MicroRNAs: Small but Key Players in Viral Infections and Immune Responses to Viral Pathogens
Abstract
Simple Summary
Abstract
1. Introduction
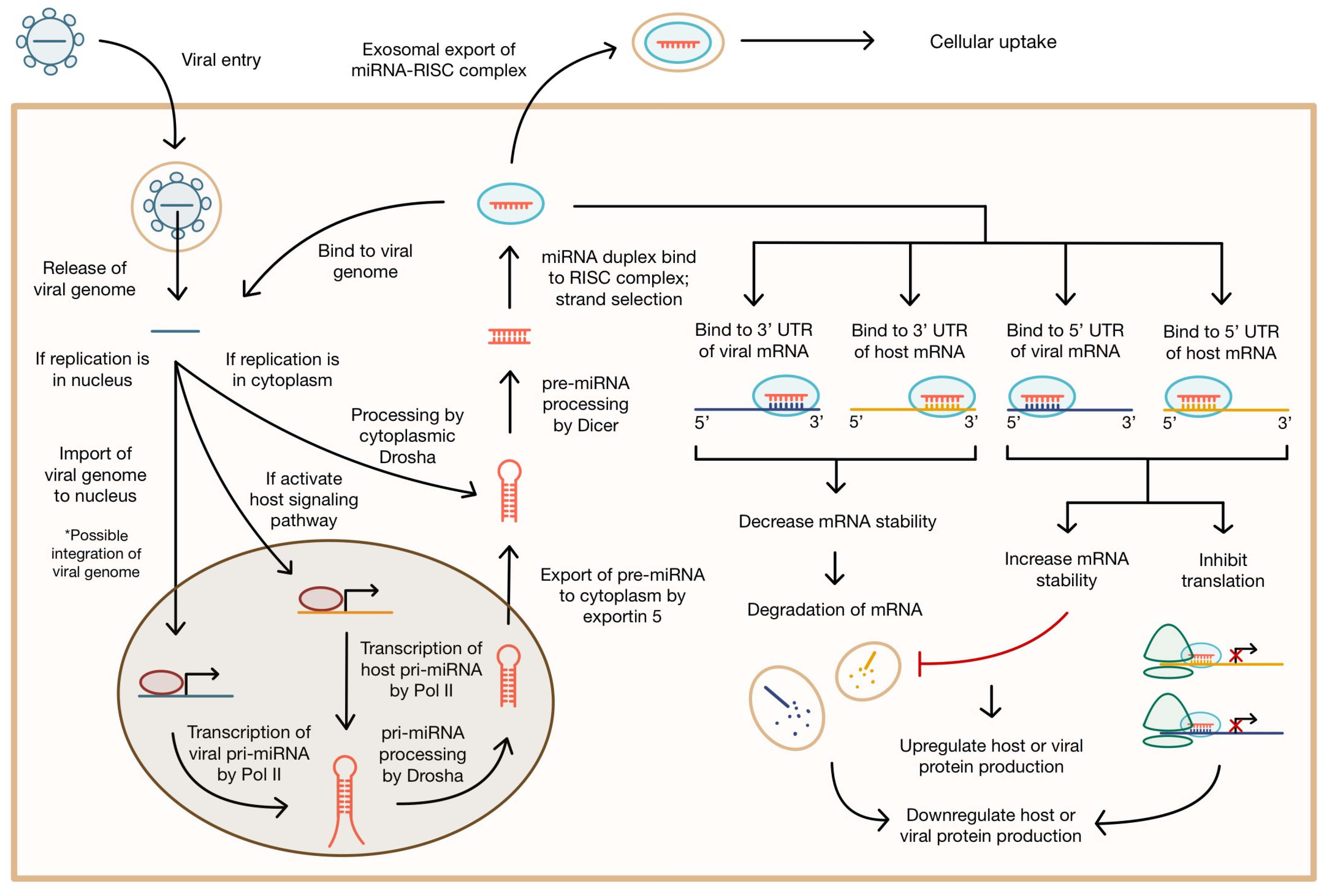
2. Modes of miRNA-Mediated Gene Regulation
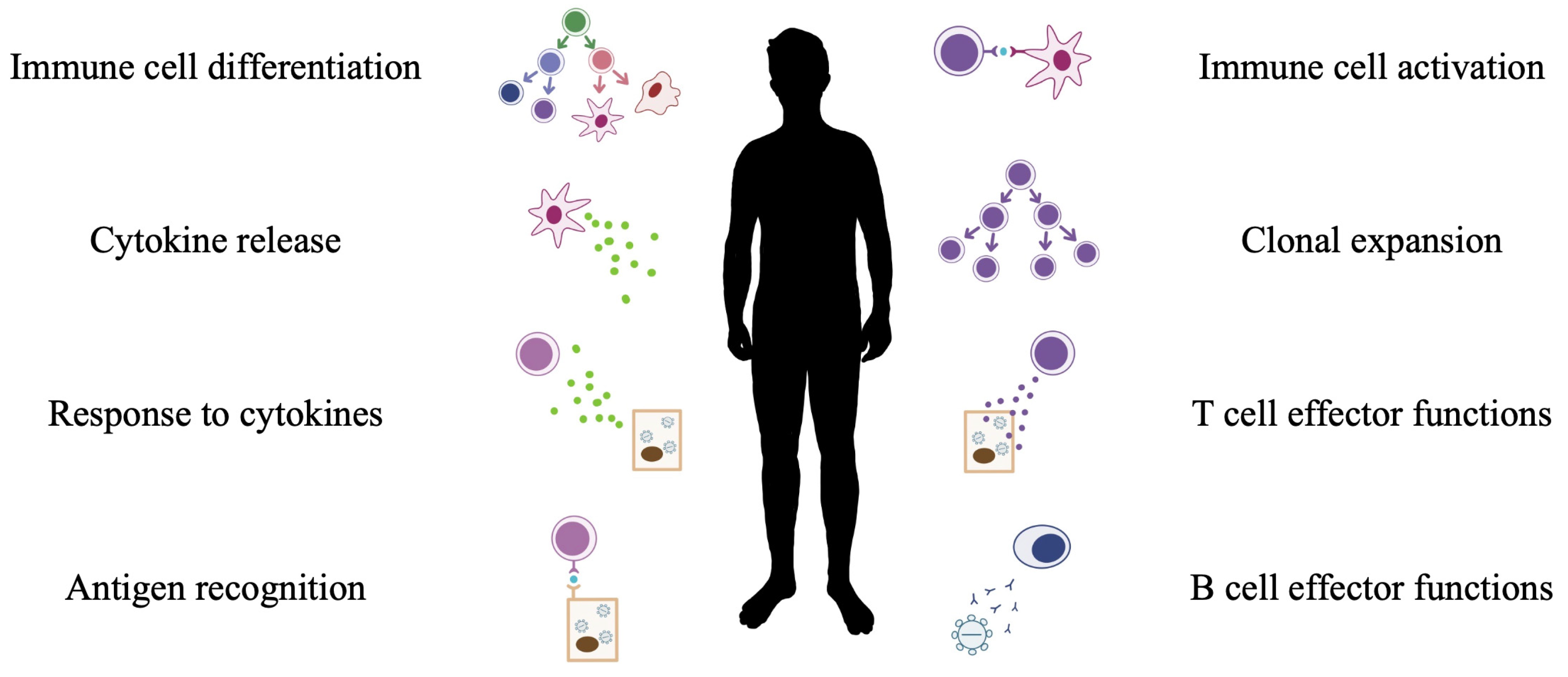
3. Host Response to Viral Pathogens
4. miRNA and Immunity
5. Pro-Viral miRNA Regulation of Viral Infection
| Virus Name | miRNA | Target(s) | Viral Effect | Viral Significance | Ref. |
|---|---|---|---|---|---|
| Epstein–Barr Virus (EBV) | miR-BART3, miR-BART19 | RIG-1 | Downregulate PRR and Type I IFN production in B cells | Innate immune evasion | [89] |
| miR-BART1; miR-BART3 | IRF-9; JAK1 | Downregulate JAK/STAT pathway response to Type I IFN and ISGs | Innate immune evasion | [89] | |
| miR-BART11; miR-BART17-3p | FOXP1; PBRM1 | Downregulate repressors of PD-L1 transcription to prevent T cell cytotoxic activity against infected cells | Immune evasion | [69] | |
| miR-BHRF1-1 | p53 gene | Downregulate p53 to decrease cell cycle arrest, prevent apoptosis, and induce proliferation | Cell survival, proliferation, tumorigenesis | [91] | |
| Herpes Simplex Virus (HSV-1) | hsa-miR-138 | HSV-1 ICP0, Oct-1, Foxc1 | High expression of miR-138 in neurons allows cell-specific repression of viral gene expression, transcription, and replication | Latency | [82] |
| hsa-miR-24 | STING | Induces the production of miR-24 to downregulate the STING pathway and decrease IFN production | Immune evasion | [77] | |
| Human Cytomegalovirus (HCMV) | miR-US33as-5p | IFNAR1 | Downregulates IFN activation of the Jak/STAT pathway and the transcription of ISGs | Immune evasion | [88] |
| miR-US5-2; miR-UL22A | NAB1; SMAD3 | Upregulate TGF-β production to decrease CD34+ HPC proliferation and myelopoiesis; downregulate TGF-β-stimulated genes | Myelosuppression; latency and reactivation | [92] | |
| miR-UL148D | ERN1 | Downregulates the JNK signaling pathway and ER stress-induced apoptosis | Host cell survival | [70] | |
| miR-UL59, UL70-3p, US4-5p, US5-1, US22-5p, US25-2-5p, US29-5p, US33-5p | ERAP1 | Viral miRNAs preferentially bind different genetic variants of ERAP1 to downregulate MHC class I antigen processing | Immune evasion | [93] | |
| Hepatitis B Virus (HBV) | hsa-miRNA-548ah | HDAC4 | Promotes miRNA-548ah expression, downregulating HDAC4 to reduce histone interactions with viral cccDNA | Viral replication; viral transcription | [86] |
| HBV-miR-3 | SOCS5 | Upregulates the JAK/STAT pathway and ISGs in hepatocytes; exosomal HBV-miR-3 triggers macrophage polarization to M1 and promotes EGFR to increase IL-6 secretion. | Innate immune activation; maintenance of chronic infection | [71] | |
| hsa-miR-192-3p | ZNF143 | Upregulates miR-192-3p in hepatocytes to downregulate ZNF143/Akt/mTOR signaling, enhancing viral replication | Viral transcription; viral replication | [43] |
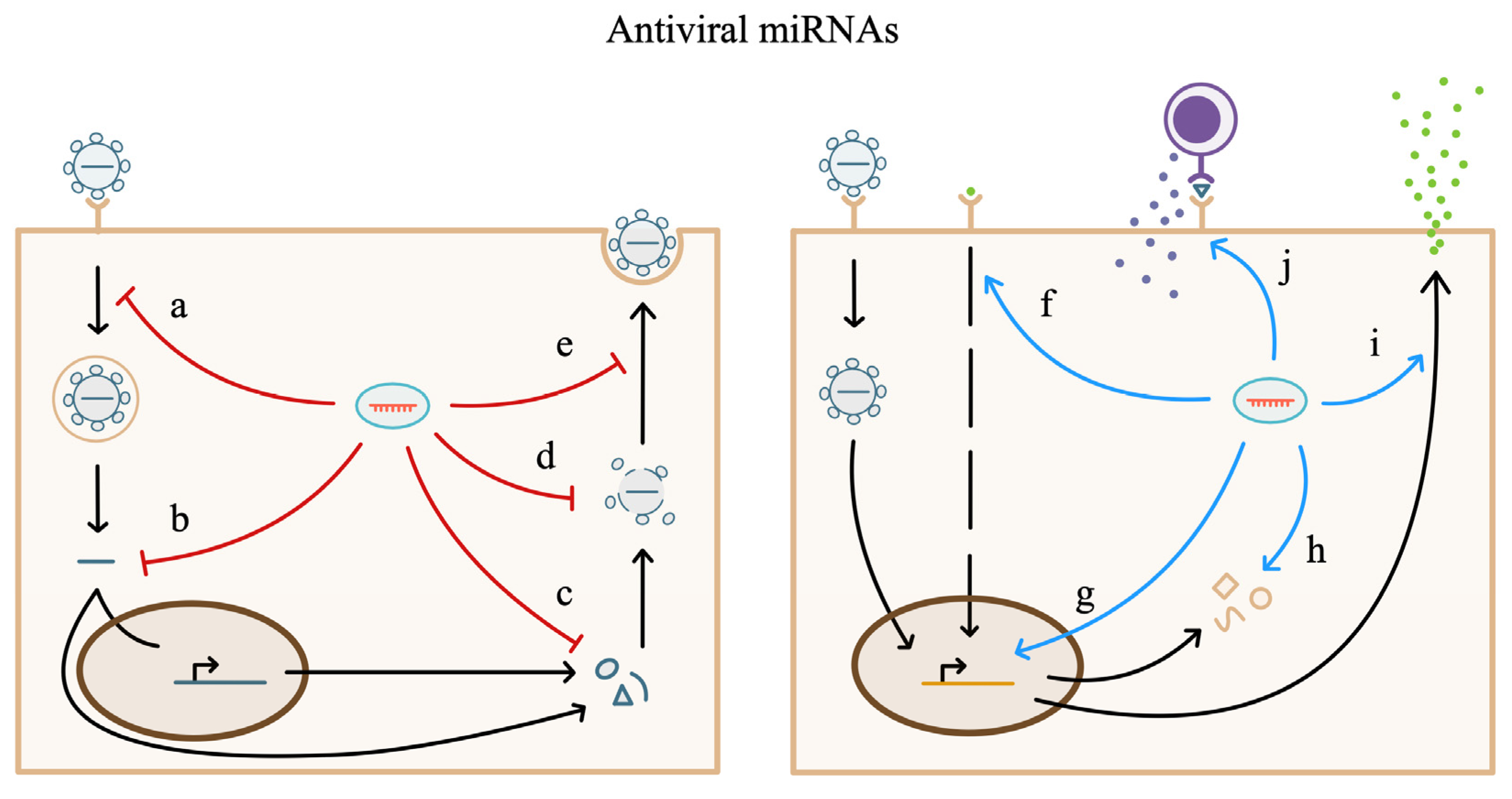
6. miRNA Regulation of the Antiviral Host Response
6.1. miRNA in the Intracellular Antiviral Response
6.2. miRNA in Cellular Signaling Pathways during Viral Infection
6.3. miRNA in Immune Cell Response to Viral Pathogens
| Virus Name | miRNA | Target(s) | Antiviral Effect | Viral Significance | Ref. |
|---|---|---|---|---|---|
| Influenza A Virus (IAV) | hsa-miR-3145 | Viral PB1 gene | Downregulates viral PB1 protein expression | Inhibits viral replication | [72] |
| hsa-miR-1307 | Viral NS1 gene | Prevents the induction of cell cycle arrest | Prevents a favorable environment for the virus | [72] | |
| hsa-miR-24, hsa-miR-124a; hsa-miR-744 | MAPK14; Myc | Suppress downstream p38 MAPK expression and activation | Inhibit viral replication | [104] | |
| Severe Acute Respiratory Syndrome-related Coronavirus (SARS-CoV-2) | hsa-miR-150-5p | Viral nsp10 gene | Downregulates the activation of downstream elements nsp14 and nsp16 | Decreases translation efficiency, immune evasion, and viral replication | [10] |
| hsa-miR-9-5p and hsa-miR-218-5p | ACE2 | Downregulate the host cell receptor for the virus | Prevent viral entry | [113] | |
| hsa-let-7d-5p, hsa-miR-494-3p, hsa-miR-382-3p, hsa-let-7e-5p, hsa-miR-181c-5p, and hsa-miR-452-5p | TMPRSS2 | Downregulate the host cell receptor for the virus | Prevent viral entry | [113] | |
| hsa-miR-1827 | CTSV | Downregulates the host protein that regulates virus entry | Prevents viral entry | [106] | |
| hsa-miR-1277-5p | CANX | Downregulates the host protein that stabilizes S protein for folding | Antigen presentation | [106] | |
| Human Immunodeficiency Virus 1 (HIV-1) | hsa-miR-17, hsa-miR-20 | PCAF | Downregulate cellular cofactor of the HIV Tat protein | Prevent viral gene expression | [111] |
| hsa-miR-28, hsa-miR-29a | HIV mRNA | Downregulate viral protein production | Prevent viral replication | [111] | |
| Hepatitis C Virus (HCV) | hsa-miR-182 | CLDN1 | Downregulates the host protein involved in the internalization of the virus | Prevents viral entry | [116] |
| Human Cytomegalovirus (HCMV) | hsa-mir-221 | SOCS1 | Downregulates the inhibitor of NF-κB phosphorylation and activation | Promotes cytokine signaling | [121] |
| Kaposi’s sarcoma-associated herpesvirus (KSHV) | hsa-miR-36 | IFITM1 | Downregulates cellular transmembrane protein | Prevents viral entry | [25] |
| Respiratory Syncytial Virus (RSV) | hsa-miR-24, hsa-miR-124a; hsa-miR-744 | MAPK14; Myc | Suppress downstream p38 MAPK expression and activation | Inhibit viral replication | [104] |
| Vesicular Stomatitis Virus (VSV) | hsa-miR-183 cluster | PP2A, TRIM27; STAT1 | Downregulate negative regulators of IRF3 phosphorylation; upregulate STAT1 | Promotes interferon production | [105] |
| Coxsackievirus (CVB3) | hsa-miR-21 | MAP2K3 | Suppresses the P38 MAPK signaling pathway | Inhibits viral release | [110] |
6.4. Considerations
7. miRNA Diagnostics and Therapeutics Targeting Viral Pathogens
7.1. miRNA Diagnostics
7.2. miRNA-Based Therapeutics
7.3. Challenges and Future Considerations
8. The miRNA Market Landscape
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Cable, J.; Heard, E.; Hirose, T.; Prasanth, K.V.; Chen, L.; Henninger, J.E.; Quinodoz, S.A.; Spector, D.L.; Diermeier, S.D.; Porman, A.M.; et al. Noncoding RNAs: Biology and applications—A Keystone Symposia report. Ann. N. Y. Acad. Sci. 2021, 1506, 118–141. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef]
- Ding, J.; Li, X.; Hu, H. MicroRNA modules prefer to bind weak and unconventional target sites. Bioinformatics 2014, 31, 1366–1374. [Google Scholar] [CrossRef][Green Version]
- Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Akula, S.M.; Bolin, P.; Cook, P.P. Cellular miR-150-5p may have a crucial role to play in the biology of SARS-CoV-2 infection by regulating nsp10 gene. RNA Biol. 2022, 19, 1–11. [Google Scholar] [CrossRef]
- Lin, Y.; Xiao, L.; Zhang, Y.; Li, P.; Wu, Y.; Lin, Y. MiR-26b-3p regulates osteoblast differentiation via targeting estrogen receptor α. Genomics 2019, 111, 1089–1096. [Google Scholar] [CrossRef]
- Friedrich, M.; Vaxevanis, C.K.; Biehl, K.; Mueller, A.; Seliger, B. Targeting the Coding Sequence: Opposing Roles in Regulating Classical and Non-Classical MHC Class I Molecules by miR-16 and miR-744. J. Immunother. Cancer 2020, 8, e000396. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, J.; Ge, Y.; Xu, Y.; Guo, M.; Mi, K.; Xu, R.; Pei, Y.; Zhang, Q.; Luan, X.; et al. SARS-CoV-2 encoded microRNAs are involved in the process of virus infection and host immune response. J. Biomed. Res. 2021, 35, 216–227. [Google Scholar] [CrossRef]
- Panigrahi, M.; Thibault, P.A.; Wilson, J.A. MicroRNA 122 Affects both the Initiation and the Maintenance of Hepatitis C Virus Infections. J. Virol. 2022, 96, e0190321. [Google Scholar] [CrossRef]
- Zhang, X.; Zuo, X.; Yang, B.; Li, Z.; Xue, Y.; Zhou, Y.; Huang, J.; Zhao, X.; Zhou, J.; Yan, Y.; et al. MicroRNA Directly Enhances Mitochondrial Translation during Muscle Differentiation. Cell 2014, 158, 607–619. [Google Scholar] [CrossRef]
- Valinezhad Orang, A.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef]
- Xiao, M.; Li, J.; Li, W.; Wang, Y.; Wu, F.; Xi, Y.; Zhang, L.; Ding, C.; Luo, H.; Li, Y.; et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017, 14, 1326–1334. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. miRNA interplay: Mechanisms and consequences in cancer. Dis. Models Mech. 2021, 14, dmm047662. [Google Scholar] [CrossRef]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. How Close Are miRNAs from Clinical Practice? A Perspective on the Diagnostic and Therapeutic Market. EJIFCC 2019, 30, 114–127. [Google Scholar]
- Siasos, G.; Bletsa, E.; Stampouloglou, P.K.; Oikonomou, E.; Tsigkou, V.; Paschou, S.A.; Vlasis, K.; Marinos, G.; Vavuranakis, M.; Stefanadis, C.; et al. MicroRNAs in cardiovascular disease. Hell. J. Cardiol. 2020, 61, 165–173. [Google Scholar] [CrossRef]
- Kataria, P.; Surela, N.; Chaudhary, A.; Das, J. MiRNA: Biological Regulator in Host-Parasite Interaction during Malaria Infection. Int. J. Environ. Res. Public Health 2022, 19, 2395. [Google Scholar] [CrossRef]
- Yuan, S.; Wu, Q.; Wang, Z.; Che, Y.; Zheng, S.; Chen, Y.; Zhong, X.; Shi, F. miR-223: An Immune Regulator in Infectious Disorders. Front. Immunol. 2021, 12, 781815. [Google Scholar] [CrossRef]
- Hussein, H.A.M.; Akula, S.M. Profiling of cellular microRNA responses during the early stages of KSHV infection. Arch. Virol. 2017, 162, 3293–3303. [Google Scholar] [CrossRef]
- Hussein, H.A.M.; Alfhili, M.A.; Pakala, P.; Simon, S.; Hussain, J.; McCubrey, J.A.; Akula, S.M. miRNAs and their roles in KSHV pathogenesis. Virus Res. 2019, 266, 15–24. [Google Scholar] [CrossRef]
- Hussein, H.A.M.; Akula, S.M. miRNA-36 inhibits KSHV, EBV, HSV-2 infection of cells via stifling expression of interferon induced transmembrane protein 1 (IFITM1). Sci. Rep. 2017, 7, 17972. [Google Scholar] [CrossRef]
- Park, J.H.; Shin, C. MicroRNA-directed cleavage of targets: Mechanism and experimental approaches. BMB Rep. 2014, 47, 417–423. [Google Scholar] [CrossRef]
- Gu, S.; Kay, M.A. How do miRNAs mediate translational repression? Silence 2010, 1, 11. [Google Scholar] [CrossRef]
- Yekta, S.; Shih, I.; Bartel, D.P. MicroRNA-Directed Cleavage of HOXB8 mRNA. Science 2004, 304, 594–596. [Google Scholar] [CrossRef]
- Faehnle, C.R.; Joshua-Tor, L. Argonautes confront new small RNAs. Curr. Opin. Chem. Biol. 2007, 11, 569–577. [Google Scholar] [CrossRef]
- Gu, K.; Mok, L.; Chong, M.M.W. Regulating gene expression in animals through RNA endonucleolytic cleavage. Heliyon 2018, 4, e00908. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and Their Regulatory Roles in Plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Xu, K.; Lin, J.; Zandi, R.; Roth, J.A.; Ji, L. MicroRNA-mediated target mRNA cleavage and 3′-uridylation in human cells. Sci. Rep. 2016, 6, 30242. [Google Scholar] [CrossRef]
- Barbato, C.; Frisone, P.; Braccini, L.; D’Aguanno, S.; Pieroni, L.; Ciotti, M.T.; Catalanotto, C.; Cogoni, C.; Ruberti, F. Silencing of Ago-2 Interacting Protein SERBP1 Relieves KCC2 Repression by miR-92 in Neurons. Cells 2022, 11, 1052. [Google Scholar] [CrossRef]
- Davis, E.; Caiment, F.; Tordoir, X.; Cavaillé, J.; Ferguson-Smith, A.; Cockett, N.; Georges, M.; Charlier, C. RNAi-Mediated Allelic trans-Interaction at the Imprinted Rtl1/Peg11 Locus. Curr. Biol. 2005, 15, 743–749. [Google Scholar] [CrossRef]
- Mauri, M.; Kirchner, M.; Aharoni, R.; Ciolli Mattioli, C.; van den Bruck, D.; Gutkovitch, N.; Modepalli, V.; Selbach, M.; Moran, Y.; Chekulaeva, M. Conservation of miRNA-mediated silencing mechanisms across 600 million years of animal evolution. Nucleic Acids Res. 2017, 45, 938–950. [Google Scholar] [CrossRef]
- Naeli, P.; Winter, T.; Hackett, A.P.; Alboushi, L.; Jafarnejad, S.M. The intricate balance between microRNA-induced mRNA decay and translational repression. FEBS J. 2023, 290, 2508–2524. [Google Scholar] [CrossRef]
- Braun, J.E.; Huntzinger, E.; Izaurralde, E. The Role of GW182 Proteins in miRNA-Mediated Gene Silencing. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2013; Volume 768, pp. 147–163. [Google Scholar]
- Fukaya, T.; Tomari, Y. MicroRNAs Mediate Gene Silencing via Multiple Different Pathways in Drosophila. Mol. Cell 2012, 48, 825–836. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, X.; Cai, Z.; Zhou, J.; Cao, R.; Zhao, Y.; Chen, Z.; Wang, D.; Ruan, W.; Zhao, Q.; et al. A novel class of microRNA-recognition elements that function only within open reading frames. Nat. Struct. Mol. Biol. 2018, 25, 1019–1027. [Google Scholar] [CrossRef]
- Mishra, R.; Kumar, A.; Ingle, H.; Kumar, H. The Interplay Between Viral-Derived miRNAs and Host Immunity During Infection. Front. Immunol. 2020, 10, 3079. [Google Scholar] [CrossRef]
- Tokorodani, M.; Ichikawa, H.; Yuasa, K.; Takahashi, T.; Hijikata, T. SV40 microRNA miR-S1-3p Downregulates the Expression of T Antigens to Control Viral DNA Replication, and TNFα and IL-17F Expression. Biol. Pharm. Bull. 2020, 43, 1715–1728. [Google Scholar] [CrossRef]
