [CrossRef]
- Yasui, Y.; Nasuda, S.; Matsuoka, Y.; Kawahara, T. The Au family, a novel short interspersed element (SINE) from Aegilops umbellulate. Theor. Appl. Genet. 2001, 102, 463–470. [Google Scholar] [CrossRef]
- Mao, H.; Wang, H. SINE_scan: An efficient tool to discover short interspersed nuclear elements (SINEs) in large-scale genomic datasets. Bioinformatics 2017, 33, 743–745. [Google Scholar] [CrossRef]
- Temin, H.M. Reverse transcription in the eukaryotic genome: Retroviruses, pararetroviruses, retrotransposons and retrotranscripts. Mol. Biol. Evol. 1985, 2, 455–468. [Google Scholar] [PubMed]
- Richert-Poggeler, K.R.; Shepherd, R.J. Petunia vein clearing virus: A plant pararetrovirus with the core sequence of an integrase function. Virology 1997, 236, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Teycheney, P.-Y.; Geering, A.D.W.; Dasgupta, I.; Hull, R.; Kreuze, J.F.; Lockhart, B.; Muller, E.; Olszewski, N.; Pappu, H.; Pooggin, M.; et al. ICTV Virus taxonomy profile: Caulimoviridae. J. Gen. Virol. 2020, 101, 1025–1026. [Google Scholar] [CrossRef]
- Jakowitsch, J.; Mette, M.F.; van Der Winden, J.; Matzke, M.A.; Matzke, A.J. Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants. Proc. Natl. Acad. Sci. USA 1999, 96, 13241–13246. [Google Scholar] [CrossRef] [PubMed]
- Geering, A.D.W.; Maumus, F.; Copetti, D.; Choisne, N.; Zwickl, D.J.; Zytnicki, M.; McTaggart, A.R.; Scalabrin, S.; Vezzulli, S.; Wing, R.A.; et al. Endogenous florendoviruses are major components of plant genomes and hallmarks of virus evolution. Nat. Commun. 2014, 5, 5269. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.S. Characterization of a Mutator system in maize. Mutat. Res. 1978, 51, 21–28. [Google Scholar] [CrossRef]
- Feschotte, C.; Pritham, E.J. DNA transposons and the evolution of the eukaryotic genomes. Annu. Rev. Genet. 2007, 41, 331–368. [Google Scholar] [CrossRef]
- Yu, Z.; Wright, S.I.; Bureau, T.E. Mutator-like elements in Arabidopsis thaliana. Structure, diversity and evolution. Genetics 2000, 156, 2019–2031. [Google Scholar] [CrossRef]
- Lisch, D. Mutator transposons. Trends Plant Sci. 2002, 7, 498–504. [Google Scholar] [CrossRef]
- Gao, D.; Caspersen, A.M.; Hu, G.; Bockelman, H.E.; Chen, X. A novel mutator-like transposable elements with unusual structure and recent transpositions in barley (Hordeum vulgare). Front. Plant Sci. 2022, 13, 904619. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Bao, Z.; Zhang, X.; Eddy, S.R.; Wessler, S.R. Pack-MULE transposable elements mediate gene evolution in plants. Nature 2004, 431, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Peterson, P.A. A mutable pale green locus in maize. Genetics 1953, 38, 682–683. [Google Scholar]
- McClintock, B. Mutations in maize and chromosomal aberrations in Neurospora. In Annual Report of the Director of the Department of Genetics, Carnegie Institution of Washington Year Book No. 53, 1953–1954; Carnegie Institution of Washington: Cold Spring, NY, USA, 1954; pp. 254–260. [Google Scholar]
- Zabala, G.; Vodkin, L. A putative autonomous 20.5 kb-CACTA transposon insertion in an F3′H. allele identifies a new CACTA transposon subfamily in Glycine max. BMC Plant Biol. 2008, 8, 124. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, S.; Nitasaka, E. Characterization of Tpn1 family in the Japanese morning glory: En/Spm-related transposable elements capturing host genes. Plant Cell Physiol. 2004, 45, 933–944. [Google Scholar] [CrossRef] [PubMed]
- McCLINTOCK, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef]
- Calvi, B.R.; Hong, T.J.; Findley, S.D.; Gelbart, W.M. Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: Hobo, Activator, and Tam3. Cell 1991, 66, 465–471. [Google Scholar] [CrossRef]
- Atkinson, P.W. hAT transposable elements. Microbiol. Spectr. 2015, 3, 773–800. [Google Scholar] [CrossRef]
