Simple Summary
Fly maggots are nutrient-rich, especially with regard to their protein content, making them an ideal replacement for the more expensive fishmeal. We aimed to analyse the effects of different treatments (i.e., fresh fly and fermented fly maggot protein feed, and fermented fly maggot protein feed with high-temperature pelleting) of fly maggot protein feed on the growth and intestinal flora of Pacific white shrimp (Litopenaeus vannamei). Fresh fly maggot protein in the feed was detrimental to shrimp growth, whereas fermented and high-temperature pelleted fly maggot protein feeds improved shrimp growth and survival. In addition, the fermented and high-temperature pelleted fly maggot protein feed increased the abundance of beneficial bacteria in shrimp intestines and reduced the growth of harmful bacteria. In contrast, fresh fly maggot proteins led to invasion by Vibrio as well as antibiotic-resistant bacteria. This study contributes to our understanding of the regulatory effects of different treatments on the growth and intestinal microflora of L. vannamei and provides a basis for the use of fly maggots as a source of protein in the context of aquaculture.
Abstract
As the intensive development of aquaculture persists, the demand for fishmeal continues to grow; however, since fishery resources are limited, the price of fishmeal remains high. Therefore, there is an urgent need to develop new sources of protein. They are rich in proteins, fatty acids, amino acids, chitin, vitamins, minerals, and antibacterial substances. Maggot meal-based diet is an ideal source of high-quality animal protein and a new type of protein-based immune enhancer with good application prospects in animal husbandry and aquaculture. In the present study, we investigated the effects of three different diets containing maggot protein on the growth and intestinal microflora of Litopenaeus vannamei. The shrimp were fed either a control feed (no fly maggot protein added), FM feed (compound feed with 30% fresh fly maggot protein added), FF feed (fermented fly maggot protein), or HT feed (high-temperature pelleted fly maggot protein) for eight weeks. The results showed that fresh fly maggot protein in the feed was detrimental to shrimp growth, whereas fermented and high-temperature-pelleted fly maggot protein improved shrimp growth and survival. The effects of different fly maggot protein treatments on the intestinal microbiota of L. vannamei also varied. Fermented fly maggot protein feed and high-temperature-pelleted fly maggot protein feed increased the relative abundance of Ruegeria and Pseudomonas, which increased the abundance of beneficial bacteria and thus inhibited the growth of harmful bacteria. In contrast, fresh fly maggot proteins alter the intestinal microbiome, disrupting symbiotic relationships between bacteria, and causing invasion by Vibrio and antibiotic-resistant bacteria. These results suggest that fresh fly maggot proteins affect the composition of intestinal microorganisms, which is detrimental to the intestinal tract of L. vannamei, whereas fermented fly maggot protein feed affected the growth of L. vannamei positively by improving the composition of intestinal microorganisms.
1. Introduction
The Pacific white shrimp Litopenaeus vannamei is a globally important aquatic species and one of the most productive shrimp species, providing a high-quality protein source for the human diet. Shrimp are omnivorous and carnivorous animals, and proteins in the compound feed are the primary nutrients that determine shrimp growth, development, and performance [1]. To meet the protein demand of aquatic animals, protein levels in general commercial formulations usually account for 25–50%. Accompanied by a continuous expansion of their feeding scale, the demand for fishmeal in the feed has increased continuously. Due to the decline in fish meal production, rising prices, and unstable supply caused by factors such as declining wild resources and climate change, the search for alternative protein sources of good quality and abundance to replace fish meal has become a popular research topic in recent years. However, with the increasing scale and intensification of farming, shrimp diseases have become more frequent, causing serious economic losses to the shrimp farming industry. Although chemical drugs and antibiotics affect disease prevention and treatment, they may lead to negative effects, such as environmental pollution, drug residues, generation of resistant strains, food safety risks, and risks to human health. Therefore, functional feed research aims to replace fish meal proteins and improve the nutrition, immunity, and organismal defence of shrimp, which are the key areas of research for aquafeed companies.
The larvae of the house fly (Musca domestica) and black gadfly (Hermetia illucens L.) (commonly known as fly maggots) have the advantages of large numbers, wide distribution, strong reproductive capacity, and low rearing costs. The crude protein content of the dry matter obtained after processing the larvae is close to fish meal (as high as 58.80%–63.89%) [2], which has approximately the same protein content as fish meal [3], and fly maggots are rich in protein, fatty acids, amino acids, chitin, vitamins, minerals, and antimicrobial active substances [2]. Maggot meal-based diets not only serve as a desirable animal protein feed source but also as a new type of protein-based immune enhancer with good application prospects in livestock and aquaculture [4,5]. Recent studies have shown that fly maggots can improve the growth performance of L. vannamei, including body composition [6] fatness, and survival [7], and increase immune enzyme activity [8] and immunity [9]. Furthermore, fly maggot protein improves appetite, weight gain, survival rate, and immune function in Trionyx sinensis [10] and tilapia (Oreochromis niloticus) [11]. Several studies [12] have shown that partially replacing fish meal with fly maggot meal as artificial bait improves the total feed consumption, feed conversion efficiency, protein efficiency ratio, specific growth rate, and survival rate of common carp (Cyprinus carpio L.). In contrast, the use of fly maggot protein as a substitute for fish meal in the rearing of clariid catfish (Clarias gariepinus) had no substantial effect on growth performance [13]. These findings suggest that fly maggots can partially replace fish meal in commercial feed to improve the growth performance and immunity of aquaculture animals. However, the effects on these functions are closely related to the manner in which the feed is used. Although the direct ingestion of fresh fly maggots is better for nutritional utilisation, some fly maggots float on the water surface and are not easily ingested, which makes them easy to pollute aquaculture water in the long term. Fly maggot powder is crushed after high-temperature drying, which destroys its active substances and immunomodulatory functions. Fly maggot antibacterial peptides are extremely expensive after being processed using extraction, separation, and drying processes [6]. Therefore, a less costly method for treating fly maggot proteins is required to fully utilise their benefits.
With the ban on antibiotics, research on alternative products has been receiving attention in the livestock, poultry, and aquaculture industries, and microbially fermented feed is one such product. Many studies have shown that the use of fermented feed can improve the content of feed proteins and vitamins [14], reduce mycotoxins and other toxic and harmful substances, and improve the safety and effectiveness of the feed and the growth performance [15]. Furthermore, it promotes the digestion and absorption of nutrients, improves the conversion rate of feed nutrients [14], balances intestinal bacteria, promotes intestinal peristalsis, and prevents diarrhoea and constipation [16]. Fermented feeds improve the diversity index of intestinal microflora and the abundance of Lactobacillus in shrimp, thereby improving the composition of the intestinal microflora of L. vannamei [17].
