Factors Influencing Pharmacokinetics of Tamoxifen in Breast Cancer Patients: A Systematic Review of Population Pharmacokinetic Models
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Literature Search
2.2. Study Selection Criteria
3. Data Extraction
Data Quality Assessment
4. Results
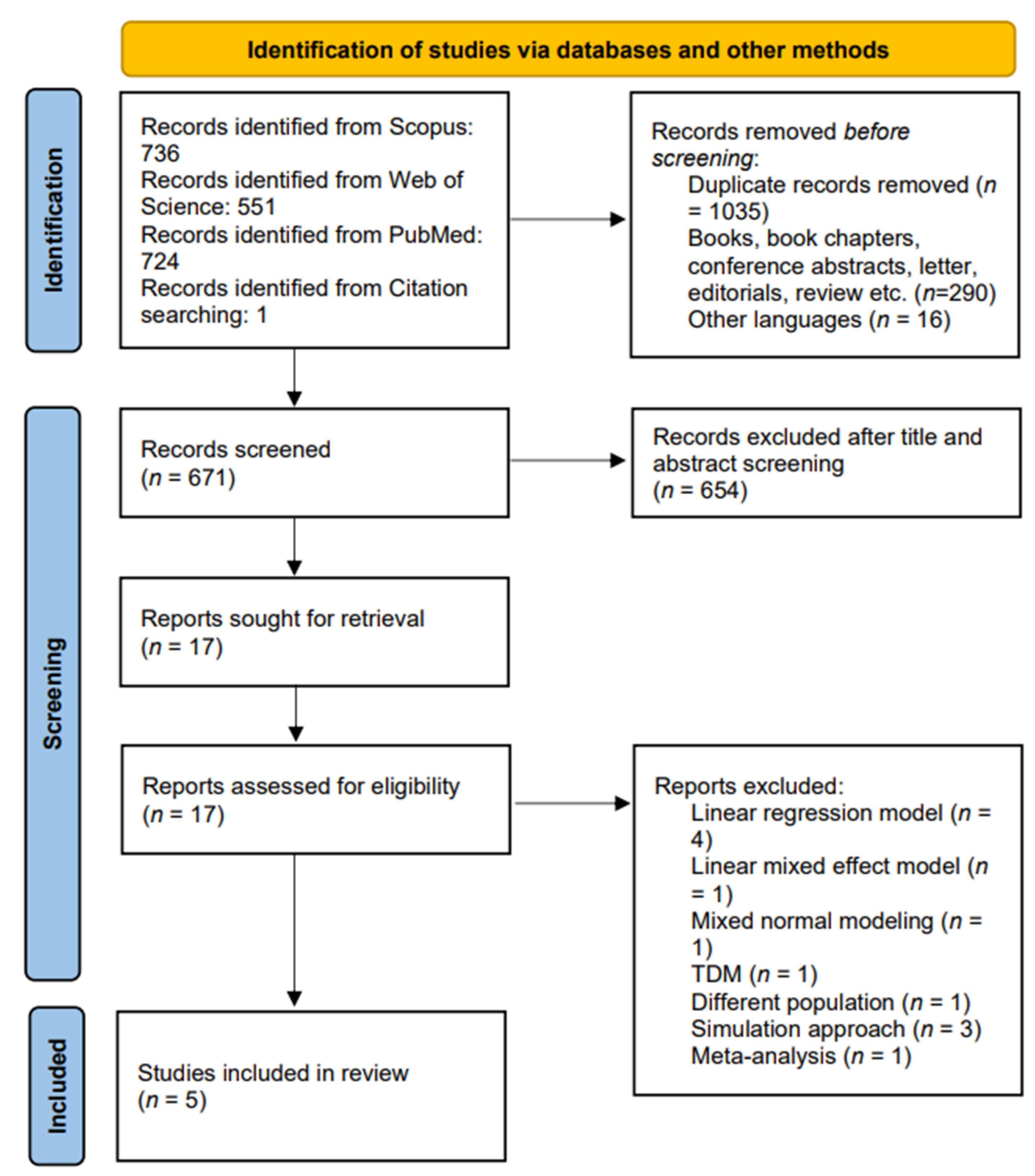
4.1. Literature Search
4.2. Quality Evaluation of Selected Literature
4.3. Population Studied and Sample Size
4.4. Sampling Procedure
4.5. CYP2D6 SNPs and Genotype
4.6. Bioanalytical Methods
4.7. Population Pharmacokinetic Modeling
4.8. Influence of CYP2D6 Phenotype on Tamoxifen Metabolism
4.9. Influence of Other Covariates on PK Parameters of Tamoxifen and Its Metabolites
4.10. External Validation
4.11. Simulation
5. Discussion
6. Limitations
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- What Is Cancer?—NCI. Available online: https://www.cancer.gov/about-cancer/understanding/what-is-cancer (accessed on 12 October 2022).
