MITF Contributes to the Body Color Differentiation of Sea Cucumbers Apostichopus japonicus through Expression Differences and Regulation of Downstream Genes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection
2.3. Real-Time Polymerase Chain Reaction (PCR) Analysis
2.4. Western Blot Analysis
2.5. In Situ Hybridization Analysis
2.6. Chromatin Immunoprecipitation Sample Preparation
2.7. Illumina Sequencing
2.8. Statistical Analysis
3. Results
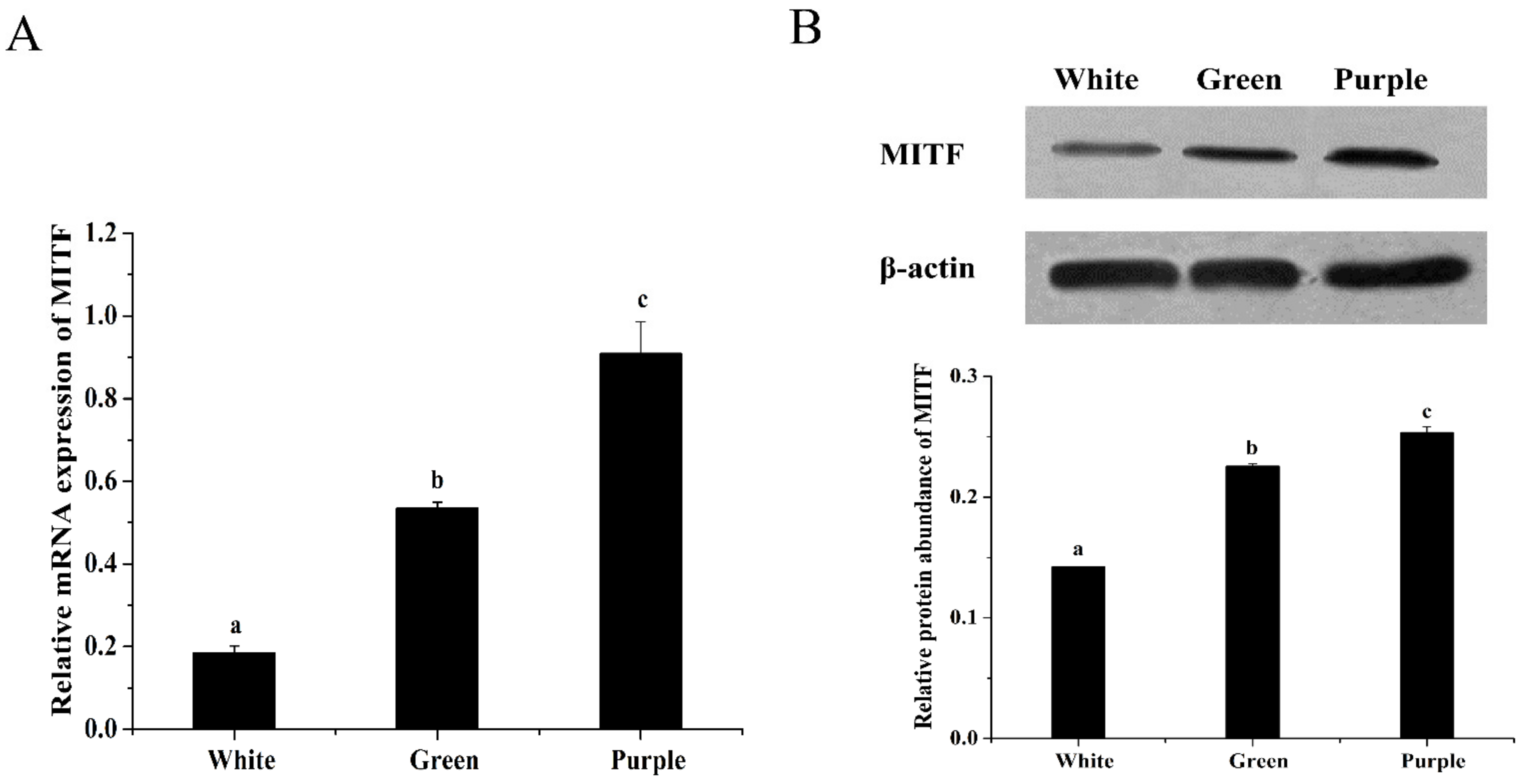
3.1. Difference in MITF Gene Expression among the Three Color Morphs of Sea Cucumbers
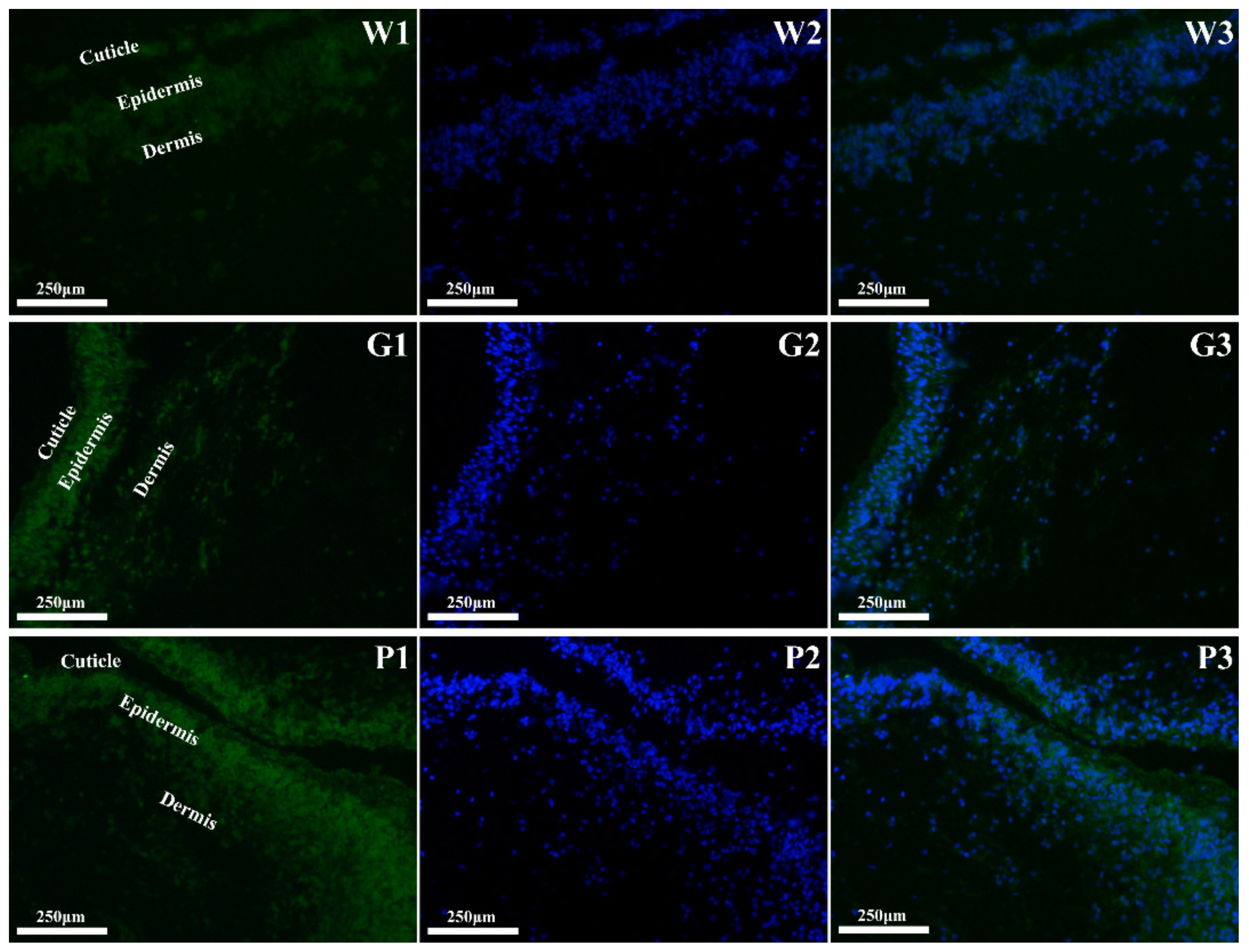
3.2. Spatial Expression of MITF at the mRNA Levels
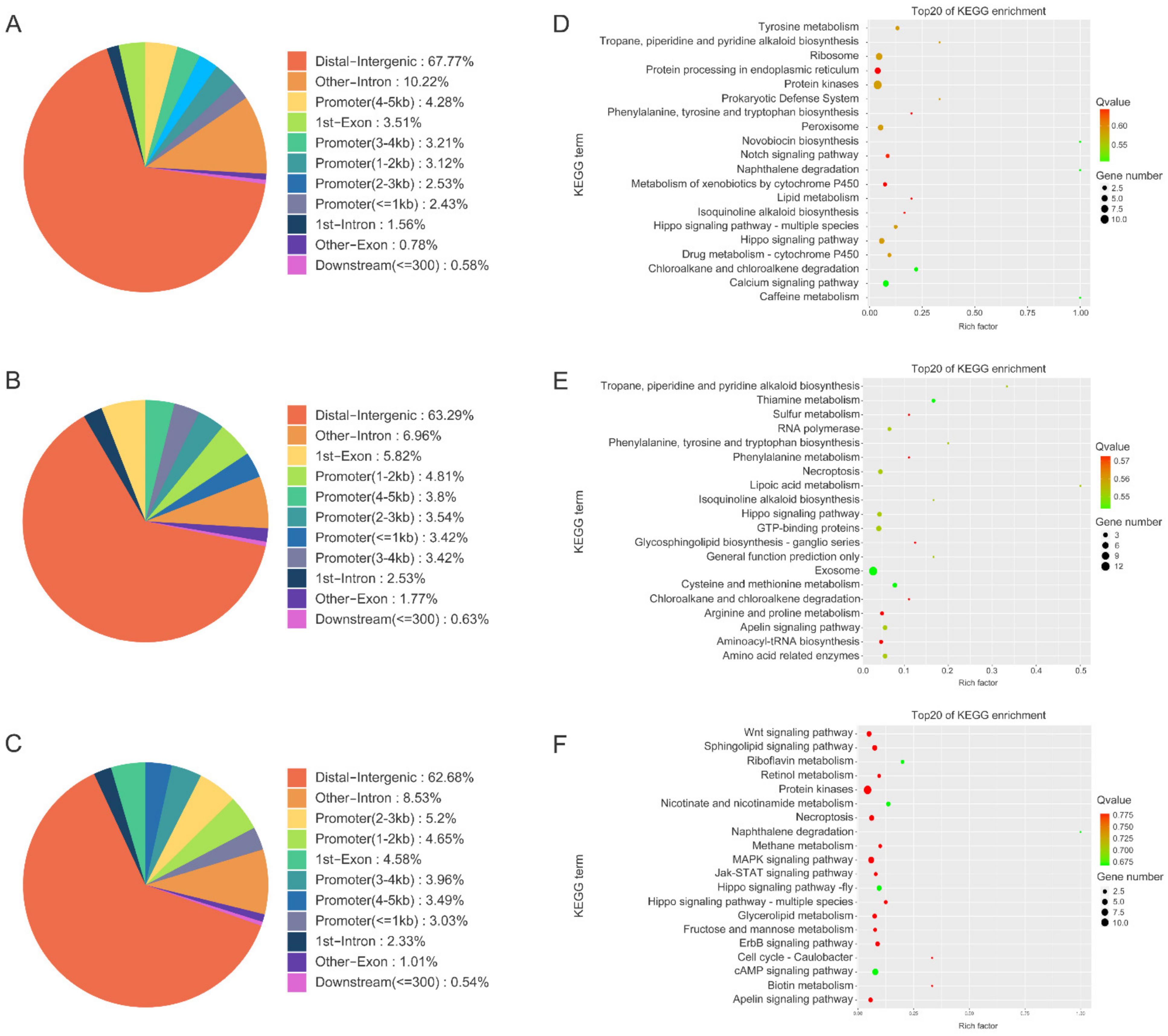
3.3. ChIP-Seq Data Analysis
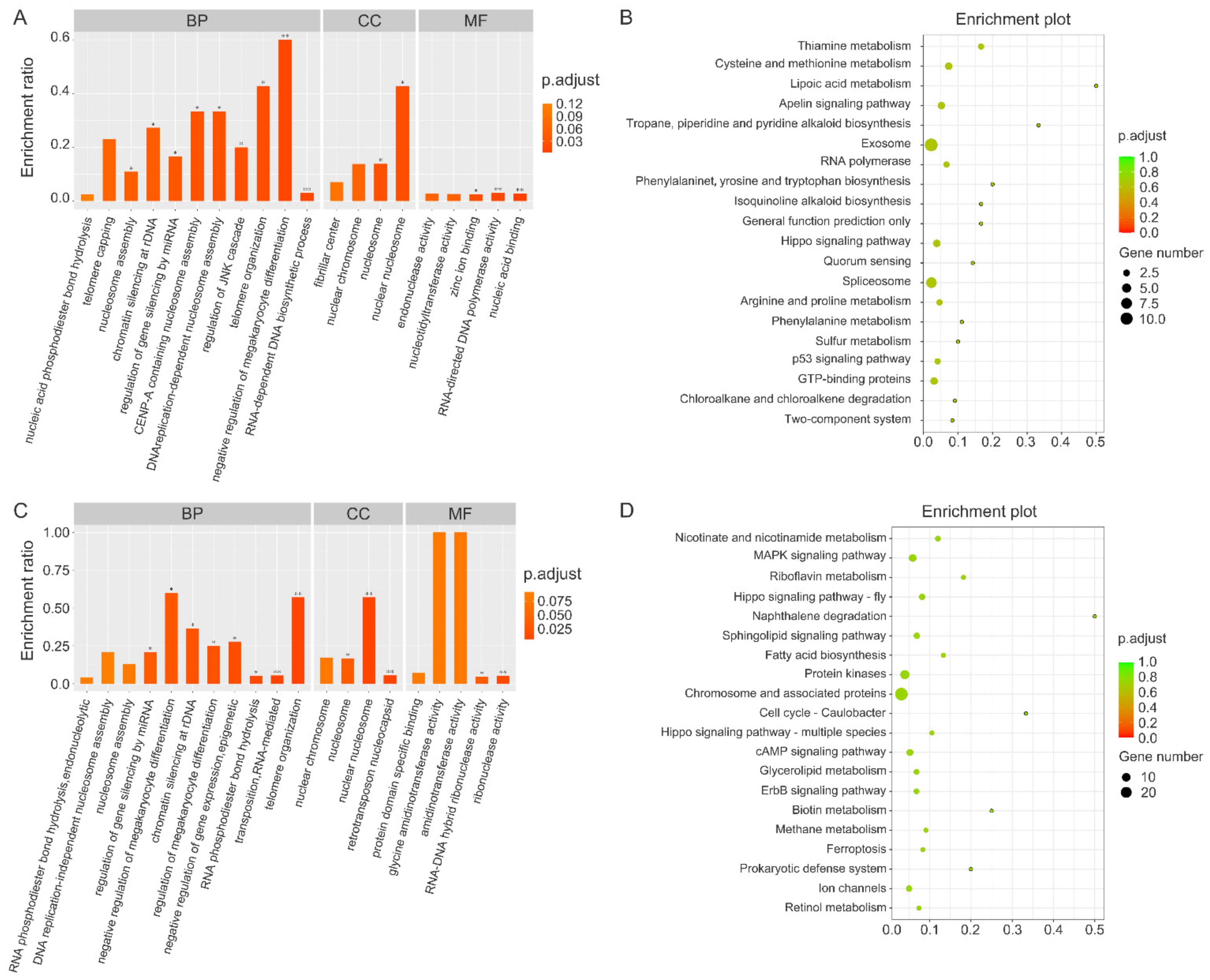
3.4. Annotation of Genes Identified by MITF ChIP
3.5. Comparison of Genes Identified by MITF ChIP in Different Color Morphs of Sea Cucumbers
3.6. Motif Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kiew, P.L.; Don, M.M. Jewel of the seabed: Sea cucumbers as nutritional and drug candidates. Int. J. Food Sci. Nutr. 2011, 63, 616–636. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Li, H.; Wang, X.; Li, X.; Liu, Y.; Sun, J.; Liu, C. A review of the immune molecules in the sea cucumber. Fish Shellfish. Immunol. 