Several factors, including the food type and intestinal microbiota, influence shrimp growth. The intestine is the most important digestive and absorptive organ of shrimp, and diet strongly alters the composition of the intestinal microbiota, which may result in the production of microbiota metabolites [18]. The intestine is inhabited by several microorganisms that influence host digestion and immunity. Stable microbial communities can promote host health by producing beneficial metabolites and immune stimulation [19]. Therefore, intestinal microbial changes and growth performance can be used to assess the effects of fly maggot protein and fermented feed. Based on previous studies, we hypothesised that different treatments with fly maggot protein might affect the growth, survival, and intestinal microbial balance of L. vannamei. In this study, we compared the effects of regular compound feed, regular compound feed supplemented with 30% fresh fly maggot protein, fly maggot protein-fermented feed, and high-temperature-pelleted fly maggot protein feed on the growth of L. vannamei and evaluated the effects of different treatment methods on the application of fly maggot protein in shrimp feed using intestinal microflora analysis. Our results provide a basis for the application of fly maggot protein in the context of L. vannamei production.
2. Methods and Materials
2.1. Shrimp and Culture Conditions
L. vannamei were collected from a semi-intensive culture pond at the Shenzhen Base, South China Sea Fisheries Research Institute of the Chinese Academy of Fishery Sciences (Shenzhen, China). Before the experiment, the shrimp were acclimated to the experimental tanks for seven days. A total of 600 shrimp of uniform size, with an average weight of 3.970 ± 0.074 g, were selected for the experiment. The shrimp were randomly allocated to 12 glass-fibre-reinforced plastic water tanks with a capacity of 500 L (1.3 m × 1.0 × 0.4 m), each containing 300 L of water and 60 shrimp. The shrimp were cultured in filtered and aerated seawater (salinity 30‰, pH 8.1–8.2, temperature 30 ± 0.5 °C, and dissolved oxygen > 6.0 mg/L) with concentrations of ammonia-N < 0.1 mg/L and nitrite-N < 0.01 mg/L. Shrimp were fed commercial feed three times daily at 5% body weight.
2.2. Diet Preparation
The basic feed for this experiment was a common compound feed, and three experimental feeds were prepared: fresh fly maggots were cleaned, decontaminated, sterilised, cleaned using an ozone cell, and milled to obtain the fly maggot protein source (BoYuJia Biotechnology, Shenzhen, China). No fish meal was added to any of the experimental feeds, and the normal feed without fish meal was mixed with 30% fresh fly maggot protein, fly maggot protein-fermented feed (feed ingredients with 30% fresh fly maggot protein fermentation treatment, no high-temperature pelleting), or fly maggot protein pellet feed (feed ingredients with 30% fresh fly maggot protein fermentation feed, high-temperature pelleting). The feed formulation (Table S1), amino acid composition (Table S2), and nutritional composition (Table S3) are available in the supplementary materials. Methionine and cystine were hydrolysed using oxidative acid hydrolysis, and the rest of the amino acids were hydrolysed via acid hydrolysis. Each treatment was tested in triplicate to obtain the average value, and the routine nutrient composition of the feeds was tested using the AOAC method [20]. Among them, the crude protein content was determined using a KjeltecTM2300 Kjeldahl nitrogen meter (FOSS, Denmark); crude fibre content was determined from the weight of dried residue after extracting the feed with acid and alkali; calcium ion content was determined using disodium ethylenediaminetetraacetic acid complexometric titration; phosphorus content was determined using spectrophotometry; and crude ash content was determined using scorching in a Marfo oven at 550 °C. Moisture content was determined using the constant temperature drying method at 105 °C. All experimental feeds were stored below −20 °C before use. The feed was removed from the −20 °C refrigerator and left at room temperature for half an hour before feeding.
2.3. Experimental Design and Sample Collection
After seven days of acclimatisation, the shrimp were randomly divided into four groups: CK, FM, FF, and HT. Three replicate tanks of 50 shrimp each were used. The CK group was fed a common compound feed, the FM group was fed a diet containing 30% fresh fly maggot protein, the FF group was fed a fermented diet containing 30% fly maggot protein, and the HT group was fed a high-temperature pelletised fermented diet containing 30% fly maggot protein. The feeding trial lasted eight weeks. The four diets were fed at 3% shrimp body weight three times a day (08:00, 12:00, and 17:00). Shrimp intake was near saturation during the feeding trials. The shrimp were fed according to their appetite, and uneaten feed and faeces were cleared from the tanks 1 h after feeding.
After the eight-week feeding trial, the growth and survival of shrimp in each tank were assessed and randomly sampled. Considering inter-individual variations, five intestines from each tank were combined into one intestinal microbial sample and one gut microbial sample. All the samples were flash-frozen in liquid nitrogen prior to analysis.
2.4. Growth Performance Analysis
At the end of the experiment, the shrimp were weighed to determine the weight gain rate (WGR), specific growth rate (SGR), and survival rate (SR). The growth performance and survival of L. vannamei in all the groups were calculated using the following equations [21]:
Weight gain rate (%) = 100 × (final weight − initial weight)/initial weight.
Specific growth rate (%/d) = 100 × (ln end weight − ln initial weight)/number of days of feeding
Survival rate (%) = 100 × (initial number of prawns − number of dead prawns)/initial number of prawns
2.5. DNA Extraction, PCR Amplification, and Sequencing
Microbial DNA from intestinal samples was extracted using the Stool Genomic DNA Extraction Kit (DP712) (TIANGEN, Beijing, China)and the magnetic bead soil method, according to the manufacturer’s protocols. A NanoDrop 2000 UV-vis spectrophotometre (Thermo Scientific, Wilmington, DE, USA) was used to determine the DNA concentration and purification. The amplification of the hypervariable V3-V4 region of the bacterial 16S rRNA gene was conducted using a thermal cycling PCR system (GeneAmp 9700, ABI, Bio-Rad, Hercules, CA, USA) and the 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGGTATCTAAT-3′) primers. The PCR cycling parameters were as follows: pre-denaturation at 95 °C for 3 min; 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, an extension at 72 °C for 45 s, and a final extension at 72 °C for 10 min (PCR instrument: ABI GeneAmp 9700, Bio-Rad, USA). The PCR reactions (20 μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA) were performed in triplicate. The PCR products were recovered using a 2% agarose gel, purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA), eluted in Tris-HCl, and detected by 2% agarose electrophoresis. The detection and quantification were conducted using QuantiFluor ~ST (Promega, Madison, WI, USA). Purified amplified segments were used for library construction following standard operating procedures for the Illumina MiSeq platform (Illumina, San Diego, CA, USA). Sequencing was performed using the Illumina MiSeq PE300 platform.