- Lo, A.K.F.; To, K.F.; Lo, K.W.; Lung, R.W.M.; Hui, J.W.Y.; Liao, G.; Hayward, S.D. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc. Natl. Acad. Sci. USA 2007, 104, 16164–16169. [Google Scholar] [CrossRef]
- Li, F.; Deng, Y.; Zhang, S.; Zhu, B.; Wang, J.; Wang, J.; Wang, X.; Zhao, Z.; Deng, W.; Mao, R.; et al. Human hepatocyte-enriched miRNA-192-3p promotes HBV replication through inhibiting Akt/mTOR signalling by targeting ZNF143 in hepatic cell lines. Emerg. Microbes Infect. 2022, 11, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Schlitzer, A.; Placek, K.; Joosten, L.A.B.; Schultze, J.L. Innate and Adaptive Immune Memory: An Evolutionary Continuum in the Host’s Response to Pathogens. Cell Host Microbe 2019, 25, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.E.; Dupuis, M.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Varadé, J.; Magadán, S.; González-Fernández, Á. Human immunology and immunotherapy: Main achievements and challenges. Cell. Mol. Immunol. 2020, 18, 805–828. [Google Scholar] [CrossRef]
- Weizman, O.; Adams, N.M.; Schuster, I.S.; Krishna, C.; Pritykin, Y.; Lau, C.; Degli-Esposti, M.A.; Leslie, C.S.; Sun, J.C.; O’sullivan, T.E. ILC1 Confer Early Host Protection at Initial Sites of Viral Infection. Cell 2017, 171, 795–808. [Google Scholar] [CrossRef]
- Yu, J.C.; Khodadadi, H.; Malik, A.; Davidson, B.; Salles, É.D.S.L.; Bhatia, J.; Hale, V.L.; Baban, B. Innate Immunity of Neonates and Infants. Front. Immunol. 2018, 9, 1759. [Google Scholar] [CrossRef]
- Yi, D.Y.; Kim, S.Y. Human Breast Milk Composition and Function in Human Health: From Nutritional Components to Microbiome and MicroRNAs. Nutrients 2021, 13, 3094. [Google Scholar] [CrossRef]
- Gong, X.; Chao, R.; Wang, P.; Huang, X.; Zhang, J.; Zhu, X.; Zhang, Y.; Yang, X.; Hou, C.; Ji, X.; et al. Interplay of transcription factors and microRNAs during embryonic hematopoiesis. Sci. China Life Sci. 2017, 60, 168–177. [Google Scholar] [CrossRef][Green Version]
- Kim, C.; Ye, Z.; Weyand, C.M.; Goronzy, J.J. miR-181a-regulated pathways in T-cell differentiation and aging. Immun. Ageing 2021, 18, 28. [Google Scholar] [CrossRef]
- Jensen, K.; Brusletto, B.S.; Aass, H.C.D.; Olstad, O.K.; Kierulf, P.; Gautvik, K.M. Transcriptional Profiling of mRNAs and microRNAs in Human Bone Marrow Precursor B Cells Identifies Subset- and Age-Specific Variations. PLoS ONE 2013, 8, e70721. [Google Scholar] [CrossRef] [PubMed]
- Cichocki, F.; Felices, M.; Mccullar, V.; Presnell, S.R.; Al-Attar, A.; Lutz, C.T.; Miller, J.S. Cutting edge: miR-181 promotes human NK cell development by regulating Notch signaling. J. Immunol. 2011, 187, 6171–6175. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Li, M.; Xu, N.; Lv, Q.; Huang, N.; He, J.; Zhang, Y. miR-181a Regulates Inflammation Responses in Monocytes and Macrophages. PLoS ONE 2013, 8, e58639. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.X.; Lee, B.; Geiger, O.; Passegger, C.; Beitzinger, M.; Romberger, J.; Stracke, A.; Högenauer, C.; Stift, A.; Stoiber, H.; et al. miR-181a Modulation of ERK-MAPK Signaling Sustains DC-SIGN Expression and Limits Activation of Monocyte-Derived Dendritic Cells. Cell Rep. 2020, 30, 3793–3805.e5. [Google Scholar] [CrossRef]
- Montagner, S.; Dehó, L.; Monticelli, S. MicroRNAs in hematopoietic development. BMC Immunol. 2014, 15, 14. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gong, J.; Yu, J.; Wang, F.; Zhang, X.; Yin, X.; Tan, Z.; Luo, Z.; Yang, G.; Shen, C.; et al. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood 2012, 119, 4992–5004. [Google Scholar] [CrossRef]
- Hardy, R.R.; Hayakawa, K. Perspectives on fetal derived CD5+ B1 B cells. Eur. J. Immunol. 2015, 45, 2978–2984. [Google Scholar] [CrossRef]
- Rajasekhar, M.; Schmitz, U.; Flamant, S.; Wong, N.L.; Bailey, C.G.; Ritchie, W.; Holst, J.; Rasko, J.E.J. Identifying microRNA determinants of human myelopoiesis. Sci. Rep. 2018, 8, 7264. [Google Scholar] [CrossRef]
- Emamgolizadeh Gurt Tapeh, B.; Mosayyebi, B.B.; Samei, M.; Beyrampour Basmenj, H.; Mohammadi, A.; Alivand, M.R.; Hassanpour, P.; Solali, S. microRNAs involved in T-cell development, selection, activation, and hemostasis. J. Cell. Physiol. 2020, 235, 8461–8471. [Google Scholar] [CrossRef]
- Berrien-Elliott, M.M.; Sun, Y.; Neal, C.; Ireland, A.; Trissal, M.C.; Sullivan, R.P.; Wagner, J.A.; Leong, J.W.; Wong, P.; Mah-Som, A.Y.; et al. MicroRNA-142 Is Critical for the Homeostasis and Function of Type 1 Innate Lymphoid Cells. Immunity 2019, 51, 479–490.e6. [Google Scholar] [CrossRef]
- Mirzaei, R.; Zamani, F.; Hajibaba, M.; Rasouli-Saravani, A.; Noroozbeygi, M.; Gorgani, M.; Hosseini-Fard, S.R.; Jalalifar, S.; Ajdarkosh, H.; Abedi, S.H.; et al. The pathogenic, therapeutic and diagnostic role of exosomal microRNA in the autoimmune diseases. J. Neuroimmunol. 2021, 358, 577640. [Google Scholar] [CrossRef]
- Cañas, J.A.; Núñez, R.; Cruz-Amaya, A.; Gómez, F.; Torres, M.J.; Palomares, F.; Mayorga, C. Epigenetics in Food Allergy and Immunomodulation. Nutrients 2021, 13, 4345. [Google Scholar] [CrossRef] [PubMed]
- Rotival, M.; Siddle, K.J.; Silvert, M.; Pothlichet, J.; Quach, H.; Quintana-Murci, L. Population variation in miRNAs and isomiRs and their impact on human immunity to infection. Genome Biol. 2020, 21, 187. [Google Scholar] [CrossRef]
- Safdar, M.; Ozaslan, M.; Mustafa, R.M.; Smail, S.W.; Khan, S.S.; Khan, M.S.; Akhtar, M.A.; Ali, H.K.; Younas, U.; Saeed, M.; et al. The severity of COVID-19 in hypertensive patients is associated with mirSNPs in the 3′ UTR of ACE2 that associate with miR-3658: In silico and in vitro studies. J. Taibah Univ. Med. Sci. 2023, 18, 1030–1047. [Google Scholar] [CrossRef]
- van der Meulen, E.; Anderton, M.; Blumenthal, M.J.; Schäfer, G. Cellular Receptors Involved in KSHV Infection. Viruses 2021, 13, 118. [Google Scholar] [CrossRef] [PubMed]
- Trobaugh, D.W.; Klimstra, W.B. MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Mol. Med. 2016, 23, 80–93. [Google Scholar] [CrossRef]
- Nanbo, A.; Furuyama, W.; Lin, Z. RNA Virus-Encoded miRNAs: Current Insights and Future Challenges. Front. Microbiol. 2021, 12, 679210. [Google Scholar] [CrossRef]
- Wang, J.; Ge, J.; Wang, Y.; Xiong, F.; Guo, J.; Jiang, X.; Zhang, L.; Deng, X.; Gong, Z.; Zhang, S.; et al. EBV miRNAs BART11 and BART17-3p promote immune escape through the enhancer-mediated transcription of PD-L1. Nat. Commun. 2022, 13, 866. [Google Scholar] [CrossRef] [PubMed]