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- What Is Breast Cancer? American Cancer Society. Available online: https://www.cancer.org/cancer/breast-cancer/about/what-is-breast-cancer.html (accessed on 12 October 2022).
- DeSantis, C.E.; Ma, J.; Sauer, A.G.; Newman, L.A.; Jemal, A. Breast Cancer Statistics, 2017, Racial Disparity in Mortality by State. CA Cancer J. Clin. 2017, 67, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.F. Breast Cancer: Current Perspectives on the Disease Status. Adv. Exp. Med. Biol. 2019, 1152, 51–64. [Google Scholar] [CrossRef]
- Cole, B.F.; Gelber, R.D.; Gelber, S.; Coates, A.S.; Goldhirsch, A. Polychemotherapy for Early Breast Cancer: An Overview of the Randomised Clinical Trials with Quality-Adjusted Survival Analysis. Lancet 2001, 358, 277–286. [Google Scholar] [CrossRef]
- Riggs, B.L.; Hartmann, L.C. Selective Estrogen-Receptor Modulators—Mechanisms of Action and Application to Clinical Practice. N. Engl. J. Med. 2003, 348, 618–629. [Google Scholar] [CrossRef]
- De Vries Schultink, A.H.M.; Zwart, W.; Linn, S.C.; Beijnen, J.H.; Huitema, A.D.R. Effects of Pharmacogenetics on the Pharmacokinetics and Pharmacodynamics of Tamoxifen. Clin. Pharm. 2015, 54, 797–810. [Google Scholar] [CrossRef]
- Lim, Y.C.; Desta, Z.; Flockhart, D.A.; Skaar, T.C. Endoxifen (4-Hydroxy-N-Desmethyl-Tamoxifen) Has Anti-Estrogenic Effects in Breast Cancer Cells with Potency Similar to 4-Hydroxy-Tamoxifen. Cancer Chemother. Pharm. 2005, 55, 471–478. [Google Scholar] [CrossRef]
- Drug Approval Package: Nolvadex (Tamoxifen Citrate) NDA #21-109. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2002/21109_Nolvadex.cfm (accessed on 18 September 2022).
- Goetz, M.P.; Suman, V.J.; Reid, J.M.; Northfelt, D.W.; Mahr, M.A.; Ralya, A.T.; Kuffel, M.; Visscher, S.A.; Kipp, B.R.; Liu, M.C.; et al. First-in-Human Phase I Study of the Tamoxifen Metabolite Z-Endoxifen in Women With Endocrine-Refractory Metastatic Breast Cancer. J. Clin. Oncol. 2017, 35, 3391. [Google Scholar] [CrossRef]
- Schroth, W.; Antoniadou, L.; Fritz, P.; Schwab, M.; Muerdter, T.; Zanger, U.M.; Simon, W.; Eichelbaum, M.; Brauch, H. Breast Cancer Treatment Outcome with Adjuvant Tamoxifen Relative to Patient CYP2D6 and CYP2C19 Genotypes. J. Clin. Oncol. 2007, 25, 5187–5193. [Google Scholar] [CrossRef]
- Ratain, M.J.; Nakamura, Y.; Cox, N.J. CYP2D6 Genotype and Tamoxifen Activity: Understanding Interstudy Variability in Methodological Quality. Clin. Pharm. 2013, 94, 185. [Google Scholar] [CrossRef]
- Mürdter, T.E.; Schroth, W.; Bacchus-Gerybadze, L.; Winter, S.; Heinkele, G.; Simon, W.; Fasching, P.A.; Fehm, T.; Eichelbaum, M.; Schwab, M.; et al. Activity Levels of Tamoxifen Metabolites at the Estrogen Receptor and the Impact of Genetic Polymorphisms of Phase i and II Enzymes on Their Concentration Levels in Plasma. Clin. Pharm. 2011, 89, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Ter Heine, R.; Binkhorst, L.; de Graan, A.J.M.; de Bruijn, P.; Beijnen, J.H.; Mathijssen, R.H.J.; Huitema, A.D.R. Population Pharmacokinetic Modelling to Assess the Impact of CYP2D6 and CYP3A Metabolic Phenotypes on the Pharmacokinetics of Tamoxifen and Endoxifen. Br. J. Clin. Pharmacol. 2014, 78, 572–586. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Spitman, A.B.; Swen, J.J.; Dezentje, V.O.; Moes, D.J.A.R.; Gelderblom, H.; Guchelaar, H.J. Clinical Pharmacokinetics and Pharmacogenetics of Tamoxifen and Endoxifen. Expert Rev. Clin. Pharmacol. 2019, 12, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Madlensky, L.; Natarajan, L.; Tchu, S.; Pu, M.; Mortimer, J.; Flatt, S.W.; Nikoloff, D.M.; Hillman, G.; Fontecha, M.R.; Lawrence, H.J.; et al. Tamoxifen Metabolite Concentrations, CYP2D6 Genotype and Breast Cancer Outcomes. Clin. Pharm. 2011, 89, 718. [Google Scholar] [CrossRef]