2.6. Bioinformatics Analysis
First, QIIME2 was used for the quality control of mouse microbiome sequencing data to detect variability in amplicon sequences [22]. Next, we obtained classification tables for species annotation by comparing the current ASV sequences with those in the Greengenes (16 S rRNA) database [23]. Each sequence in the Silva 132 database was analysed for classification using the RDP classifier algorithm, and only results with a confidence level above 70% were considered. Based on the results of a recent study, the mi-biome diversity of four groups of experimental animals was analysed by evaluating α-diversity indices (including Chao1, Simpson, and Shannon indices) as well as β-diversity metrics (using the principal coordinate analysis method) [24]. Linear discriminant analysis (LDA) and effect size (LEfSe) analyses were performed to identify factors with notable effects by identifying bacterial taxa that differed between the groups. In this analysis, the LDA score threshold of 3.0. was considered significant [25]. In addition, network analysis was used to reveal the differences in the microbiota among the four shrimp groups [4,26,27]. Ultimately, using the PICRUSt and MetaCyc databases for annotation, we predicted the potential function of KEGG homologues (KO) that may be present in shrimp microbiota [28]. Unless otherwise indicated, the parameters used in these analyses were set to default [23].
2.7. Correlations between Intestine Bacteria and the Growth of Shrimp
Redundancy analysis (RDA) was performed using the R package ‘vegan’ to reveal potential associations between microbial communities and related environmental factors. Co-occurrence analysis was performed by calculating Spearman’s rank correlations among the taxa and network diagrams were used to show the associations among the taxa.
2.8. Statistical Analysis
The data were expressed as the mean ± SD of each variable. Statistical significance was determined using a one-way analysis of variance (ANOVA) and Duncan’s multiple range test (SPSS version 22.0). Statistical significance was set at p < 0.05. significant.
3. Results
3.1. The Growth and Survival of the Shrimp
Compared to the CK group, there were significant differences in WGR, SGR, and SR among the test groups (Table 1). In terms of weight gain rate, the HT group had a significantly higher WGR than the FM and FF groups (p < 0.05) but lower than that of the CK group (p > 0.05). The HT group had the highest SR (p < 0.05).

Table 1.
Changes in the growth performance and survival rate of Litopenaeus vannamei.
3.2. Intestine Microbial Richness and Diversity
A total of 920,715 good-quality sequences, averaging 50,591 sequences per sample, were obtained by analysing the intestine of L. vannamei. Venn diagram analysis showed that all groups had the same 169 operational taxonomic units (OTUs); the number of unique OTUs was the second highest in the HT and FM groups, and the lowest in the FF group (Figure 1A).
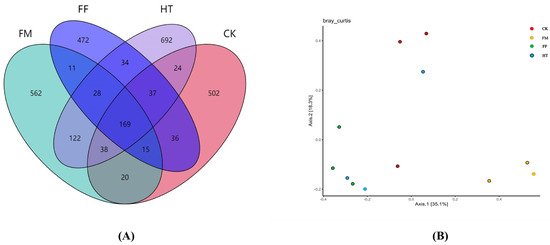
Figure 1.
Diversity and richness of intestinal microbial variability in Litopenaeus vannamei. (A) Venn diagram; (B) PCoA analysis on the basis of Bray-Curtis distance.
The alpha richness and plurality of intestinal microorganisms were evaluated using the Chao1, Simpson, and Shannon indices. Alpha diversity analysis showed that the Simpson and Shannon indices were significantly different among the groups (Table 2). The CK group had the lowest Chao1 index, and there were no significant differences among the experimental groups. In addition, the Shannon index of species diversity was lower in all the experimental groups compared to the HT group. There was no significant difference in the Simpson index between the FM and FF groups; however, the Simpson indices were similar and significantly higher in the CK and HT groups than in the FM and FF groups. The Shannon index of species diversity was lower in all the experimental groups than in the HT group. The Shannon index of species diversity was not significantly different between the FM and FF groups but was significantly lower in the HT group. The microbial samples from the FM and FF groups were clearly distinguishable based on the PCoA plot. The CK group could only be distinguished from the FM group and not from the other experimental groups (Figure 1B).

Table 2.
Changes in intestinal microbial alpha-diversity of Litopenaeus vannamei.
3.3. Intestinal Microbial Composition
Twenty bacterial phyla were identified in each group (Figure 2A). The bacterial composition of all groups consisted mainly of Proteobacteria, Bacteroidetes, Firmicutes, Tenericutes, and Verrucomicrobia. The bacterial composition of the intestine was dominated by Proteobacteria in all groups, with higher abundances in the FM (92.07%) and FF (91.07%) groups than in the CK group (70.33%). Notably, compared with the MC group, fly maggot supplementation decreased the relative abundance of Firmicutes.
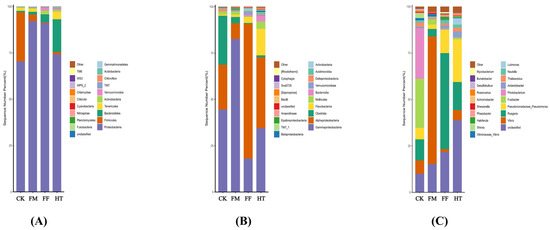
Figure 2.
Changes in the composition of the intestinal microbial community of Litopenaeus vannamei. (A) relative bacterial abundance at the phylum level; (B) relative bacterial abundance at the class level; (C) relative bacterial abundance at the genus level.
At the class level, intestinal bacterial composition was dominated by Alphaproteobacteria and Gammaproteobacteria (Figure 2B). Gammaproteobacteria was more abundant in the FM group than in the CK group. The abundance of Alphaproteobacteria in the FF group (72.76%) was lower than that in the CK group, whereas the groups in the CK group were divergent with no common dominant species.
At the genus level, compared to the CK group, Ruegeria abundance in the FF group (51.67%) and Vibrio in the FM group (68.80%) increased, and Pseudomonas, Antarctobacter, Nautella, and Lutimonas were higher in the HT group. Ruegeria abundance was higher in the FF group (51.67%) than in the HT group (15.14%) (Figure 2C).
3.4. Differential Analysis of Intestinal Microbiota
LEfSe was used to analyse the differential abundances of the three bacterial groups. Four bacterial taxa, Clostridiaceae (family), Campylobacteraceae (family), Camplobacterales (order), and Epsilonproteobacteria (family), were enriched in the FM group, three bacterial taxa, Actinomycetales (phylum), Actinobacteria (class), and Actinobacteria (phylum), were enriched in the FF group, and only one bacterial taxon, Peptostreptococcaceae (family), was enriched in the HT group (Figure 3A). Among the differential bacteria with LDA scores above 3.5, Clostridiaceae (family) dominated the FM group, whereas Actinomycetales (phylum), Actinobacteria (class), and Actinobacteria (phylum) dominated the FF group (Figure 3B).
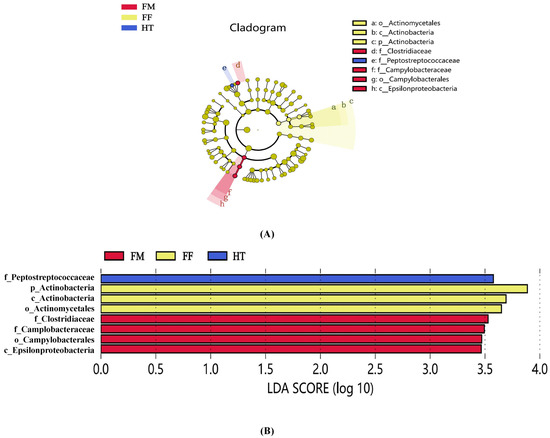
Figure 3.