- Pandeya, A.; Khalko, R.K.; Singh, S.; Kumar, M.; Gosipatala, S.B. Hcmv-miR-UL148D regulates the staurosporine-induced apoptosis by targeting the Endoplasmic Reticulum to Nucleus signaling 1(ERN1). PLoS ONE 2022, 17, e0275072. [Google Scholar] [CrossRef]
- Zhao, X.; Sun, L.; Mu, T.; Yi, J.; Ma, C.; Xie, H.; Liu, M.; Tang, H. An HBV-encoded miRNA activates innate immunity to restrict HBV replication. J. Mol. Cell Biol. 2020, 12, 263–276. [Google Scholar] [CrossRef]
- Liao, Y.; Guo, S.; Liu, G.; Qiu, Z.; Wang, J.; Yang, D.; Tian, X.; Qiao, Z.; Ma, Z.; Liu, Z. Host Non-Coding RNA Regulates Influenza A Virus Replication. Viruses 2021, 14, 51. [Google Scholar] [CrossRef]
- Haneklaus, M.; Gerlic, M.; Kurowska-Stolarska, M.; Rainey, A.; Pich, D.; Mcinnes, I.B.; Hammerschmidt, W.; O’Neill, L.A.J.; Masters, S.L. Cutting edge: miR-223 and EBV miR-BART15 Regulate the NLRP3 Inflammasome and IL-1β Production. J. Immunol. 2012, 189, 3795–3799. [Google Scholar] [CrossRef]
- Pascut, D.; Hoang, M.; Nguyen, N.N.Q.; Pratama, M.Y.; Tiribelli, C. HCV Proteins Modulate the Host Cell miRNA Expression Contributing to Hepatitis C Pathogenesis and Hepatocellular Carcinoma Development. Cancers 2021, 13, 2485. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jiang, J.; Peng, Z. MicroRNA-mediated interactions between host and hepatitis C virus. World J. Gastroenterol. WJG 2016, 22, 1487–1496. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, M.; Pandhare, J.; Dash, C. Are microRNAs Important Players in HIV-1 Infection? An Update. Viruses 2018, 10, 110. [Google Scholar] [CrossRef]
- Sharma, N.; Wang, C.; Kessler, P.; Sen, G.C. Herpes simplex virus 1 evades cellular antiviral response by inducing microRNA-24, which attenuates STING synthesis. PLoS Pathog. 2021, 17, e1009950. [Google Scholar] [CrossRef]
- Rosato, P.; Anastasiadou, E.; Garg, N.; Lenze, D.; Boccellato, F.; Vincenti, S.; Severa, M.; Coccia, E.M.; Bigi, R.; Cirone, M.; et al. Differential regulation of miR-21 and miR-146a by Epstein–Barr virus-encoded EBNA2. Leukemia 2012, 26, 2343–2352. [Google Scholar] [CrossRef]
- Jiang, H.; Bai, L.; Ji, L.; Bai, Z.; Su, J.; Qin, T.; Wang, G.; Balasubramaniam, V.; Wang, X.; Cui, M.; et al. Degradation of MicroRNA miR-466d-3p by Japanese Encephalitis Virus NS3 Facilitates Viral Replication and Interleukin-1β Expression. J. Virol. 2020, 94, e00294-20. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Peng, Q.; Qian, F.; You, Q.; Feng, L.; Hu, S.; Liu, W.; Huang, L.; Shu, X.; Sun, B. HIV-1 Vpr protein upregulates microRNA-210-5p expression to induce G2 arrest by targeting TGIF2. PLoS ONE 2021, 16, e0261971. [Google Scholar] [CrossRef]
- Liao, T.; Chen, Y.; Hsieh, S.; Tang, K.; Chen, D.; Yang, Y.; Liu, H.; Yang, S. Hepatitis C Virus-Induced Exosomal MicroRNAs and Toll-Like Receptor 7 Polymorphism Regulate B-Cell Activating Factor. mBio 2021, 12, e0276421. [Google Scholar] [CrossRef]
- Sun, B.; Yang, X.; Hou, F.; Yu, X.; Wang, Q.; Oh, H.S.; Raja, P.; Pesola, J.M.; Vanni, E.A.H.; Mccarron, S.; et al. Regulation of host and virus genes by neuronal miR-138 favours herpes simplex virus 1 latency. Nat. Microbiol. 2021, 6, 682–696. [Google Scholar] [CrossRef]
- Nahand, J.S.; Mahjoubin-Tehran, M.; Moghoofei, M.; Pourhanifeh, M.H.; Mirzaei, H.R.; Asemi, Z.; Khatami, A.; Bokharaei-Salim, F.; Mirzaei, H.R.; Hamblin, M.R. Exosomal miRNAs: Novel players in viral infection. Epigenomics 2020, 12, 353–370. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.R. Immunomodulatory roles of human herpesvirus-encoded microRNA in host-virus interaction. Rev. Med. Virol. 2020, 30, e2081. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Murray, S.C.; Staitieh, B.S.; Spearman, P.; Guidot, D.M. HIV Impairs Alveolar Macrophage Function via MicroRNA-144-Induced Suppression of Nrf2. Am. J. Med. Sci. 2021, 361, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Xing, T.; Zhu, J.; Xian, J.; Li, A.; Wang, X.; Wang, W.; Zhang, Q. miRNA-548ah promotes the replication and expression of hepatitis B virus by targeting histone deacetylase 4. Life Sci. 2019, 219, 199–208. [Google Scholar] [CrossRef]
- Othumpangat, S.; Beezhold, D.H.; Umbright, C.M.; Noti, J.D. Influenza Virus-Induced Novel miRNAs Regulate the STAT Pathway. Viruses 2021, 13, 967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Song, X.; Ma, P.; Lv, L.; Zhang, Y.; Deng, J.; Zhang, Y. Human Cytomegalovirus miR-US33as-5p Targets IFNAR1 to Achieve Immune Evasion During Both Lytic and Latent Infection. Front. Immunol. 2021, 12, 8364. [Google Scholar] [CrossRef]
- Bouvet, M.; Voigt, S.; Tagawa, T.; Albanese, M.; Chen, Y.-F.A.; Chen, Y.; Fachko, D.N.; Pich, D.; Göbel, C.; Skalsky, R.L.; et al. Multiple Viral microRNAs Regulate Interferon Release and Signaling Early during Infection with Epstein-Barr Virus. mBio 2021, 12, e03440-20. [Google Scholar] [CrossRef]
- Wakabayashi, K.; Machitani, M.; Tachibana, M.; Sakurai, F.; Mizuguchi, H. A MicroRNA Derived from Adenovirus Virus-Associated RNAII Promotes Virus Infection via Posttranscriptional Gene Silencing. J. Virol. 2019, 93, e01265-18. [Google Scholar] [CrossRef]
- Xu, D.; Kong, Y.; Wang, L.; Zhu, H.; Wu, J.; Xia, Y.; Li, Y.; Qin, S.; Fan, L.; Li, J.; et al. EBV-miR-BHRF1-1 Targets p53 Gene: Potential Role in Epstein-Barr Virus Associated Chronic Lymphocytic Leukemia. Cancer Res. Treat. 2019, 52, 492–504. [Google Scholar] [CrossRef]
- Hancock, M.H.; Crawford, L.B.; Pham, A.H.; Mitchell, J.; Struthers, H.M.; Yurochko, A.D.; Caposio, P.; Nelson, J.A. Human Cytomegalovirus miRNAs Regulate TGF-β to Mediate Myelosuppression while Maintaining Viral Latency in CD34+ Hematopoietic Progenitor Cells. Cell Host Microbe 2020, 27, 104–114.e4. [Google Scholar] [CrossRef] [PubMed]
- Melaiu, O.; D’Amico, S.; Tempora, P.; Lucarini, V.; Fruci, D. Impact of Natural Occurring ERAP1 Single Nucleotide Polymorphisms within miRNA-Binding Sites on HCMV Infection. Int. J. Mol. Sci. 2020, 21, 5861. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Lin, X.; Yang, X.; Lu, G.; Zhang, Q.; Zhang, C. MicroRNA-132-3p suppresses type I IFN response through targeting IRF1 to facilitate H1N1 influenza A virus infection. Biosci. Rep. 2019, 39, BSR20192769. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Banerjea, A.C. SARS-CoV-2 Spike Targets USP33-IRF9 Axis via Exosomal miR-148a to Activate Human Microglia. Front. Immunol. 2021, 12, 656700. [Google Scholar] [CrossRef]