- Ring, A.; Dowsett, M. Mechanisms of Tamoxifen Resistance. Endocr. Relat. Cancer 2004, 11, 643–658. [Google Scholar] [CrossRef]
- Dorssers, L.C.J.; van der Flier, S.; Brinkman, A.; van Agthoven, T.; Veldscholte, J.; Berns, E.M.J.J.; Klijn, J.G.M.; Beex, L.V.A.M.; Foekens, J.A. Tamoxifen Resistance in Breast Cancer. Drugs 2012, 61, 1721–1733. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F. Polymorphism of Human Cytochrome P450 2D6 and Its Clinical Significance: Part i. Clin. Pharm. 2009, 48, 689–723. [Google Scholar] [CrossRef]
- Zhou, S.F. Polymorphism of Human Cytochrome P450 2D6 and Its Clinical Significance. Clin. Pharmacokinet. 2012, 48, 761–804. [Google Scholar] [CrossRef]
- Lim, H.S.; Lee, H.J.; Lee, K.S.; Lee, E.S.; Jang, I.J.; Ro, J. Clinical Implications of CYP2D6 Genotypes Predictive of Tamoxifen Pharmacokinetics in Metastatic Breast Cancer. J. Clin. Oncol. 2007, 25, 3837–3845. [Google Scholar] [CrossRef]
- Xu, Y.; Sun, Y.; Yao, L.; Shi, L.; Wu, Y.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Fan, T.; et al. Association between CYP2D6 *10 Genotype and Survival of Breast Cancer Patients Receiving Tamoxifen Treatment. Ann. Oncol. 2008, 19, 1423–1429. [Google Scholar] [CrossRef]
- Park, H.S.; Choi, J.Y.; Lee, M.J.; Park, S.; Yeo, C.W.; Lee, S.S.; Shin, J.G.; Park, B.W. Association between Genetic Polymorphisms of CYP2D6 and Outcomes in Breast Cancer Patients with Tamoxifen Treatment. J. Korean Med. Sci. 2011, 26, 1007. [Google Scholar] [CrossRef] [PubMed]
- Mould, D.R.; Upton, R.N. Basic Concepts in Population Modeling, Simulation, and Model-Based Drug Development—Part 2: Introduction to Pharmacokinetic Modeling Methods. CPT Pharmacomet. Syst Pharm. 2013, 2, e38. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. Syst. Rev. 2021, 10, n71. [Google Scholar] [CrossRef]
- Jamsen, K.M.; McLeay, S.C.; Barras, M.A.; Green, B. Reporting a Population Pharmacokinetic-Pharmacodynamic Study: A Journal’s Perspective. Clin. Pharm. 2014, 53, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Kanji, S.; Hayes, M.; Ling, A.; Shamseer, L.; Chant, C.; Edwards, D.J.; Edwards, S.; Ensom, M.H.H.; Foster, D.R.; Hardy, B.; et al. Reporting Guidelines for Clinical Pharmacokinetic Studies: The ClinPK Statement. Clin. Pharm. 2015, 54, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.; Raju, A.P.; Chaithra; Sekhar, M.S.; Varma, M.; Saravu, K.; Banerjee, M.; Sv, C.S.; Mallayasamy, S.; Rao, M. Influence of N-Acetyltransferase 2 (NAT2) Genotype/Single Nucleotide Polymorphisms on Clearance of Isoniazid in Tuberculosis Patients: A Systematic Review of Population Pharmacokinetic Models. Eur. J. Clin. Pharm. 2022, 2, 1535–1553. [Google Scholar] [CrossRef]
- Muda, M.R.; Harun, S.N.; Sulaiman, S.A.S.; Ghadzi, S.M.S. Population Pharmacokinetics Analyses of Rifampicin in Adult and Children Populations: A Systematic Review. Br. J. Clin. Pharm. 2022, 88, 3132–3152. [Google Scholar] [CrossRef]