Between-group variability of intestinal microorganisms of Litopenaeus vannamei. (A) In the LEfSe cladogram, the diameter of each circle is proportional to the bacterial taxon abundance. (B) LDA scores of LEfSe-PICRUSt, only the taxon with LDA values > 3.5 are shown. The lowercase letters in front of the strain name represent the taxonomic level at which the strain is located: k for kingdom, p for phylum, c for class, o for order, f for family, g for genus, and s for species.
3.5. The Correlation Network and Metabolic Analyses of Intestinal Microbiota
Microbial community interactions involve assemblages of microorganisms and their environments [29] that influence community stability and ecological characteristics [30]. In the bacterial phylum-based correlation network, Proteobacteria was negatively associated with Bacteroidetes, whereas Firmicutes was positively associated with Cyanobacteria. (Figure 4A). Vibrio and Reugeria had the highest abundance and were negatively correlated in the Bacteroidetes correlation network (Figure 4B).
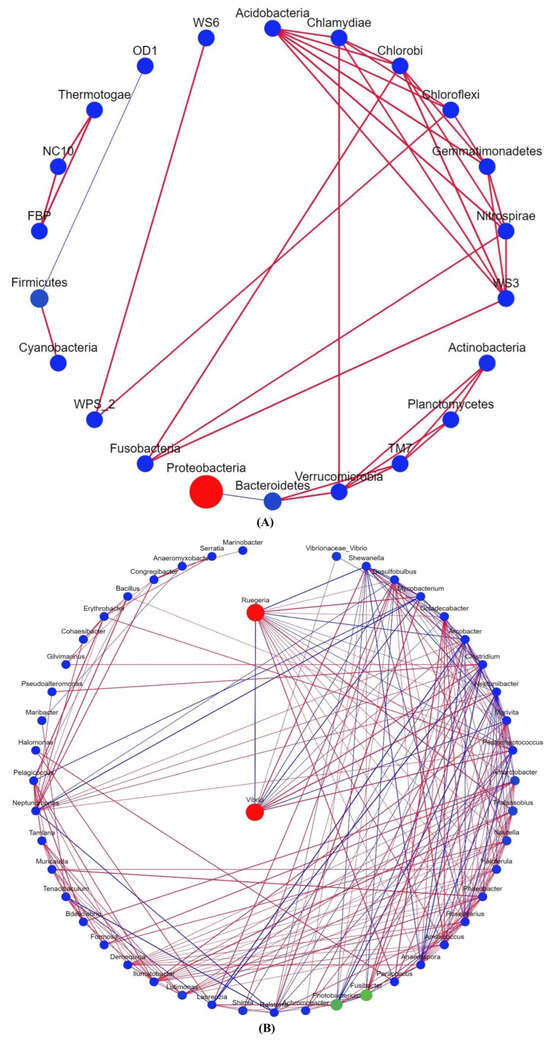
Figure 4.
Correlation network analyses of intestinal microbial of Litopenaeus vannamei. (A) The correlation network based on bacterial phyla. (B) The correlation network of bacteria genera. Circles indicate the species, size indicates its relative abundance, different colours indicate different species phylum classifications, the lines connecting the circles denote the significant correlation between the two species (p < 0.05), red lines represent positive correlations, and blue lines represent negative correlations. The thicker the line, the greater the absolute value of the correlation coefficient.
3.6. Treatment with Fly Maggot Protein Could Affect the Microbiome Function
Based on the random forest KEGG classification, the predicted functions of the intestinal microflora of L. vannamei were analysed using the PICRUSt 2. Functional categories were similar across the groups. Analysis of level 2 and 3 biometabolism in the KEGG functional annotation revealed 20 subfunctional components (Figure 5A,B).
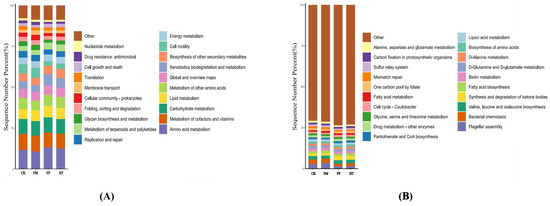
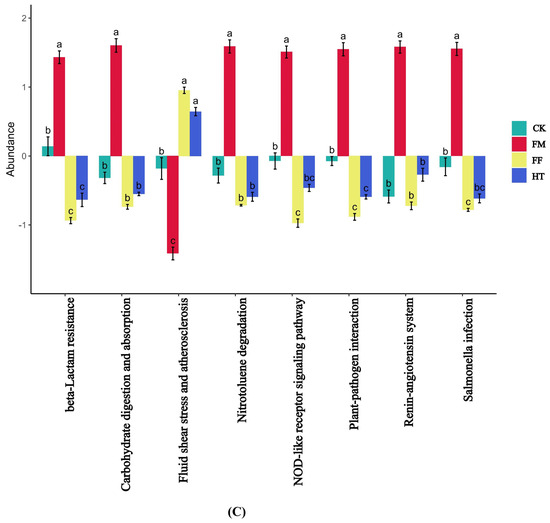
Figure 5.
Analysis of bacterial metabolism in the intestinal KEGG pathway of Litopenaeus vannamei based on fermented feed and fly maggot protein: (A) results of KEGG level 2 analysis; (B) results of KEGG level 3 analysis; (C) results of the predicted variability of the pathway groups under KEGG level 3. Different letters are used to show significant differences (p < 0.05).
Microbial metabolic functions affect L. vannamei growth. In this study, under the triple annotation depth of KEGG, the FF and HT groups showed some similarities in the metabolic functions of the intestinal microorganisms of shrimp after the intake of different treatments of fly maggot protein-fermented feeds. An analysis of microbial metabolic pathways in the intestine of shrimp in the FM group showed that there were significant differences between the shrimp that ingested fresh fly maggot protein and the remaining three groups in the above pathways. The metabolic pathways that were significantly upregulated were “beta-Lactam resistance”, “carbohydrate digestion and absorption”, “nitrotohuene degradation”, “NOD-like receptor signalling pathways”, “plant-pathogen interactions”, “renin-angiotensin system”, and “salmonella infection”, and “fluid shear stress and atherosclerosis” were significantly downregulated metabolic pathways. In contrast, the metabolic function of the intestinal microbes was not significantly altered in the FF and HT groups (Figure 5C).
3.7. Correlations between Intestine Bacteria and the Growth of Shrimp
The correlation between microorganisms with weight gain rate (WG) and SR was analysed using correlation heat maps. SR was positively correlated with the abundance of Lactobacillus, Microbulbifer, Rubellimicrobium, Marinoscillum, and Oscillospira and negatively correlated with Glaciecola, Hahella, and Bacteriovorax abundance. WG positively correlated with Burkholderia abundance and negatively correlated with Acidaminobacter abundance (Figure 6).