- Arisan, E.D.; Dart, D.A.; Grant, G.H.; Dalby, A.D.; Kancagi, D.D.; Turan, R.D.; Yurtsever, B.; Karakus, G.S.; Ovali, E.; Lange, S.; et al. microRNA 1307 Is a Potential Target for SARS-CoV-2 Infection: An in Vitro Model. ACS Omega 2022, 7, 38003–38014. [Google Scholar] [CrossRef]
- Madrid-Elena, N.; Serrano-Villar, S.; Gutiérrez, C.; Sastre, B.; Morín, M.; Luna, L.; Martín, L.; Santoyo-López, J.; López-Huertas, M.R.; Moreno, E.; et al. Selective miRNA inhibition in CD8+ cytotoxic T lymphocytes enhances HIV-1 specific cytotoxic responses. Front. Immunol. 2022, 13, 998368. [Google Scholar] [CrossRef] [PubMed]
- Aydemir, M.N.; Aydemir, H.B.; Korkmaz, E.M.; Budak, M.; Cekin, N.; Pinarbasi, E. Computationally predicted SARS-COV-2 encoded microRNAs target NFKB, JAK/STAT and TGFB signaling pathways. Gene Rep. 2021, 22, 101012. [Google Scholar] [CrossRef]
- Li, X.; Zou, X. An overview of RNA virus-encoded microRNAs. ExRNA 2019, 1, 37. [Google Scholar] [CrossRef]
- Komori, C.; Takahashi, T.; Nakano, Y.; Ui-Tei, K. TRBP–Dicer interaction may enhance HIV-1 TAR RNA translation via TAR RNA processing, repressing host-cell apoptosis. Biol. Open 2020, 9, bio050435. [Google Scholar] [CrossRef]
- Harwig, A.; Jongejan, A.; van Kampen, A.H.C.; Berkhout, B.; Das, A.T. Tat-dependent production of an HIV-1 TAR-encoded miRNA-like small RNA. Nucleic Acids Res. 2016, 44, 4340–4353. [Google Scholar] [CrossRef]
- Bernard, M.A.; Zhao, H.; Yue, S.C.; Anandaiah, A.; Koziel, H.; Tachado, S.D. Novel HIV-1 MiRNAs Stimulate TNFα Release in Human Macrophages via TLR8 Signaling Pathway. PLoS ONE 2014, 9, e106006. [Google Scholar] [CrossRef]
- Zabrodskaya, Y.; Plotnikova, M.; Gavrilova, N.; Lozhkov, A.; Klotchenko, S.; Kiselev, A.; Burdakov, V.; Ramsay, E.; Purvinsh, L.; Egorova, M.; et al. Exosomes Released by Influenza-Virus-Infected Cells Carry Factors Capable of Suppressing Immune Defense Genes in Naïve Cells. Viruses 2022, 14, 2690. [Google Scholar] [CrossRef]
- McCaskill, J.L.; Ressel, S.; Alber, A.; Redford, J.; Power, U.F.; Schwarze, J.; Dutia, B.M.; Buck, A.H. Broad-Spectrum Inhibition of Respiratory Virus Infection by MicroRNA Mimics Targeting p38 MAPK Signaling. Mol. Ther. Nucleic Acids 2017, 7, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Singaravelu, R.; Ahmed, N.; Quan, C.; Srinivasan, P.; Ablenas, C.J.; Roy, D.G.; Pezacki, J.P. A conserved miRNA-183 cluster regulates the innate antiviral response. J. Biol. Chem. 2019, 294, 19785–19794. [Google Scholar] [CrossRef]
- Khurana, P.; Gupta, A.; Sugadev, R.; Sharma, Y.K.; Varshney, R.; Ganju, L.; Kumar, B. nSARS-Cov-2, pulmonary edema and thrombosis: Possible molecular insights using miRNA-gene circuits in regulatory networks. ExRNA 2020, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Zawislak, C.L.; Beaulieu, A.M.; Loeb, G.B.; Karo, J.; Canner, D.; Bezman, N.A.; Lanier, L.L.; Rudensky, A.Y.; Sun, J.C. Stage-specific regulation of natural killer cell homeostasis and response against viral infection by microRNA-155. Proc. Natl. Acad. Sci. USA 2013, 110, 6967–6972. [Google Scholar] [CrossRef]
- Dudda, J.; Salaun, B.; Ji, Y.; Palmer, D.; Monnot, G.; Merck, E.; Boudousquie, C.; Utzschneider, D.; Escobar, T.; Perret, R.; et al. MicroRNA-155 Is Required for Effector CD8+ T Cell Responses to Virus Infection and Cancer. Immunity 2013, 38, 742–753. [Google Scholar] [CrossRef] [PubMed]
- Goncalves-Alves, E.; Saferding, V.; Schliehe, C.; Benson, R.; Kurowska-Stolarska, M.; Brunner, J.S.; Puchner, A.; Podesser, B.K.; Smolen, J.S.; Redlich, K.; et al. MicroRNA-155 Controls T Helper Cell Activation During Viral Infection. Front. Immunol. 2019, 10, 1367. [Google Scholar] [CrossRef]
- He, F.; Xiao, Z.; Yao, H.; Li, S.; Feng, M.; Wang, W.; Liu, Z.; Liu, Z.; Wu, J. The protective role of microRNA-21 against coxsackievirus B3 infection through targeting the MAP2K3/P38 MAPK signaling pathway. J. Transl. Med. 2019, 17, 335. [Google Scholar] [CrossRef]
- Guo, L.; Xu, X.L.; Zhou, L.L.; Zhou, R.L.; Wang, X.; Li, J.X.; Liu, J.; Liu, H.; Zhang, B.; Ho, W. Human Intestinal Epithelial Cells Release Antiviral Factors That Inhibit HIV Infection of Macrophages. Front. Immunol. 2018, 9, 247. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, T.; Gao, S.; Koparde, V.N.; Gonzalez, M.; Spouge, J.L.; Serquiña, A.P.; Lurain, K.; Ramaswami, R.; Uldrick, T.S.; Yarchoan, R.; et al. Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc. Natl. Acad. Sci. USA 2018, 115, 12805–12810. [Google Scholar] [CrossRef] [PubMed]
- Pierce, J.B.; Simion, V.; Icli, B.; Pérez-Cremades, D.; Cheng, H.S.; Feinberg, M.W. Computational Analysis of Targeting SARS-CoV-2, Viral Entry Proteins ACE2 and TMPRSS2, and Interferon Genes by Host MicroRNAs. Genes 2020, 11, 1354. [Google Scholar] [CrossRef]
- Lodge, R.; Bellini, N.; Laporte, M.; Salahuddin, S.; Routy, J.; Ancuta, P.; Costiniuk, C.T.; Jenabian, M.; Cohen, É.A. Interleukin-1β Triggers p53-Mediated Downmodulation of CCR5 and HIV-1 Entry in Macrophages through MicroRNAs 103 and 107. mBio 2020, 11, e02314-20. [Google Scholar] [CrossRef]
- Smith, J.L.; Jeng, S.; McWeeney, S.K.; Hirsch, A.J. A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection. J. Virol. 2017, 91, e02388-16. [Google Scholar] [CrossRef] [PubMed]
- Riad, S.E.; Elhelw, D.S.; Shawer, H.; El-Ekiaby, N.; Salah, A.; Zekri, A.; Esmat, G.; Amleh, A.; Abdelaziz, A.I. Disruption of Claudin-1 Expression by miRNA-182 Alters the Susceptibility to Viral Infectivity in HCV Cell Models. Front. Genet. 2018, 9, 93. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Meng, F.; Shi, J.; Deng, G.; Zeng, X.; Ge, J.; Li, Y.; Liu, L.; Chen, P.; Jiang, Y.; et al. A Novel Intronic Circular RNA Antagonizes Influenza Virus by Absorbing a microRNA That Degrades CREBBP and Accelerating IFN-β Production. mBio 2021, 12, e0101721. [Google Scholar] [CrossRef]
- Barbu, M.G.; Condrat, C.E.; Thompson, D.C.; Bugnar, O.L.; Cretoiu, D.; Toader, O.D.; Suciu, N.; Voinea, S.C. MicroRNA Involvement in Signaling Pathways During Viral Infection. Front. Cell Dev. Biol. 2020, 8, 143. [Google Scholar] [CrossRef]
- Banerjee, A.; Chawla-Sarkar, M.; Mukherjee, A. Rotavirus-Mediated Suppression of miRNA-192 Family and miRNA-181a Activates Wnt/β-Catenin Signaling Pathway: An In Vitro Study. Viruses 2022, 14, 558. [Google Scholar] [CrossRef]
- Lin, X.; Yu, S.; Ren, P.; Sun, X.X.; Jin, M. Human microRNA-30 inhibits influenza virus infection by suppressing the expression of SOCS1, SOCS3, and NEDD4. Cell. Microbiol. 2019, 22, e13150. [Google Scholar] [CrossRef]