2015, 44, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Keesing, J.K.; Liu, D. A Review of Sea Cucumber Aquaculture, Ranching, and Stock Enhancement in China. Rev. Fish. Sci. Aquac. 2016, 24, 326–341. [Google Scholar] [CrossRef]
- Xing, L.; Sun, L.; Liu, S.; Zhang, L.; Yang, H. Comparative metabolomic analysis of the body wall from four varieties of the sea cucumber Apostichopus japonicus. Food Chem. 2021, 352, 129339. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Dong, S.; Gao, Q.; Wang, F.; Tian, X. Comparative study on nutrient composition and growth of green and red sea cucumber, Apostichopus japonicus (Selenka, 1867), under the same culture conditions. Aquac. Res. 2011, 44, 317–320. [Google Scholar] [CrossRef]
- Zhang, Y.-P.; Wang, Z.-D.; Guo, Y.-S.; Liu, L.; Yu, J.; Zhang, S.; Liu, S.-J.; Liu, C.-W. Morphological Characters and Transcriptome Profiles Associated with Black Skin and Red Skin in Crimson Snapper (Lutjanus erythropterus). Int. J. Mol. Sci. 2015, 16, 26991–27004. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhang, L.; Liu, S.; Ru, X.; Xing, L.; Cao, X.; Zhang, T.; Yang, H. The effect of salinity on the growth, energy budget and physiological performance of green, white and purple color morphs of sea cucumber, Apostichopus japonicus. Aquaculture 2015, 437, 297–303. [Google Scholar] [CrossRef]