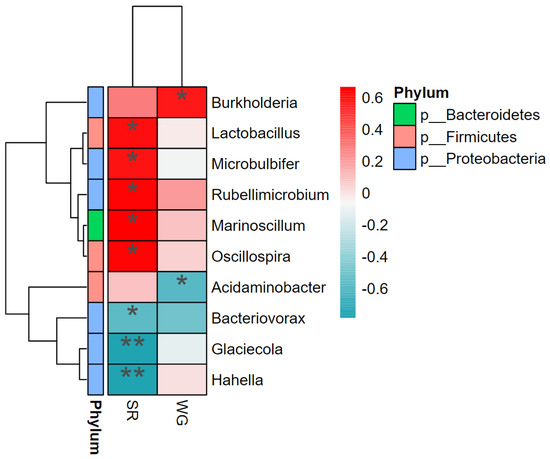
Figure 6.
Heat map of the interrelationship between microbial species and environmental factors (SR and WG) at the genus level. * Denotes significant correlation (*p < 0.05; **p < 0.01).
4. Discussion
The application of fly maggot protein in aquatic animals has been reported to enhance the survival and growth of aquatic animals and improve intestinal microbiota. However, the effects of fly maggot protein from different treatment methods on aquatic animals still need to be investigated. In the present study, high-temperature pelleted fly maggot protein-fermented feed increased the survival and weight gain rates of L. vannamei significantly. Regarding the application of fly maggot protein on L. vannamei, existing studies have also shown that fly maggot protein can improve the SR of L. vannamei [7]. However, in this study, although fly maggot protein was present in all the feeds of the experimental groups, the results obtained from the different treatments differed. In the present study, fly maggot protein was fermented at high temperatures after fermentation, ensuring that the same amount of fly maggot protein was supplied to the diet. Zhang et al. [31] reported that fermented feed promoted growth. The WG of shrimp fed either fly maggot protein-fermented feed or high-temperature pelleted fly maggot protein-fermented feed was significantly higher than that of the shrimp that were fed fresh fly maggot protein. This phenomenon suggests that fly maggot protein can significantly promote shrimp growth after fermentation and high-temperature treatments.
The functional activity and stability of the intestinal microbiota are important for shrimp health because they perform many functions related to immunity and pathogen resistance [32]. Previous studies have shown that the addition of fly maggot protein powder to fattening pig feed can inhibit the growth of harmful intestinal bacteria, promote the proliferation of beneficial bacteria, and effectively improve the digestion and absorption of nutrients [33]. These studies indicate that fly maggot proteins can alter the composition of intestinal microorganisms. Similarly, fly maggot proteins altered the relative abundance of the dominant intestinal bacteria in L. vannamei in this study, including an increase in the level of Proteobacteria in all experimental groups, Bacteroidetes in the HT group, and a decrease in the level of Firmicutes in all experimental groups. These results suggest that the changes in the relative abundance of the dominant bacteria produced by the fly maggot Proteobacteria varied among the treatments.
Some potentially beneficial bacterial genera, especially health-related host bacteria, differed between the groups. One group of potential probiotics is Roseobacter, which produces tropodithietic acid, which can kill or reduce the growth of several Vibrio pathogens in multiple aquaculture systems [34,35,36,37]. Kang et al. [38] found that Pseudomonas aeruginosa UCBPP-PA14 significantly lyses Microcystis aeruginosa and degrades microcystins. In this study, after the ingestion of fermented and high-temperature fly maggot protein feed by the shrimp, the relative abundances of Ruegeria and Pseudomonas were elevated, suggesting that fermented fly maggot protein feed can inhibit the growth of harmful bacteria by enhancing the abundance of beneficial bacteria and inhibiting the growth of harmful bacteria. In contrast, certain potentially harmful bacterial genera differed between the groups. Vibrio is a Gram-negative bacterium, and Vibrio parahaemolyticus infestation increases intestinal permeability and inhibits glucose and amino acid uptake by L. vannamei [39]. In the present study, the relative abundance of Vibrio was elevated in the FM group, suggesting that the ingestion of fresh fly maggot protein by shrimp may disrupt intestinal cell membrane permeability and be detrimental to nutrient uptake, thereby inhibiting shrimp growth. In addition, based on the bacterial correlation network, there was a positive correlation between the beneficial bacteria mentioned above, whereas Vibrio, a harmful bacterium, was negatively correlated with the beneficial bacteria. This was similar to the conclusions of Faust et al. [26]. Bacteria may cooperate to build biofilms that lead to antibiotic resistance among their members, resulting in mutually beneficial symbiotic relationships; conversely, negative interactions occur when metabolic by-products of one microorganism alter the environment to the detriment of other microorganisms [26]. Therefore, it is predicted that fly maggot protein-fermented feed can maintain the homeostasis of beneficial bacteria in the intestine of L. vannamei, which is beneficial for intestinal health. However, fresh fly maggot proteins alter the intestinal microbiome and disrupt the symbiotic relationship between the bacteria, resulting in the invasion of Vibrio and other pathogenic bacteria that may damage the intestine and affect shrimp health.
The metabolic functions of microorganisms influence L. vannamei growth. In this study, the results of KEGG pathway analysis based on the depth of triple annotation showed that significant differences in intestinal microbial metabolic functions occurred after the shrimp were fed different fly maggot protein diets. In this study, we found that the fly maggot protein significantly enhanced beta-lactam resistance in the intestinal microorganisms of L. vannamei. Beta-lactam antibiotics are widely used to inhibit bacterial growth by inhibiting the synthesis of bacterial cell walls [40] and destroying bacterial structure [41]. Antibiotic resistance genes are considered novel environmental contaminants [42] with a long-lasting high transmission frequency and harmfulness. In contrast, the beta-lactam resistance of the intestinal microorganisms was significantly reduced after feeding with fly maggot protein-fermented feed and fermented feed pelleted at high temperatures. Therefore, feeding fresh fly maggot proteins increases the abundance of antibiotic-resistant bacteria in the intestines of L. vannamei, which is detrimental to human health and ecological safety. In contrast, fermented feed significantly reduced the relative abundance of antibiotic-resistant bacteria in the shrimp, thereby mitigating environmental harm.