- Chen, Y.T.; Wang, C.Y.; Yin, Y.W.; Li, Z.R.; Lin, W.W.; Zhu, M.; Jiao, Z. Population Pharmacokinetics of Oxcarbazepine: A Systematic Review. Br. J. Clin. Pharmacol. 2021, 14,, 853–864. [Google Scholar] [CrossRef]
- Mueller-Schoell, A.; Klopp-Schulze, L.; Schroth, W.; Mürdter, T.; Michelet, R.; Brauch, H.; Huisinga, W.; Joerger, M.; Neven, P.; Koolen, S.L.W.; et al. Obesity Alters Endoxifen Plasma Levels in Young Breast Cancer Patients: A Pharmacometric Simulation Approach. Clin. Pharm. 2020, 108, 661–670. [Google Scholar] [CrossRef]
- Klopp-Schulze, L.; Mueller-Schoell, A.; Neven, P.; Koolen, S.L.W.; Mathijssen, R.H.J.; Joerger, M.; Kloft, C.; De Cock, P.A.; Gotta, V.; Blanco, J.S.P.; et al. Integrated Data Analysis of Six Clinical Studies Points Toward Model-Informed Precision Dosing of Tamoxifen. Front. Pharmacol. 2020, 11, 283. [Google Scholar] [CrossRef]
- Puszkiel, A.; Arellano, C.; Vachoux, C.; Evrard, A.; le Morvan, V.; Boyer, J.C.; Robert, J.; Delmas, C.; Dalenc, F.; Debled, M.; et al. Model-Based Quantification of Impact of Genetic Polymorphisms and Co-Medications on Pharmacokinetics of Tamoxifen and Six Metabolites in Breast Cancer. Clin. Pharm. 2021, 109, 1244–1255. [Google Scholar] [CrossRef] [PubMed]
- Dahmane, E.B.A. Tamoxifen Pharmacokinetics and Pharmacogenetics in Endocrine Sensitive Breast Cancer Patients. Univ. De Genève, Geneva, Switzerland, 26 November 2013. [Google Scholar]
- Goetz, M.P.; Sangkuhl, K.; Guchelaar, H.J.; Schwab, M.; Province, M.; Whirl-Carrillo, M.; Symmans, W.F.; McLeod, H.L.; Ratain, M.J.; Zembutsu, H.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin. Pharm. 2018, 103, 770–777. [Google Scholar] [CrossRef]
- Ahmad, A.; Shahabuddin, S.; Sheikh, S.; Kale, P.; Krishnappa, M.; Rane, R.C.; Ahmad, I. Endoxifen, a New Cornerstone of Breast Cancer Therapy: Demonstration of Safety, Tolerability, and Systemic Bioavailability in Healthy Human Subjects. Clin. Pharm. 2010, 88, 814–817. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.D.; Comen, E.A.; Reiner, A.S.; Orlow, I.; Leong, S.F.; Liang, X.; Mellemkjær, L.; Knight, J.A.; Lynch, C.F.; John, E.M.; et al. CYP2D6 Phenotype, Tamoxifen, and Risk of Contralateral Breast Cancer in the WECARE Study. Breast Cancer Res. 2018, 20, 149. [Google Scholar] [CrossRef]
- Lammers, L.A.; Mathijssen, R.H.J.; van Gelder, T.; Bijl, M.J.; de Graan, A.J.M.; Seynaeve, C.; van Fessem, M.A.; Berns, E.M.; Vulto, A.G.; van Schaik, R.H.N. The Impact of CYP2D6-Predicted Phenotype on Tamoxifen Treatment Outcome in Patients with Metastatic Breast Cancer. Br. J. Cancer 2010, 103, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Beverage, J.N.; Sissung, T.M.; Sion, A.M.; Danesi, R.; Figg, W.D. CYP2D6 Polymorphisms and the Impact on Tamoxifen Therapy. J. Pharm. Sci. 2007, 96, 2224–2231. [Google Scholar] [CrossRef]
- Schroth, W.; Winter, S.; Mürdter, T.; Schaeffeler, E.; Eccles, D.; Eccles, B.; Chowbay, B.; Khor, C.C.; Tfayli, A.; Zgheib, N.K.; et al. Improved Prediction of Endoxifen Metabolism by CYP2D6 Genotype in Breast Cancer Patients Treated with Tamoxifen. Front. Pharm. 2017, 8, 582. [Google Scholar] [CrossRef]
- Kelly, C.M.; Pritchard, K.I. CYP2D6 Genotype as a Marker for Benefit of Adjuvant Tamoxifen in Postmenopausal Women: Lessons Learned. JNCI J. Natl. Cancer Inst. 2012, 104, 427–428. [Google Scholar] [CrossRef]