- Choe, S.; Ohshima, Y. on the morphological and ecological differences between two commercial forms, “green” and “red”, of the japanese common sea cucumber, Stichopus japonicus Selenka. Nippon Suisan Gakkaishi 1961, 27, 97–106. [Google Scholar] [CrossRef]
- Jo, J.; Park, J.; Lee, H.-G.; Kern, E.M.; Cheon, S.; Jin, S.; Park, J.-K.; Cho, S.-J.; Park, C. Comparative transcriptome analysis of three color variants of the sea cucumber Apostichopus japonicus. Mar. Genom. 2016, 28, 21–24. [Google Scholar] [CrossRef]
- Kanno, M.; Suyama, Y.; Li, Q.; Kijima, A. Microsatellite Analysis of Japanese Sea Cucumber, Stichopus (Apostichopus) japonicus, Supports Reproductive Isolation in Color Variants. Mar. Biotechnol. 2006, 8, 672–685. [Google Scholar] [CrossRef]
- Kanno, M.; Kijima, A. Quantitative and qualitative evaluation on the color variation of the Japanese sea cucumber Stichopus japonicus. Suisanzoshoku 2002, 50, 63–69. [Google Scholar]
- Xing, L.; Sun, L.; Liu, S.; Li, X.; Miao, T.; Zhang, L.; Yang, H. Comparison of pigment composition and melanin content among white, light-green, dark-green, and purple morphs of sea cucumber, Apostichopus japonicus. Acta Oceanol. Sin. 2017, 36, 45–51. [Google Scholar] [CrossRef]