The intestinal tract is inhabited by numerous microorganisms that affect the host’s digestion and immunity. Stable microbial communities can promote host health by producing beneficial metabolites and immune stimulation [19]. In the present study, the growth of L. vannamei was assessed based on survival and WG rates. Beneficial bacterial genera play an important role in the healthy growth of shrimp. We found that the relative abundances of Lactobacillus, Micrococcus, Erythrobacter, Marinoscillum, and Oscillospira were significantly and positively correlated with survival rates. For example, lactic acid produced by Lactobacillus, a probiotic in fermented feeds, is an osmotic agent for the outer membrane of Gram-negative pathogens and increases the susceptibility of pathogenic bacteria to antimicrobial molecules [43]; on the other hand, the culture extracts of Microbulbifer also exhibit unique broad-spectrum antimicrobial activity against Gram-positive and Gram-negative bacteria and fungi [44]. Second, Rubellimobium spp. are representatives of the Roseobacter clade in Rhodobacteraceae [45], and as a probiotic for aquaculture, Roseobacter maintains its anti-pathogenic activity against pathogenic aquaculture cultures (live feeds and fish eggs/larvae) in the live antagonistic activity of Vibrio vulnificus [37]. Finally, Oscillospira produces short-chain fatty acids (SCFAs), which have been identified as next-generation probiotic candidates because of their positive effects on specific diseases, such as metabolic disorders related to obesity [46]. On the other hand, Marinoscillum, a member of the family “Flexibacteraceae” and the phylum “Bacteroidetes” [47,48], can inhibit the growth of Vibrio [49,50]. In contrast, Glaciecola, Hahella, and Bacteriovorax were significantly negatively correlated with survival. Glaciecola and Hahella are members of the “Gammaprotobacteria” class [51,52], but due to the production of metabolites that can specifically kill harmful algae [53], the role of Hahella in aquaculture requires further analysis. In addition, Bacteriovorax is a member of the phylum “Proteobacteria” based on the similarity of affinities [54]. The negative correlation between Bacteriovorax and shrimp survival is also consistent with the finding that Proteobacteria often act as the dominant bacteria in diseased shrimp [55].
Burkholderia was positively correlated with WG, and members of Burkholderia have both benefits and drawbacks, first as plant and human pathogens [56,57], but also through their own enzymes to degrade certain contaminants [58]. Therefore, the role of Burkholderia in L. vannamei needs to be analysed in greater detail. Based on the results of this study, Burkholderia cepacia has growth-promoting effects. The abundance of Acidaminobacter was negatively correlated with WG. Acidaminobacter is a member of Gammaproteobacteria [59], which includes pathogenic bacteria in aquaculture environments, such as Vibrio, which are detrimental to shrimp health and growth.
These findings also corroborate that probiotic bacteria are positively correlated with the survival and weight gain rates of the bacteria, whereas harmful bacteria are negatively correlated with the survival and weight gain rates of L. vannamei. In addition, by combining the intestinal microbial composition of each group, we found that most probiotic bacteria, such as Roseobacter, were present in the FF and HT groups, suggesting that protein-fermented fly maggot feed increased the relative abundance of beneficial bacteria in the intestinal tract of L. vannamei, which was beneficial to the health of the hosts. In contrast, most harmful bacteria, such as Vibrio, were from the FM group, which also suggests that fresh fly maggot protein can disrupt the homeostasis of the intestinal tract of shrimp, resulting in an increase in the abundance of harmful bacteria, thus jeopardising the health of shrimp. In addition, the abundance of Vibrio anguillarum in shrimp intestines increases linearly with the progression of acute hepatopancreatic necrotic disease [60], suggesting that fresh fly maggot proteins lead to an increase in the number of harmful bacteria, which may be detrimental to shrimp health. However, there are some limitations to this study. For example, the intestinal gut structure was not investigated, and the effects of fly maggot proteins produced in the intestines under different treatments were not analysed from a physiological point of view. In addition, gene expression and enzyme activities related to the healthy growth and development of L. vannamei were not explored in this study. Therefore, future studies should analyse the effects of fly maggot proteins on L. vannamei under different treatments from multiple perspectives, including gene expression, enzyme activity, and histology.
5. Conclusions
Overall, we found that fly maggot protein improved the growth and survival of L. vannamei after fermentation and high-temperature treatment. In addition, different treatments with fly maggot protein altered the relative abundance of dominant bacteria, particularly beneficial substance-producing bacteria (Ruegeria and Pseudomonas) and pathogenic bacteria (Vibrio). Different treatments with fly maggot protein also affected the metabolic functions of microorganisms in the shrimp intestines, and the resulting differences in the relative abundance of beneficial and harmful intestinal microorganisms were significantly correlated with weight gain and survival rates. These results suggest that differently treated fly maggot proteins have different effects on the microbial composition of the intestinal tract of L. vannamei, and rebuilt protein-fermented feeds are ideal for use in the context of feeding shrimp in a commercial setting.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology12111433/s1, Table S1: Feed formulation for each group; Table S2: Amino acid composition of each feed group; Table S3: Nutritional composition of groups of feeds.
Author Contributions
J.H.; conceived and designed the experiments. X.L. and L.Y.; writing—original draft, writing—review and editing. S.J. (Shigui Jiang); software and validation. F.Z.; supervision. S.J. (Song Jiang); methodology and software. Y.L.; data curation. X.C. and Q.Y.; visualization and investigation. Y.D.: data curation. All authors have read and agreed to the published version of the manuscript.
Funding
National Key R & D Program of China (2022YFD2400104), Rural Revitalization Strategy Special Fund Seed Industry Revitalization Project of Guangdong Province (2022SPY02001, 2022SPY00002, 2022SJS02001), China Agriculture Research System of MOF and MARA (CARS-48), Central Public-interest Scientific Institution Basal Research Fund, CAFS (NO. 2023TD34).