- Yan, B.; Ma, H.; Jiang, S.; Shi, J.; Yang, Z.; Zhu, W.; Kong, C.; Chen, L.; Yan, H.; Ma, C. microRNA-221 restricts human cytomegalovirus replication via promoting type I IFN production by targeting SOCS1/NF-κB pathway. Cell Cycle 2019, 18, 3072–3084. [Google Scholar] [CrossRef]
- Ni, F.; Guo, C.; Sun, R.; Fu, B.; Yang, Y.; Wu, L.; Ren, S.; Tian, Z.; Wei, H. MicroRNA transcriptomes of distinct human NK cell populations identify miR-362-5p as an essential regulator of NK cell function. Sci. Rep. 2015, 5, 9993. [Google Scholar] [CrossRef]
- King, J.K.; Tran, T.M.; Paing, M.H.; Yin, Y.; Jaiswal, A.K.; Tso, C.; Roy, K.; Casero, D.; Rao, D.S. Regulation of T-independent B-cell responses by microRNA-146a. Front. Immunol. 2022, 13, 984302. [Google Scholar] [CrossRef] [PubMed]
- Baumjohann, D.; Heissmeyer, V. Posttranscriptional Gene Regulation of T Follicular Helper Cells by RNA-Binding Proteins and microRNAs. Front. Immunol. 2018, 9, 1794. [Google Scholar] [CrossRef]
- Cho, S.; Wu, C.; Yasuda, T.; Cruz, L.O.; Khan, A.A.; Lin, L.; Nguyen, D.T.; Miller, M.; Lee, H.; Kuo, M.; et al. miR-23~27~24 clusters control effector T cell differentiation and function. J. Exp. Med. 2016, 213, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Wigton, E.J.; Ansel, K.M. Noncoding RNAs in B cell responses. RNA Biol. 2021, 18, 633–639. [Google Scholar] [CrossRef]
- Grimaldi, A.; Pietropaolo, G.; Stabile, H.; Kosta, A.; Capuano, C.; Gismondi, A.; Santoni, A.; Sciumè, G.; Fionda, C. The Regulatory Activity of Noncoding RNAs in ILCs. Cells 2021, 10, 2742. [Google Scholar] [CrossRef]
- Nanbakhsh, A.; Malarkannan, S. The Role of microRNAs in NK Cell Development and Function. Cells 2021, 10, 2020. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martin, R.; Wang, G.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Kumar Patel, S.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022, 601, 446–451. [Google Scholar] [CrossRef]
- Groot, M.; Lee, H. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells 2020, 9, 1044. [Google Scholar] [CrossRef]
- Liu, X.; Ma, L.; Schekman, R. Selective sorting of microRNAs into exosomes by phase-separated YBX1 condensates. eLife 2021, 10, e71982. [Google Scholar] [CrossRef]
- Loureiro, D.; Tout, I.; Narguet, S.; Benazzouz, S.M.; Mansouri, A.; Asselah, T. miRNAs as Potential Biomarkers for Viral Hepatitis B and C. Viruses 2020, 12, 1440. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liang, H.; Chen, X.; Ke, Y.; Zhou, Z.; Yang, M.; Zen, K.; Yang, R.; Liu, C.; Zhang, C. An Ebola virus-encoded microRNA-like fragment serves as a biomarker for early diagnosis of Ebola virus disease. Cell Res. 2016, 26, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Moreno, R.; Torre-Cisneros, J.; Cantisán, S. Human cytomegalovirus (HCMV)-encoded microRNAs: Potential biomarkers and clinical applications. RNA Biol. 2021, 18, 2194–2202. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Fu, Y.; Liu, Y.; Wu, H.; Ma, P.; Zeng, W.; Zhang, T.; Lian, S.; Wu, H. Potential Application of MicroRNA Profiling to the Diagnosis and Prognosis of HIV-1 Infection. Front. Microbiol. 2018, 9, 3185. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Benítez, I.D.; Pinilla, L.; Carratalá, A.; Moncusí-Moix, A.; Gort-Paniello, C.; Molinero, M.; González, J.; Torres, G.; Bernal, M.; et al. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl. Res. J. Lab. Clin. Med. 2021, 236, 147–159. [Google Scholar] [CrossRef]
- Zhang, G.; Zong, J.; Lin, S.; Verhoeven, R.J.A.; Tong, S.; Chen, Y.; Ji, M.; Cheng, W.; Tsao, S.; Lung, M.; et al. CirculatingEpstein–Barr virus microRNAs miR-BART7and miR-BART13as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int. J. Cancer 2015, 136, 301. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yu, L.; Gao, X.; Hu, J.; Wang, J.; Dai, Z.; Wang, J.; Zhang, Z.; Lu, S.; Huang, X.; et al. Plasma MicroRNA Panel to Diagnose Hepatitis B Virus–Related Hepatocellular Carcinoma. J. Clin. Oncol. 2011, 29, 4781–4788. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, X.; Chen, X.; Guo, Z.; Liu, J.; Hao, J.; Zhang, J. Real-Time Monitoring of miRNA Function in Pancreatic Cell Lines Using Recombinant AAV-Based miRNA Asensors. PLoS ONE 2013, 8, e66315. [Google Scholar] [CrossRef] [PubMed]
- Chockalingam, A.K.; Hamed, S.; Goodwin, D.G.; Rosenzweig, B.A.; Pang, E.; Boyne, M.T., II; Patel, V. The Effect of Oseltamivir on the Disease Progression of Lethal Influenza A Virus Infection: Plasma Cytokine and miRNA Responses in a Mouse Model. Dis. Markers 2016, 2016, 9296457. [Google Scholar] [CrossRef]
- Ceccarelli, G.; Fratino, M.; Selvaggi, C.; Giustini, N.; Serafino, S.; Schietroma, I.; Corano Scheri, G.; Pavone, P.; Passavanti, G.; Alunni Fegatelli, D.; et al. A pilot study on the effects of probiotic supplementation on neuropsychological performance and microRNA-29a-c levels in antiretroviral-treated HIV-1-infected patients. Brain Behav. 2017, 7, e00756. [Google Scholar] [CrossRef]
- Sabbatinelli, J.; Giuliani, A.; Matacchione, G.; Latini, S.; Laprovitera, N.; Pomponio, G.; Ferrarini, A.; Svegliati Baroni, S.; Pavani, M.; Moretti, M.; et al. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech. Ageing Dev. 2021, 193, 111413. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cheng, P.; Nie, S.; Cui, W. miR-370 mimic inhibits replication of Japanese encephalitis virus in glioblastoma cells. Neuropsychiatr. Dis. Treat. 2016, 12, 2411–2417. [Google Scholar] [CrossRef]
- Zhang, F.; Sun, X.; Zhu, Y.; Qin, W. Downregulation of miR-146a inhibits influenza A virus replication by enhancing the type I interferon response in vitro and in vivo. Biomed. Pharmacother. 2019, 111, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.L.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Ottosen, S.; Parsley, T.B.; Yang, L.; Zeh, K.; van Doorn, L.; van der Veer, E.; Raney, A.K.; Hodges, M.R.; Patick, A.K. In Vitro Antiviral Activity and Preclinical and Clinical Resistance Profile of Miravirsen, a Novel Anti-Hepatitis C Virus Therapeutic Targeting the Human Factor miR-122. Antimicrob. Agents Chemother. 2015, 59, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Stelma, F.; Ree, M.H.; Sinnige, M.J.; Brown, A.; Swadling, L.; Vree, J.M.L.; Willemse, S.B.; Valk, M.; Grint, P.; Neben, S.; et al. A single dose of anti-miR-122, RG-101, in CHC patients results in NK cell normalization with no effect on HCV-specific CD8+ T cell function. Hepatology 2017, 66, 57–68. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Shafaati, M.; Jamalidoust, M.; Kargar, M.; Arefian, E.; Kafilzadeh, F. Downregulation of hepatitis C virus replication by miR-196a using lentiviral vectors. Microbiol. Immunol. 2021, 65, 161–170. [Google Scholar] [CrossRef]