- De Graan, A.J.M.; Teunissen, S.F.; de Vos, F.Y.F.L.; Loos, W.J.; van Schaik, R.H.N.; de Jongh, F.E.; de Vos, A.I.; van Alphen, R.J.; van der Holt, B.; Verweij, J.; et al. Dextromethorphan as a Phenotyping Test to Predict Endoxifen Exposure in Patients on Tamoxifen Treatment. J. Clin. Oncol. 2011, 29, 3240–3246. [Google Scholar] [CrossRef]
- Binkhorst, L.; van Gelder, T.; Mathijssen, R.H.J. Individualization of Tamoxifen Treatment for Breast Carcinoma. Clin. Pharm. 2012, 92, 431–433. [Google Scholar] [CrossRef]
- Nowell, S.; Sweeney, C.; Winters, M.; Stone, A.; Lang, N.P.; Hutchins, L.F.; Kadlubar, F.F.; Ambrosone, C.B. Association Between Sulfotransferase 1A1 Genotype and Survival of Breast Cancer Patients Receiving Tamoxifen Therapy. JNCI J. Natl. Cancer Inst. 2002, 94, 1635–1640. [Google Scholar] [CrossRef]
- Nowell, S.A.; Ahn, J.; Rae, J.M.; Scheys, J.O.; Trovato, A.; Sweeney, C.; MacLeod, S.L.; Kadlubar, F.F.; Ambrosone, C.B. Association of Genetic Variation in Tamoxifen-Metabolizing Enzymes with Overall Survival and Recurrence of Disease in Breast Cancer Patients. Breast Cancer Res. Treat. 2005, 91, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Lien, E.A.; Søiland, H.; Lundgren, S.; Aas, T.; Steen, V.M.; Mellgren, G.; Gjerde, J. Serum Concentrations of Tamoxifen and Its Metabolites Increase with Age during Steady-State Treatment. Breast Cancer Res. Treat. 2013, 141, 243. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H.; Pike, M.C.; Williams, L.D.; Spicer, D.; Tseng, C.C.; Churchwell, M.I.; Doerge, D.R. Tamoxifen, Soy, and Lifestyle Factors in Asian American Women with Breast Cancer. J. Clin. Oncol. 2007, 25, 3024–3030. [Google Scholar] [CrossRef] [PubMed]
- Peyrade, F.; Frenay, M.; Etienne, M.C.; Ruch, F.; Guillemare, C.; François, E.; Namer, M.; Ferrero, J.M.; Milano, G. Age-Related Difference in Tamoxifen Disposition. Clin. Pharm. 1996, 59, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Antunes, M.v.; da Fontoura Timm, T.A.; de Oliveira, V.; Staudt, D.E.; Raymundo, S.; Gössling, G.; Biazús, J.v.; Cavalheiro, J.A.; Rosa, D.D.; Wallemacq, P.; et al. Influence of CYP2D6 and CYP3A4 Phenotypes, Drug Interactions, and Vitamin D Status on Tamoxifen Biotransformation. Drug Monit. 2015, 37, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Antunes, M.V.; de Oliveira, V.; Raymundo, S.; Staudt, D.E.; Gössling, G.; Biazús, J.V.; Cavalheiro, J.A.; Rosa, D.D.; Mathy, G.; Wallemacq, P.; et al. CYP3A4*22 Is Related to Increased Plasma Levels of 4-Hydroxytamoxifen and Partially Compensates for Reduced CYP2D6 Activation of Tamoxifen. Pharmacogenomics 2015, 16, 601–617. [Google Scholar] [CrossRef] [PubMed]
- Sendur, M.A.N.; Aksoy, S.; Ozdemir, N.Y.; Zengin, N.; Yazici, O.; Sever, A.R.; Altundag, K. Effect of Body Mass Index on the Efficacy of Adjuvant Tamoxifen in Premenopausal Patients with Hormone Receptor-Positive Breast Cancer. J. Buon. 2016, 21, 27–34. [Google Scholar]
- Pirl, W.F. Evidence Report on the Occurrence, Assessment, and Treatment of Depression in Cancer Patients. JNCI Monogr. 2004, 2004, 32–39. [Google Scholar] [CrossRef]
- Zainal, N.Z.; Nik-Jaafar, N.R.; Baharudin, A.; Sabki, Z.A.; Ng, C.G. Prevalence of Depression in Breast Cancer Survivors: A Systematic Review of Observational Studies. Asian Pac. J. Cancer Prev. 2013, 14, 2649–2656. [Google Scholar] [CrossRef]