Institutional Review Board Statement
All experiments in this study were approved by the Animal Care and Use Committee of the South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (no. SCSFRI96-253), and performed according to the regulations and guidelines established by this committee.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and supplementary materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jobling, M. National Research Council (NRC): Nutrient requirements of fish and shrimp. Aquac. Int. 2012, 20, 601–602. [Google Scholar] [CrossRef]
- Liu, C.H.; Chen, Z.H. Research progress on nutritional component and feeding valueof maggot protein. J. South. Agric. 2012, 43, 705–709. [Google Scholar]
- Henry, M.; Gasco, L.; Piccolo, G.; Fountoulaki, E. Review on the use of insects in the diet of farmed fish: Past and future. Anim. Feed Sci. Technol. 2015, 203, 1–22. [Google Scholar] [CrossRef]
- Andries, J.P.M.; Heyden, Y.V. Improved multi-class discrimination by Common-Subset-of-Independent-Variables Partial-Least-Squares Discriminant Analysis. Talanta 2021, 234, 122595. [Google Scholar] [CrossRef]
- Elahi, U.; Ma, Y.B.; Wu, S.G.; Wang, J.; Zhang, H.J.; Qi, G.H. Growth performance, carcass characteristics, meat quality and serum profile of broiler chicks fed on housefly maggot meal as a replacement of soybean meal. J. Anim. Physiol. Anim. Nutr. 2020, 104, 1075–1084. [Google Scholar] [CrossRef]
- Cheng, X.M.; Chen, Z.T.; Yan, Z.B.; Zhao, P.Y.; Wang, F.H.; Li, G.D. Effects of Housefly Protein on Growth Performance, Immunity and Muscular Composition in Pacific White Leg Shrimp Litopenaeus vannamei. Fish. Sci. 2018, 37, 324–329. [Google Scholar]
- Cao, J.M.; Yan, J.; Wang, G.X.; Huang, Y.H.; Zhang, R.B.; Zhou, T.T.; Liu, Q.F.; Sun, Z.W. Effects of replacement of fish meal with housefly maggot meal ondigestive enzymes, transaminases activities and hepatopancreashistological structure of Litopenaeus vannamei. South China Fish. Sci. 2012, 8, 72–79. [Google Scholar]
- Wang, W.; Feng, J.; Wang, Z.; Sun, G.; Bao, Z. Preliminary study on anti-baculovirus mechanism of feeding housefly larvae (Musca domestia) and population infection model of outbreaking epidemic disease of shrimp (Panaeus chinensis). Ying Yong Sheng Tai Xue Bao 2002, 13, 728–730. [Google Scholar]
- Liu, L.B.; Li, S.D.; Chen, J.H.; Zhang, S.C.; Wang, H. Effect of Fresh Housefly Larva on Growth and Immunity in Pacific White Leg Shrimp Litopenaeus vannamei. Fish. Sci. 2010, 29, 721–724. [Google Scholar]
- Cheng, X.M.; Huang, Q.C.; Wang, F.H.; Li, G.D. Effects of Dietary Housefly Protein on Growth Performance and Nutrional Quality of Soft Shelled Turtle Trionyx sinensis. Fish. Sci. 2018, 37, 51–58. [Google Scholar] [CrossRef]
- Kurniawan, D.R.; Arief, M.; Agustono; Lamid, M. Effect of maggot (Hermetia illucens) flour in commercial feed on protein retention, energy retention, protein content, and fat content in tilapia (Oreochromis niloticus). IOP Conf. Ser. Earth Environ. Sci. 2018, 137, 012072. [Google Scholar] [CrossRef]
- Herawati, V.E.; Pinandoyo; Darmanto, Y.S.; Hutabarat, J. Growth Performance and Nutrient Content of Carp (Cyprinus Carpio) With the Feeding of Maggot Meal Substitution Cultivated in Different Media. IOP Conf. Ser. Earth Environ. Sci. 2019, 246, 012003. [Google Scholar] [CrossRef]
- Fasakin, E.A.; Balogun, A.M.; Ajayi, O.O. Evaluation of full-fat and defatted maggot meals in the feeding of clariid catfish Clarias gariepinus fingerlings. Aquac. Res. 2003, 34, 733–738. [Google Scholar] [CrossRef]
- Feng, J.; Liu, X.; Xu, Z.R.; Lu, Y.P.; Liu, Y.Y. The effect of Aspergillus oryzae fermented soybean meal on growth performance, digestibility of dietary components and activities of intestinal enzymes in weaned piglets. Anim. Feed Sci. Technol. 2007, 134, 295–303. [Google Scholar] [CrossRef]
- Nemati, Z.; Karimi, A.; Besharati, M. Effects of Aflatoxin B1 and Yeast Cell Wall Supplementation on the Growth Performance of Broilers. In Proceedings of the 2015 International Conference on Innovations in Chemical and Agricultural Engineering (ICICAE’2015), Kuala Lumpur, Malaysia, 8–9 February 2015. [Google Scholar]
- Chambers, J.R.; Gong, J. The intestinal microbiota and its modulation for Salmonella control in chickens. Food Res. Int. 2011, 44, 3149–3159. [Google Scholar] [CrossRef]
- Yuan, C.Y.; Meng, Y.; Bi, J.C.; Cui, Q.M. Effects of fermented feed on digestive enzyme activities and intestinal microflora of Penaeus vannamei. Feed Ind. 2018, 39, 24–28. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Bäckhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef]
- Levy, M.; Blacher, E.; Elinav, E. Microbiome, metabolites and host immunity. Curr. Opin. Microbiol. 2017, 35, 8–15. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of the Association of Official Analytical Chemists, 16th ed.; Association of Official Analytical Chemists: Arlington, VA, USA, 1995. [Google Scholar]
- Zheng, L.; Xie, S.; Zhuang, Z.; Liu, Y.; Tian, L.; Niu, J. Effects of yeast and yeast extract on growth performance, antioxidant ability and intestinal microbiota of juvenile Pacific white shrimp (Litopenaeus vannamei). Aquaculture 2021, 530, 735941. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. GigaScience 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Avena, L.; Castell, F.; Gaudilliere, A.; Mélot, C. Random Forests and Networks Analysis. J. Stat. Phys. 2017, 173, 985–1027. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Konopka, A. What is microbial community ecology? ISME J. 2009, 3, 1223–1230. [Google Scholar] [CrossRef]
- Williams, R.J.; Howe, A.; Hofmockel, K.S. Demonstrating microbial co-occurrence pattern analyses within and between ecosystems. Front. Microbiol. 2014, 5, 358. [Google Scholar] [CrossRef]
- Zhang, M.Z.; Pan, L.Q.; Fan, D.P.; He, J.J.; Su, C.; Gao, S.; Zhang, M.Y. Study of fermented feed by mixed strains and their effects on the survival, growth, digestive enzyme activity and intestinal flora of Penaeus vannamei. Aquaculture 2021, 530, 735703. [Google Scholar] [CrossRef]
- Duan, Y.F.; Wang, Y.; Liu, Q.S.; Dong, H.B.; Li, H.; Xiong, D.L.; Zhang, J.S. Changes in the intestine microbial, digestion and immunity of Litopenaeus vannamei in response to dietary resistant starch. Sci. Rep. 2019, 9, 6464. [Google Scholar] [CrossRef]
- Li, B.; Zeng, Q.; Song, Y.; Gao, Z.; Jiang, L.; Ma, H.; He, J. The effect of fly maggot in pig feeding diets on growth performance and gut microbial balance in Ningxiang pigs. J. Anim. Physiol. Anim. Nutr. 2020, 104, 1867–1874. [Google Scholar] [CrossRef] [PubMed]