- Sadegh Ehdaei, B.; Pirouzmand, A.; Shabani, M.; Mirzaei, A.; Moghim, S. Cellular miR-101-1 Reduces Efficiently the Replication of HSV-1 in HeLa Cells. Intervirology 2021, 64, 88–95. [Google Scholar] [CrossRef]
- Shabani, M.; Nasr Esfahani, B.; Sadegh Ehdaei, B.; Moghim, S.; Mirzaei, A.; Sharifi, M.; Mouhebat, L. Inhibition of herpes simplex virus type 1 replication by novel hsa-miR-7704 in vitro. Res. Pharm. Sci. 2019, 14, 167–174. [Google Scholar] [CrossRef]
- Ma, L.; Shen, C.; Cohen, É.A.; Xiong, S.; Wang, J. miRNA-1236 Inhibits HIV-1 Infection of Monocytes by Repressing Translation of Cellular Factor VprBP. PLoS ONE 2014, 9, e99535. [Google Scholar] [CrossRef]
- Panda, M.; Kalita, E.; Singh, S.; Kumar, K.; Rao, A.; Prajapati, V.K. MiRNA-SARS-CoV-2 dialogue and prospective anti-COVID-19 therapies. Life Sci. 2022, 305, 120761. [Google Scholar] [CrossRef] [PubMed]
- Karothia, D.; Kumar Dash, P.; Parida, M.; Bhagyawant, S.S.; Kumar, J.S. Vector derived artificial miRNA mediated inhibition of West Nile virus replication and protein expression. Gene 2020, 729, 144300. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021, 28, 127–138. [Google Scholar] [CrossRef]
- Lee, Y.; Chang, C.; Yeh, Y.; Huang, C.F.; Lin, F.; Huang, J.; Hsieh, C.; Wang, J.; Liu, H. Honeysuckle Aqueous Extracts Induced let-7a Suppress EV71 Replication and Pathogenesis In Vitro and In Vivo and Is Predicted to Inhibit SARS-CoV-2. Viruses 2021, 13, 308. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhou, Z.; Jiang, X.; Zheng, Y.; Chen, X.; Fu, Z.; Xiao, G.; Zhang, C.; Zhang, L.; Yi, Y. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020, 6, 54. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, X.; Liu, J.; Dong, L.; Chen, Q.; Liu, J.; Kong, H.; Zhang, Q.; Qi, X.; Hou, D.; et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015, 25, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhou, Y.; Jiang, X.; Wang, Y.; Chen, X.; Xiao, G.; Zhang, C.; Yi, Y.; Zhang, L.; Li, L. Decreased HD-MIR2911 absorption in human subjects with the SIDT1 polymorphism fails to inhibit SARS-CoV-2 replication. Cell Discov. 2020, 6, 63. [Google Scholar] [CrossRef]
- Zmievskaya, E.; Valiullina, A.; Ganeeva, I.; Petukhov, A.; Rizvanov, A.; Bulatov, E. Application of CAR-T Cell Therapy beyond Oncology: Autoimmune Diseases and Viral Infections. Biomedicines 2021, 9, 59. [Google Scholar] [CrossRef]
- Schaible, P.; Bethge, W.; Lengerke, C.; Haraszti, R.A. RNA Therapeutics for Improving CAR T-cell Safety and Efficacy. Cancer Res. 2023, 83, 354–362. [Google Scholar] [CrossRef]
- Renrick, A.N.; Thounaojam, M.C.; de Aquino, M.T.P.; Chaudhuri, E.; Pandhare, J.; Dash, C.; Shanker, A. Bortezomib Sustains T Cell Function by Inducing miR-155-Mediated Downregulation of SOCS1 and SHIP1. Front. Immunol. 2021, 12, 607044. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, J.; Zheng, G.; Wang, Q.; Li, X.; Feng, Y.; Shang, F.; He, S.; Jiang, Q.; Shi, B.; et al. Co-Expression of miR155 or LSD1 shRNA Increases the Anti-Tumor Functions of CD19 CAR-T Cells. Front. Immunol. 2022, 12, 811364. [Google Scholar] [CrossRef]
- Wirges, A.; Bunse, M.; Joedicke, J.J.; Blanc, E.; Gudipati, V.; Moles, M.W.; Shiku, H.; Beule, D.; Huppa, J.B.; Höpken, U.E.; et al. EBAG9 silencing exerts an immune checkpoint function without aggravating adverse effects. Mol. Ther. 2022, 30, 3358–3378. [Google Scholar] [CrossRef] [PubMed]
- Mohebbi, M.; Ding, L.; Malmberg, R.L.; Cai, L. Human MicroRNA Target Prediction via Multi-Hypotheses Learning. J. Comput. Biol. 2021, 28, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhu, X.; Zhang, L.; Liang, Z.; Li, Z. Combined embedding model for MiRNA-disease association prediction. BMC Bioinform. 2021, 22, 161. [Google Scholar] [CrossRef] [PubMed]
- Uthayopas, K.; de Sá, A.G.C.; Alavi, A.; Pires, D.E.V.; Ascher, D.B. TSMDA: Target and symptom-based computational model for miRNA-disease-association prediction. Mol. Ther. Nucleic Acids 2021, 26, 536–546. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, 1470. [Google Scholar] [CrossRef]
- Ding, Y.; Lei, X.; Liao, B.; Wu, F. MLRDFM: A multi-view Laplacian regularized DeepFM model for predicting miRNA-disease associations. Brief. Bioinform. 2022, 23, bbac079. [Google Scholar] [CrossRef]
- Wang, S.; Wang, C.; Huang, L.; Miao, L.; Chen, X. Dual-Network Collaborative Matrix Factorization for predicting small molecule-miRNA associations. Brief. Bioinform. 2022, 23, bbab500. [Google Scholar] [CrossRef]
- Wang, X.; Yu, C.; Li, L.; You, Z.; Huang, W.; Li, Y.; Ren, Z.; Guan, Y. KGDCMI: A New Approach for Predicting circRNA-miRNA Interactions From Multi-Source Information Extraction and Deep Learning. Front. Genet. 2022, 13, 958096. [Google Scholar] [CrossRef]
- Diallo, I.; Ho, J.; Laffont, B.; Laugier, J.; Benmoussa, A.; Lambert, M.; Husseini, Z.; Soule, G.; Kozak, R.; Kobinger, G.P.; et al. Altered microRNA Transcriptome in Cultured Human Liver Cells upon Infection with Ebola Virus. Int. J. Mol. Sci. 2021, 22, 3792. [Google Scholar] [CrossRef] [PubMed]
- Farr, R.J.; Godde, N.; Cowled, C.; Sundaramoorthy, V.; Green, D.; Stewart, C.; Bingham, J.; O’Brien, C.M.; Dearnley, M. Machine Learning Identifies Cellular and Exosomal MicroRNA Signatures of Lyssavirus Infection in Human Stem Cell-Derived Neurons. Front. Cell. Infect. Microbiol. 2021, 11, 783140. [Google Scholar] [CrossRef] [PubMed]
- Müller Coan, B.G.; Cesarman, E.; Acencio, M.L.; Elgui de Oliveira, D. Latent Membrane Protein 1 (LMP1) from Epstein–Barr Virus (EBV) Strains M81 and B95.8 Modulate miRNA Expression When Expressed in Immortalized Human Nasopharyngeal Cells. Genes 2022, 13, 353. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.; Khurana, S.; Saini, D.; Rajput, S.; Thakur, C.J.; Singh, J.; Jaswal, A.; Kapoor, Y.; Kumar, V.; Saini, A. In silico analysis of genomic landscape of SARS-CoV-2 and its variant of concerns (Delta and Omicron) reveals changes in the coding potential of miRNAs and their target genes. Gene 2023, 853, 147097. [Google Scholar] [CrossRef]