- Ximenez, J.P.B.; de Andrade, J.M.; Marques, M.P.; Coelho, E.B.; Suarez-Kurtz, G.; Lanchote, V.L. Hormonal Status Affects Plasma Exposure of Tamoxifen and Its Main Metabolites in Tamoxifen-Treated Breast Cancer Patients. BMC Pharm. Toxicol. 2019, 20, 81. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.-S. Evolving Role of Modeling and Simulation in Drug Development. Transl Clin. Pharm. 2019, 27, 19. [Google Scholar] [CrossRef] [PubMed]
| Quality Criteria | Ter Heine et al. 2014 [15] | Schoell et al. 2020 [32] | Schulze et al. 2020 [33] | Puszkiel et al. 2021 [34] | Dahmane et al. [35] | Compliance Rate of Each Criterion (%) |
|---|---|---|---|---|---|---|
| Title | ||||||
| The title identifies the drug(s) and patient population(s) studied | × | √ | × | √ | √ | 60 |
| Abstract | ||||||
| Name of the drug(s) studied | √ | √ | √ | √ | √ | 100 |
| Patient population studied | √ | √ | √ | √ | × | 80 |
| Primary objective(s) | √ | √ | √ | √ | √ | 100 |
| Major findings | √ | √ | √ | √ | √ | 100 |
| Background/introduction | ||||||
| Study rationale | √ | √ | √ | √ | √ | 100 |
| Specific objectives/hypothesis | √ | √ | √ | √ | √ | 100 |
| Methods | ||||||
| Ethics approval | √ | √ | √ | √ | √ | 100 |
| Eligibility criteria of study participants | √ | √ | √ | √ | 80 | |
| Co-administration or food | × | × | × | √ | × | 20 |
| Dosing/frequency/formulation | × | √ | √ | √ | √ | 80 |
| Sampling time and frequency | √ | × | √ | × | √ | 60 |
| Type of sample | √ | √ | √ | √ | √ | 100 |
| Bioanalytical method | √ | √ | √ | √ | √ | 100 |
| Statistical method and software used | × | × | × | √ | × | 20 |
| Modeling software | √ | × | × | √ | √ | 60 |
| Modeling assumptions made | × | × | √ | √ | √ | 60 |
| Estimation method(s) used | √ | × | √ | × | √ | 60 |
| Structural model | √ | √ | √ | √ | √ | 100 |
| Covariates tested | √ | √ | √ | √ | √ | 100 |
| Covariate analysis strategy | √ | √ | √ | √ | √ | 100 |
| Residual error model | √ | √ | √ | √ | √ | 100 |
| Methods for final model evaluation | √ | √ | √ | × | √ | 80 |
| External model validation | NA | √ | NA | NA | NA | 100 |
| Model selection criteria (OFV/AIC, etc.) | √ | √ | √ | √ | √ | 100 |
| Number of study subjects | × | √ | √ | √ | √ | 80 |
| Number of samples used for analyses | × | √ | √ | × | √ | 60 |
| Equations for all model structures and covariate relationships | √ | × | √ | × | × | 40 |
| Results | ||||||
| Demographics details and clinical variables | √ | √ | √ | √ | √ | 100 |
| Concentration vs. time plot | × | × | × | × | × | 0 |
| Schematic of the final model | √ | √ | √ | √ | √ | 100 |
| Table of final model parameters | √ | √ | √ | √ | √ | 100 |
| Summary of the model building process and the derived final model | √ | √ | √ | √ | √ | 100 |
| Final model evaluation plots | √ | √ | √ | √ | √ | 100 |
| A description of simulation results or scenarios (if applicable) | NA | √ | √ | √ | × | 75 |
| Discussion/conclusion | ||||||
| Study limitations | √ | × | × | √ | × | 40 |
| Study findings | √ | √ | √ | √ | √ | 100 |
| Total compliance rate of each study (%) | 77.1 | 75.6 | 83.3 | 80.5 | 80.5 |
| Variables | Ter Heine et al. 2014 [15] | Schoell et al. 2020 [32] | Schulze et al. 2020 [33] | Puszkiel et al. 2021 [34] | Dahmane et al. [35] |