- Rubin, E.; Lithwick, G.; Levy, A.A. Structure and evolution of the hAT transposon superfamily. Genetics 2001, 158, 949–957. [Google Scholar] [CrossRef]
- Essers, L.; Adolphs, R.H.; Kunze, R. A highly conserved domain of the maize activator transposase is involved in dimerization. Plant Cell 2000, 12, 211–224. [Google Scholar] [CrossRef]
- Zhang, X.; Feschotte, C.; Zhang, Q.; Jiang, N.; Eggleston, W.B.; Wessler, S.R. P Instability Factor: An Active Maize Transposon System Associated with the Amplification of Tourist-like MITEs and a New Superfamily of Transposases. Proc. Natl. Acad. Sci. USA 2001, 98, 12572–12577. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica 1999, 107, 27–37. [Google Scholar] [CrossRef]
- Jiang, N.; Bao, Z.; Zhang, X.; Hirochika, H.; Eddy, S.; McCouch, S.R.; Wessler, S.R. An active DNA transposon family in rice. Nature 2003, 421, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Velanis, C.N.; Perera, P.; Thomson, B.; de Leau, E.; Liang, S.C.; Hartwig, B.; Förderer, A.; Thornton, H.; Arede, P.; Chen, J.; et al. The domesticated transposase ALP2 mediates formation of a novel Polycomb protein complex by direct interaction with MSI1, a core subunit of Polycomb Repressive Complex 2 (PRC2). PLoS Genet. 2020, 16, e1008681. [Google Scholar] [CrossRef]
- Mao, D.; Tao, S.; Li, X.; Gao, D.; Tang, M.; Liu, C.; Wu, D.; Bai, L.; He, Z.; Wang, X.; et al. The Harbinger transposon-derived gene PANDA epigenetically coordinates panicle number and grain size in rice. Plant Biotechnol. J. 2022, 20, 1154–1166. [Google Scholar] [CrossRef] [PubMed]
- Emmons, S.W.; Yesner, L.; Ruan, K.; Katzenberg, D. Evidence for a transposon in Caenorhabditis elegans. Cell 1983, 32, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, J.W.; Medhora, M.M.; Hartl, D.L. Molecular structure of a somatically unstable transposable element in Drosophila. Proc. Natl. Acad. Sci. USA 1986, 83, 8684–8688. [Google Scholar] [CrossRef] [PubMed]
- Dupeyron, M.; Baril, T.; Bass, C.; Hayward, A. Phylogenetic analysis of the Tc1/mariner superfamily reveals the unexplored diversity of pogo-like elements. Mob. DNA 2020, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, G. Tc1-like transposable elements in plant genomes. Mob. DNA 2014, 5, 17. [Google Scholar] [CrossRef]
- Wells, D.J. Tdd-4, a DNA transposon of Dictyostelium that encodes proteins similar to LTR retroelement integrases. Nucleic Acids Res. 1999, 27, 2408–2415. [Google Scholar] [CrossRef] [PubMed]
- Glockner, G.; Szafranski, K.; Winckler, T.; Dingermann, T.; Quail, M.A.; Cox, E.; Eichinger, L.; Noegel, A.A.; Rosenthal, A. The complex repeats of Dictyostelium discoideum. Genome Res. 2001, 11, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Zhao, D.; Abernathy, B.; Iwata-Otsubo, A.; Herrera-Estrella, A.; Jiang, N.; Jackson, S.A. Dynamics of a novel highly repetitive CACTA family in common bean (Phaseolus vulgaris). G3 2016, 6, 2091–2101. [Google Scholar] [CrossRef] [PubMed]
- Boutanaev, A.M.; Osbourn, A.E. Multigenome analysis implicates miniature inverted-repeat transposable elements (MITEs) in metabolic diversification in eudicots. Proc. Natl. Acad. Sci. USA 2018, 115, E6650–E6658. [Google Scholar] [CrossRef] [PubMed]
- Jurka, J.; Kapitonov, V.V. PIFs Meet Tourists and Harbingers: A Superfamily Reunion. Proc. Natl. Acad. Sci. USA 2001, 98, 12315–12316. [Google Scholar] [CrossRef] [PubMed]
- Grzebelus, D.; Lasota, S.; Gambin, T.; Kucherov, G.; Gambin, A. Diversity and structure of PIF/Harbinger-like elements in the genome of Medicago truncatula. BMC Genom. 2007, 8, 409. [Google Scholar] [CrossRef]
- Deprá, M.; Ludwig, A.; Valente, V.L.; Loreto, E.L. Mar, a MITE family of hAT transposons in Drosophila. Mob. DNA 2012, 3, 13. [Google Scholar] [CrossRef]