- Coenen-Stass, A.M.L.; Pauwels, M.J.; Hanson, B.; Martin Perez, C.; Conceição, M.; Wood, M.J.A.; Mäger, I.; Roberts, T.C. Extracellular microRNAs exhibit sequence-dependent stability and cellular release kinetics. RNA Biol. 2019, 16, 696–706. [Google Scholar] [CrossRef]
- Traber, G.M.; Yu, A. RNAi-Based Therapeutics and Novel RNA Bioengineering Technologies. J. Pharmacol. Exp. Ther. 2023, 384, 133–154. [Google Scholar] [CrossRef]
- Maepa, M.B.; Ely, A.; Grayson, W.; Arbuthnot, P. Sustained Inhibition of HBV Replication In Vivo after Systemic Injection of AAVs Encoding Artificial Antiviral Primary MicroRNAs. Mol. Ther. Nucleic Acids 2017, 7, 190–199. [Google Scholar] [CrossRef]
- van den Berg, F.; Limani, S.W.; Mnyandu, N.; Maepa, M.B.; Ely, A.; Arbuthnot, P. Advances with RNAi-Based Therapy for Hepatitis B Virus Infection. Viruses 2020, 12, 851. [Google Scholar] [CrossRef]
- Jie, J.; Liu, D.; Wang, Y.; Wu, Q.; Wu, T.; Fang, R. Generation of MiRNA sponge constructs targeting multiple MiRNAs. J. Clin. Lab. Anal. 2022, 36, e24527. [Google Scholar] [CrossRef]
- Zempleni, J.; Aguilar-Lozano, A.; Sadri, M.; Sukreet, S.; Manca, S.; Wu, D.; Zhou, F.; Mutai, E. Biological Activities of Extracellular Vesicles and Their Cargos from Bovine and Human Milk in Humans and Implications for Infants. J. Nutr. 2017, 147, 3–10. [Google Scholar] [CrossRef]
- Bellini, N.; Lodge, R.; Pham, T.N.Q.; Jain, J.; Murooka, T.T.; Herschhorn, A.; Bernard, N.F.; Routy, J.; Tremblay, C.L.; Cohen, É.A. MiRNA-103 downmodulates CCR5 expression reducing human immunodeficiency virus type-1 entry and impacting latency establishment in CD4+ T cells. iScience 2022, 25, 105234. [Google Scholar] [CrossRef] [PubMed]
- Da Luz, L.T.; Nascimento, B.; Shankarakutty, A.K.; Rizoli, S.; Adhikari, N.K. Effect of thromboelastography (TEG®) and rotational thromboelastometry (ROTEM®) on diagnosis of coagulopathy, transfusion guidance and mortality in trauma: Descriptive systematic review. Crit. Care 2014, 18, 518. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. MicroRNA (miRNA) Market by Products (Instruments, Kits, Reagents & Consumables), by Research & Tools (qRT-PCR, Biomarkers), by Application (Cancer, Infectious Diseases, Immunological Disorder), by End-Use, and by Region Forecast to 2032. 2023. Available online: https://www.reportsanddata.com/report-detail/microrna-mirna-market (accessed on 15 August 2023).
- Anonymous. MicroRNA Market Size, Share & Trends Analysis Report by Products & Services (Profiling, Localization, & Quantification), by Application (Cancer, Neurological Disease,) by End Use, by Region, and Segment Forecasts, 2023–2030. 2021. Available online: https://www.grandviewresearch.com/industry-analysis/microrna-market (accessed on 14 August 2023).
- Anonymous. MicroRNA Market (by Products: Instruments, Consumables; by Services: Service Type, Specimen; by Application: Cancer, Infectious Diseases Immunological Disorder, Cardiovascular Disease, Neurological Disease, Others; by End Use: Biotechnology & Pharmaceutical Companies, Academic & Government Research Institutes, Other End-Users)—Global Industry Analysis, Size, Share, Growth, Trends, Regional Outlook, and Forecast 2023–2032. 2023. Available online: https://www.precedenceresearch.com/microrna-market (accessed on 15 August 2023).
| Virus Name | miRNA | Target(s) | Viral Effect | Viral Significance | Ref. |
|---|---|---|---|---|---|
| Influenza A Virus (IAV) | hsa-miR-132-3p | IRF1 | Upregulates miR-132-3p to inhibit Type I IFN production and downregulates ISGs | Immune evasion, host cell survival, and viral replication | [94] |
| put-hsa-miR-34 | STAT3 | Downregulates STAT3/IL-6-mediated antiviral response and upregulates the NF-κB pathway | Viral replication; prevention of immune and inflammatory responses | [87] | |
| Severe Acute Respiratory Syndrome-related Coronavirus (SARS-CoV-2) | MR-147-3p | EXOC7; RAD9A; TFE3 | Downregulates exocytosis; cell death and apoptosis; lipid and glucose metabolism, and TGF-β-induced transcription | Exocytosis; host cell survival; metabolism and transcription of host genes | [13] |
| MR359-5p | FOXO3; GCPR1 | Downregulates autophagy and dysregulates oxidative damage responses; binds 5′UTR to upregulate GPCR1 and viral propagation | Host cell survival; viral pathogenesis | [13] | |
| hsa-miR-148a; hsa-miR-590 | USP33; IRF9 | Higher exosomal loading of miR-148a and miR-590 in infected cells; downregulate USP33 and IRF9 in macrophages to upregulate NF-kB, TNFα, and IFNβ pathways | Hyperinflammation | [95] | |
| hsa-miR-150-5p | Viral nsp10 gene | Downregulates miR-150-5p to prevent decreased viral gene expression | Translational efficiency, viral replication, and immune evasion | [10] | |
| Human Immunodeficiency Virus 1 (HIV-1) | hsa-miR-144 | Nrf2 | Upregulates miR-144 to downregulate antioxidant response and impair alveolar macrophage phagocytosis | Immune evasion | [85] |
| hsa-miR-210-5p | TGIF2 | Induces miR-210-5p production to downregulate TGIF2 and promote G2 cell cycle arrest | Cell cycle arrest | [80] | |
| Hepatitis C Virus (HCV) | hsa-miR-122 | TLR7 | Induces host miRNA and exosomal transport to macrophages to activate TLR7, inducing the NF-κB pathway and upregulating B cell activating factor (BAFF) | Autoimmune response | [81] |
| hsa-miR-122 | HCV genome | Liver-specific miRNA increases the stability of the viral genome and promotes viral translation | Viral replication and gene expression | [14] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauer, A.N.; Majumdar, N.; Williams, F.; Rajput, S.; Pokhrel, L.R.; Cook, P.P.; Akula, S.M. MicroRNAs: Small but Key Players in Viral Infections and Immune Responses to Viral Pathogens. Biology 2023, 12, 1334. https://doi.org/10.3390/biology12101334
Bauer AN, Majumdar N, Williams F, Rajput S, Pokhrel LR, Cook PP, Akula SM. MicroRNAs: Small but Key Players in Viral Infections and Immune Responses to Viral Pathogens. Biology. 2023; 12(10):1334. https://doi.org/10.3390/biology12101334
Chicago/Turabian StyleBauer, Anais N., Niska Majumdar, Frank Williams, Smit Rajput, Lok R. Pokhrel, Paul P. Cook, and Shaw M. Akula. 2023. "MicroRNAs: Small but Key Players in Viral Infections and Immune Responses to Viral Pathogens" Biology 12, no. 10: 1334. https://doi.org/10.3390/biology12101334
APA StyleBauer, A. N., Majumdar, N., Williams, F., Rajput, S., Pokhrel, L. R., Cook, P. P., & Akula, S. M. (2023). MicroRNAs: Small but Key Players in Viral Infections and Immune Responses to Viral Pathogens. Biology, 12(10), 1334. https://doi.org/10.3390/biology12101334