|---|---|---|---|---|---|
| No. of subjects | 40 | 452 | 468 | 928 | 97 |
| No. of samples per patient | 9 | 1–9 | 1–27 | 7 | 5 |
| Total no. of samples | 680 (349 + 331) | NA | 3554 | 27,433 | 457 |
| Bio-analytical method | UPLC-MS/MS | HPLC-MS/MS, | HPLC-MS/MS, | UPLC-MS/MS | HPLC-MS/MS |
| UPLC-MS/MS | UPLC-MS/MS | ||||
| Type of sample | Plasma | Serum | Serum | Plasma | Plasma |
| Plasma | Plasma | ||||
| Age (years) | 53 (22–71) | 64 (25–95) | 64 (25–95) | 48 (25–84) | 50 (32–78) |
| Height (m) | 1.69 (1.56–1.79) | NA | NA | NA | 1.65 (1.51–1.83) |
| Weight (kg) | 72.7 (48.5–114) | 70 (42–150) | NA | 64 (40–131) | 65 (47–116) |
| Tamoxifen dose (%) | |||||
| 20 mg QD | 70 | 98.9 | 96 | 100 | 100 |
| 40 mg QD | 30 | 1.1 | 4 | NA | |
| CYP2D6 phenotype (%) | |||||
| Ultrarapid metabolizer (UM) | 2.5 | NA | 1 | 3.7 | 3 |
| Normal metabolizer (NM) | 50 | 53.5 (including UM) | 78 | 83.3 | 62 |
| Intermediate metabolizer (IM) | 45 | 34.5 | 8 | 8.6 | 31 |
| Poor metabolizer (PM) | 2.5 | 5.53 | 6 | 4.4 | 4 |
| Missing | NA | 6.42 | 7 | NA | NA |
| Menopause status (%) | NA | NA | NA | NA | |
| Pre-menopause | 51.5 | ||||
| Post-menopause | 48.4 | ||||
| Missing |
| Study and Year | Model Structure | External Validation | Residual Variability | Parameter Estimates | Significant Covariates |
|---|---|---|---|---|---|
| Ter Heine et al. 2014 [15] | Two-compartment model with first-order absorption and elimination | No | Proportional error | Ka (1/h)—1.90 | CYP2D6 phenotype |
| Tlag (h)—0.455 | CYP3A4 | ||||
| Q1 (l/h)—61.8 | |||||
| Vd tamoxifen (l)—753 | |||||
| CLTAM (l/h)—9.34 | |||||
| CLMET (l/h)—0.324 | |||||
| Clendo(l/hr) = 5.1 | |||||
| Vdendo(l) = 400 | |||||
| Theta2D6,1 = 0.262 | |||||
| Theta3A4,1 = 0.157 | |||||
| Schoell et al. 2020 [32] | Two-compartment model with first-order absorption and elimination | Yes | Proportional error | Ka (1/h)—1.08 (Fixed) | Age |
| Tlag (h)—0.442 (Fixed) | Body weight | ||||
| VTAM/F (l)—912 (Fixed) | CYP2D6 phenotype Activity score | ||||
| CL30/F (l/h)—5.10 (Fixed) | |||||
| VENDX/F (l)—400 (Fixed) | |||||
| CL20/F (l/h)—5.07 | |||||
| CL23/F (l/h)—0.459 | |||||
| CL20/F_Age: −0.17 | |||||
| CL20/F_Bodyweight: 0.284 | |||||
| CL23/F_AS: 0: −0.759 | |||||
| CL23/F_AS: 0.5: −0.598 | |||||
| CL23/F_AS: 1: −0.347 | |||||
| CL23/F_AS: 1.5: −0.16 | |||||
| CL23/F_AS: 2.5–3: 0.302 | |||||
| Schulze et al. 2020 [33] | Two-compartment model with first-order absorption and elimination | No | Proportional error | Ka (1/h)—1.78 | Age |
| Tlag (h)—0.389 | CYP2D6 phenotype | ||||
| VTAM/F (l)—1120 | Co-medication (Rifampicin/SSRI) | ||||
| CL30/F (l/h)—5.10 (Fixed) | |||||
| VENDX/F (l)—400 (Fixed) | |||||
| CL20/F (l/h)—5.77 | |||||
| CL23/F (l/h)—0.493 | |||||
| Vtam/F_Rif: 0.581 | |||||
| CL20/F_Rif: 6.51 | |||||
| CL20/F_Age: −0.886 | |||||
| CL23/F_AS: 0: −0.722 | |||||
| CL23/F_AS: 0.5: −0.510 | |||||
| CL23/F_AS: 1: −0.323 | |||||
| CL23/F_AS: 1.5: −0.211 | |||||
| CL23/F_AS: 2.5–3: 0.533 | |||||
| CL23_SSRI: −0.654 | |||||
| CL23_Rif: 1.18 | |||||
| Puszkiel et al. 2021 [34] | Seven-compartment model with first-order absorption and elimination | No | Proportional error | Ka (1/h)—0.90 (Fixed) | CYP3A4*22 genotype |
| VTAM (l)—1380 | Age | ||||
| KTAM/NDT (1/h)—5.20 × 10−3 | CYP2D6 phenotype | ||||
| Effect of CYP3A4*22 genotype: 0.773 | CYP2C19*2 genotype | ||||