- Warburton, P.E.; Giordano, J.; Cheung, F.; Gelfand, Y.; Benson, G. Inverted repeat structure of the human genome: The X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004, 14, 1861–1869. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 2001, 98, 8714–8719. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Fefelova, N.; Caronna, J.; He, L.; Dooner, H.K. The Polychromatic Helitron Landscape of the Maize Genome. Proc. Natl. Acad. Sci. USA 2009, 106, 19916–19921. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.; Su, W.; Liao, Y.; Chougule, K.; Agda, J.R.A.; Hellinga, A.J.; Lugo, C.S.B.; Elliott, T.A.; Ware, D.; Peterson, T.; et al. Benchmarking Transposable Element Annotation Methods for Creation of a Streamlined, Comprehensive Pipeline. Genome Biol. 2019, 20, 275. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef]
- Riehl, K.; Riccio, C.; Miska, E.A.; Hemberg, M. TransposonUltimate: Software for transposon classification, annotation and detection. Nucleic Acids Res. 2022, 50, e64. [Google Scholar] [CrossRef] [PubMed]
- Baril, T.; Imrie, R.M.; Hayward, A. Earl Grey: A fully automated user-friendly transposable element annotation and analysis pipeline. bioRxiv 2022. [CrossRef]
- Llorens, C.; Futami, R.; Covelli, L.; Domínguez-Escribá, L.; Viu, J.M.; Tamarit, D.; Aguilar-Rodríguez, J.; Vicente-Ripolles, M.; Fuster, G.; Bernet, G.P.; et al. The Gypsy Database (GyDB) of mobile genetic elements: Release 2.0. Nucleic Acids Res. 2011, 39, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Flutre, T.; Duprat, E.; Feuillet, C.; Quesneville, H. Considering transposable element diversification in de novo annotation approaches. PLoS ONE 2011, 6, 0016526. [Google Scholar] [CrossRef]
- Hayward, A.; Gilbert, C. Transposable elements. Curr. Biol. 2022, 32, R904–R909. [Google Scholar] [CrossRef]
- Neumann, P.; Novák, P.; Hoštáková, N.; Macas, J. Systematic Survey of Plant LTR-Retrotransposons Elucidates Phylogenetic Relationships of Their Polyprotein Domains and Provides a Reference for Element Classification. Mob. DNA 2019, 10, 1. [Google Scholar] [CrossRef]
- Goubert, C.; Craig, R.J.; Bilat, A.F.; Peona, V.; Vogan, A.A.; Protasio, A.V. A beginner’s guide to manual curation of transposable elements. Mob. DNA 2022, 13, 7. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- SanMiguel, P.; Tikhonov, A.; Jin, Y.K.; Motchoulskaia, N.; Zakharov, D.; Melake-Berhan, A.; Springer, P.S.; Edwards, K.J.; Lee, M.; Avramova, Z.; et al. Nested retrotransposons in the intergenic regions of the maize genome. Science 1996, 274, 765–768. [Google Scholar] [CrossRef]
- Kronmiller, B.A.; Wise, R.P. TEnest: Automated chronological annotation and visualization of nested plant transposable elements. Plant Physiol. 2008, 146, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Buisine, N.; Quesneville, H.; Colot, V. Improved detection and annotation of transposable elements in sequenced genomes using multiple reference sequence sets. Genomics 2008, 91, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Tanskanen, J.; Chang, W.; Antonius, K.; Sela, H.; Peleg, O.; Schulman, A.H. Cassandra retrotransposons carry independently transcribed 5S RNA. Proc. Natl. Acad. Sci. USA 2008, 105, 5833–5838. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, D. Introduction of Plant Transposon Annotation for Beginners. Biology 2023, 12, 1468. https://doi.org/10.3390/biology12121468
Gao D. Introduction of Plant Transposon Annotation for Beginners. Biology. 2023; 12(12):1468. https://doi.org/10.3390/biology12121468
Chicago/Turabian StyleGao, Dongying. 2023. "Introduction of Plant Transposon Annotation for Beginners" Biology 12, no. 12: 1468. https://doi.org/10.3390/biology12121468
APA StyleGao, D. (2023). Introduction of Plant Transposon Annotation for Beginners. Biology, 12(12), 1468. https://doi.org/10.3390/biology12121468