- D’Alvise, P.W.; Lillebø, S.; Prol-Garcia, M.J.; Wergeland, H.I.; Nielsen, K.F.; Bergh, Ø.; Gram, L. Phaeobacter gallaeciensis reduces Vibrio anguillarum in cultures of microalgae and rotifers, and prevents vibriosis in cod larvae. PLoS ONE 2012, 7, e43996. [Google Scholar] [CrossRef] [PubMed]
- Grotkjær, T.; Bentzon-Tilia, M.; D’Alvise, P.; Dierckens, K.; Bossier, P.; Gram, L. Phaeobacter inhibens as probiotic bacteria in non-axenic Artemia and algae cultures. Aquaculture 2016, 462, 64–69. [Google Scholar] [CrossRef]
- Rasmussen, B.B.; Erner, K.E.; Bentzon-Tilia, M.; Gram, L. Effect of TDA-producing Phaeobacter inhibens on the fish pathogen Vibrio anguillarum in non-axenic algae and copepod systems. Microb. Biotechnol. 2018, 11, 1070–1079. [Google Scholar] [CrossRef] [PubMed]
- Sonnenschein, E.C.; Jimenez, G.; Castex, M.; Gram, L. The Roseobacter-Group Bacterium Phaeobacter as a Safe Probiotic Solution for Aquaculture. Appl. Environ. Microbiol. 2021, 87, e0258120. [Google Scholar] [CrossRef]
- Kang, Y.-H.; Park, C.-S.; Han, M.-S. Pseudomonas aeruginosa UCBPP-PA14 a useful bacterium capable of lysing Microcystis aeruginosa cells and degrading microcystins. J. Appl. Phycol. 2012, 24, 1517–1525. [Google Scholar] [CrossRef]
- Jiao, L.F.; Dai, T.M.; Zhong, S.Q.; Jin, M.; Sun, P.; Zhou, Q.C. Vibrio parahaemolyticus infection impaired intestinal barrier function and nutrient absorption in Litopenaeus vannamei. Fish Shellfish Immunol. 2020, 99, 184–189. [Google Scholar] [CrossRef]
- Georgopapadakou, N.H.; Liu, F.Y. Penicillin-binding proteins in bacteria. Antimicrob. Agents Chemother. 1980, 18, 148–157. [Google Scholar] [CrossRef]
- Yocum, R.R.; Waxman, D.J.; Rasmussen, J.R.; Strominger, J.L. Mechanism of penicillin action: Penicillin and substrate bind covalently to the same active site serine in two bacterial D-alanine carboxypeptidases. Proc. Natl. Acad. Sci. USA 1979, 76, 2730–2734. [Google Scholar] [CrossRef]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef]
- Alakomi, H.L.; Skyttä, E.; Saarela, M.; Mattila-Sandholm, T.; Latva-Kala, K.; Helander, I.M. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 2000, 66, 2001–2005. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.R.U.; Harunari, E.; Oku, N.; Akasaka, K.; Igarashi, Y. Bulbimidazoles A-C, Antimicrobial and Cytotoxic Alkanoyl Imidazoles from a Marine Gammaproteobacterium Microbulbifer Species. J. Nat. Prod. 2020, 83, 1295–1299. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, A.; Riedel, T.; Gronow, S.; Petersen, J.; Klenk, H.P.; Göker, M. Genome sequence of the reddish-pigmented Rubellimicrobium thermophilum type strain (DSM 16684(T)), a member of the Roseobacter clade. Stand. Genom. Sci. 2013, 8, 480–490. [Google Scholar] [CrossRef][Green Version]
- Yang, J.; Yang, Y. Metal—Organic Frameworks for Biomedical Applications. Small 2020, 16, 1906846. [Google Scholar] [CrossRef]
- Seo, H.S.; Kwon, K.K.; Yang, S.H.; Lee, H.S.; Bae, S.S.; Lee, J.H.; Kim, S.J. Marinoscillum gen. nov., a member of the family ‘Flexibacteraceae’, with Marinoscillum pacificum sp. nov. from a marine sponge and Marinoscillum furvescens nom. rev., comb. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 1204–1208. [Google Scholar] [CrossRef]
- Garrity, G.M.; Holt, J.G. The Road Map to the Manual. In Bergey’s Manual® of Systematic Bacteriology: Volume One: The Archaea and the Deeply Branching and Phototrophic Bacteria; Boone, D.R., Castenholz, R.W., Garrity, G.M., Eds.; Springer: New York, NY, USA, 2001; pp. 119–166. [Google Scholar]
- Wexler, A.G.; Goodman, A.L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2017, 2, 17026. [Google Scholar] [CrossRef] [PubMed]
- Holdeman, L.V.; Moore, W.E.C. New Genus, Coprococcus, Twelve New Species, and Emended Descriptions of Four Previously Described Species of Bacteria from Human Feces. Int. J. Syst. Evol. Microbiol. 1974, 24, 260–277. [Google Scholar] [CrossRef]
- Bowman, J.P.; McCammon, S.A.; Brown, J.L.; McMeekin, T.A. Glaciecola punicea gen. nov., sp. nov. and Glaciecola pallidula gen. nov., sp. nov.: Psychrophilic bacteria from Antarctic sea-ice habitats. Int. J. Syst. Evol. Microbiol. 1998, 48, 1213–1222. [Google Scholar] [CrossRef]
- Lee, H.K.; Chun, J.; Moon, E.Y.; Ko, S.H.; Lee, D.S.; Lee, H.S.; Bae, K.S. Hahella chejuensis gen. nov., sp. nov., an extracellular-polysaccharide-producing marine bacterium. Int. J. Syst. Evol. Microbiol. 2001, 51, 661–666. [Google Scholar] [CrossRef]
- Jeong, H.; Yim, J.H.; Lee, C.; Choi, S.H.; Park, Y.K.; Yoon, S.H.; Hur, C.G.; Kang, H.Y.; Kim, D.; Lee, H.H.; et al. Genomic blueprint of Hahella chejuensis, a marine microbe producing an algicidal agent. Nucleic Acids Res. 2005, 33, 7066–7073. [Google Scholar] [CrossRef]
- Baer, M.L.; Ravel, J.; Chun, J.; Hill, R.T.; Williams, H.N. A proposal for the reclassification of Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax gen. nov. as Bacteriovorax stolpii comb. nov. and Bacteriovorax starrii comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 1, 219–224. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, J.; Dai, W.; Qiu, Q.; Dong, C.; Zhang, J.; Xiong, J. Contrasting Ecological Processes and Functional Compositions between Intestinal Bacterial Community in Healthy and Diseased Shrimp. Microb. Ecol. 2016, 72, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Naughton, L.M.; An, S.Q.; Hwang, I.; Chou, S.H.; He, Y.Q.; Tang, J.L.; Ryan, R.P.; Dow, J.M. Functional and genomic insights into the pathogenesis of Burkholderia species to rice. Environ. Microbiol. 2016, 18, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Dursun, A.; Zenciroglu, A.; Karagol, B.S.; Hakan, N.; Okumus, N.; Gol, N.; Tanir, G. Burkholderia gladioli sepsis in newborns. Eur. J. Pediatr. 2012, 171, 1503–1509. [Google Scholar] [CrossRef]
- Pérez-Pantoja, D.; Nikel, P.I.; Chavarría, M.; de Lorenzo, V. Endogenous stress caused by faulty oxidation reactions fosters evolution of 2,4-dinitrotoluene-degrading bacteria. PLoS Genet. 2013, 9, e1003764. [Google Scholar] [CrossRef]
- Meijer, W.G.; Nienhuis-Kuiper, M.E.; Hansen, T.A. Fermentative bacteria from estuarine mud: Phylogenetic position of Acidaminobacter hydrogenoformans and description of a new type of gram-negative, propionigenic bacterium as Propionibacter pelophilus gen. nov., sp. nov. Int. J. Syst. Bacteriol. 1999, 49 Pt 3, 1039–1044. [Google Scholar] [CrossRef]
- Shen, H.; Song, T.; Lu, J.; Qiu, Q.; Chen, J.; Xiong, J. Shrimp AHPND Causing Vibrio anguillarum Infection: Quantitative Diagnosis and Identifying Antagonistic Bacteria. Mar. Biotechnol. 2021, 23, 964–975. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).