| Effect of age: −0.298 | CYP2B6*6/*6 genotype, | ||||
| KTAM/4-OHTAM (1/h)—3.72 × 10−5 | Co-medication (CYP2D6 inhibitors) | ||||
| Effect of CYP2D6 IM or PM phenotype: 0.768 | Body weight | ||||
| Effect of CYP2D6 missing phenotype: 1.25 | |||||
| Effect of CYP2C19*2 genotype: 0.866 | |||||
| Effect of age: −0.547 | |||||
| KTAM/4’-OHTAM (1/h)—6.16 × 10−8 | |||||
| KTAM/NOX-TAM (1/h)—2.48 × 10−7 | |||||
| Effect of CYP2B6*6/*6 genotype: 0.766 | |||||
| Effect of age: −0.296 | |||||
| KNDT/ENDO: | |||||
| CYP2D6 UM (h−1): 6.87 × 10−4 | |||||
| CYP2D6 NM (h−1): 5.42 × 10−4 | |||||
| CYP2D6 IM (h−1): 2.86 × 10−4 | |||||
| CYP2D6 PM (h−1): 0.88 × 10−4 | |||||
| Missing CYP2D6 phenotype(h−1): 6.04 × 10−4 | |||||
| Effect of weak/moderate CYP2D6 inhibitor in NM and UM: 0.680 | |||||
| Effect of potent CYP2D6 inhibitor in NM and UM: 0.434 | |||||
| Effect of age: −0.480 | |||||
| KNDT/Z’-ENDO (1/h)—4.08 × 10−7 | |||||
| K4-OHTAM/ENDO (1/h)—1.81 × 10−3 | |||||
| Ke,NDT (1/h)—2.46 × 10−3 | |||||
| Effect of CYP3A4*22 genotype: 0.812 | |||||
| Effect of body weight: 0.245 | |||||
| Ke,ENDO (1/h)—7.93 × 10−3 | |||||
| Ke,4′-OHTAM (1/h)—2.01 × 10−6 (Fixed) | |||||
| Ke,NOX-TAM (1/h)—1.77 × 10−6 (Fixed) | |||||
| Ke,Z’-ENDO (1/h)—1.08 × 10−5 (Fixed) | |||||
| Dahmane et al. [35] | Four-compartment model with first-order absorption and elimination | No | Proportional error | CLTAM/F (l/h)—5.8 | Age |
| θAge: 0.5 | Metabolic ratio | ||||
| θMR: 0.16 | Compliance | ||||
| θCompliance: 0.09 | CYP2D6 phenotype | ||||
| V2/F (l)—724 | Co-medication (CYP2D6 inhibitor) | ||||
| Ka (1/h)—0.7 (Fixed) | |||||
| K23 (1/h)—7.07 × 10−3 | |||||
| θMR: 0.07 | |||||
| K24 (1/h)—5.49 × 10−5 | |||||
| θCYP2D6 PM/IM: 0.26 | |||||
| K35 (1/h)—2.84 × 10−4 | |||||
| θCYP2D6 PM: 0.96 | |||||
| θCYP2D6 IM: 0.56 | |||||
| θpotent 2D6 inhibitor: 0.85 | |||||
| θmoderate 2D6 inhibitor: 0.41 | |||||
| K45 (1/h)—0.015 | |||||
| CLNDT/F (l/h)—3.4 | |||||
| CL4-OHTAM/F (l/h)—2.9 | |||||
| CLEND/F (l/h)—6.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dilli Batcha, J.S.; Raju, A.P.; Matcha, S.; Raj S., E.A.; Udupa, K.S.; Gota, V.; Mallayasamy, S. Factors Influencing Pharmacokinetics of Tamoxifen in Breast Cancer Patients: A Systematic Review of Population Pharmacokinetic Models. Biology 2023, 12, 51. https://doi.org/10.3390/biology12010051
Dilli Batcha JS, Raju AP, Matcha S, Raj S. EA, Udupa KS, Gota V, Mallayasamy S. Factors Influencing Pharmacokinetics of Tamoxifen in Breast Cancer Patients: A Systematic Review of Population Pharmacokinetic Models. Biology. 2023; 12(1):51. https://doi.org/10.3390/biology12010051
Chicago/Turabian StyleDilli Batcha, Jaya Shree, Arun Prasath Raju, Saikumar Matcha, Elstin Anbu Raj S., Karthik S. Udupa, Vikram Gota, and Surulivelrajan Mallayasamy. 2023. "Factors Influencing Pharmacokinetics of Tamoxifen in Breast Cancer Patients: A Systematic Review of Population Pharmacokinetic Models" Biology 12, no. 1: 51. https://doi.org/10.3390/biology12010051
APA StyleDilli Batcha, J. S., Raju, A. P., Matcha, S., Raj S., E. A., Udupa, K. S., Gota, V., & Mallayasamy, S. (2023). Factors Influencing Pharmacokinetics of Tamoxifen in Breast Cancer Patients: A Systematic Review of Population Pharmacokinetic Models. Biology, 12(1), 51. https://doi.org/10.3390/biology12010051