- Xing, L.; Sun, L.; Liu, S.; Wan, Z.; Li, X.; Miao, T.; Zhang, L.; Bai, Y.; Yang, H. Growth, histology, ultrastructure and expression of MITF and astacin in the pigmentation stages of green, white and purple morphs of the sea cucumber, Apostichopus japonicus. Aquac. Res. 2017, 49, 177–187. [Google Scholar] [CrossRef]
- Seberg, H.E.; Van Otterloo, E.; Cornell, R.A. Beyond MITF: Multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma. Pigment Cell Melanoma Res. 2017, 30, 454–466. [Google Scholar] [CrossRef]
- Raposo, G.; Marks, M.S. Melanosomes—Dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 2007, 8, 786–797. [Google Scholar] [CrossRef]
- Edea, Z.; Kim, K.-S. MITF gene locus is associated with coat color variation of Ethiopian cattle populations adapted to different altitude environments. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Lin, R.; Lin, W.; Zhou, S.; Chen, Q.; Pan, J.; Miao, Y.; Zhang, M.; Huang, Z.; Xiao, T. Integrated analysis of mRNA expression, CpG island methylation, and polymorphisms in the MITF gene in ducks (Anas platyrhynchos). BioMed Res. Int. 2019, 2019, 8512467. [Google Scholar] [CrossRef]
- Hauswirth, R.; Haase, B.; Blatter, M.; Brooks, S.A.; Burger, D.; Drögemüller, C.; Gerber, V.; Henke, D.; Janda, J.; Jude, R.; et al. Mutations in MITF and PAX3 Cause “Splashed White” and Other White Spotting Phenotypes in Horses. PLoS Genet. 2012, 8, 404–412. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, J.; Fu, W.; Xu, W.; Zhang, H.; Chen, S.; Liu, W.; Peng, L.; Xiao, Y. Comparative Transcriptome and DNA methylation analyses of the molecular mechanisms underlying skin color variations in Crucian carp (Carassius carassius L.). BMC Genet. 2017, 18, 1–12. [Google Scholar] [CrossRef]
- Deardorff, M.A.; Bando, M.; Nakato, R.; Watrin, E.; Itoh, T.; Minamino, M.; Saitoh, K.; Komata, M.; Katou, Y.; Clark, D.; et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 2012, 489, 313–317. [Google Scholar] [CrossRef]
- Zuin, J.; Franke, V.; van Ijcken, W.; Van Der Sloot, A.A.; Krantz, I.D.; Van Der Reijden, M.I.J.A.; Nakato, R.; Lenhard, B.; Wendt, K.S. A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS. PLoS Genet. 2014, 10, e1004153. [Google Scholar] [CrossRef]
- Izumi, K.; Nakato, R.; Zhang, Z.; Edmondson, A.C.; Noon, S.; Dulik, M.C.; Rajagopalan, R.; Venditti, C.P.; Gripp, K.; Samanich, J.; et al. Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat. Genet. 2015, 47, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, T.S.; Xu, Z.; Zhang, X.; Wang, L.; Gimble, J.M.; Lander, E.S.; Rosen, E.D. Comparative Epigenomic Analysis of Murine and Human Adipogenesis. Cell 2010, 143, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Jeppsson, K.; Carlborg, K.K.; Nakato, R.; Berta, D.G.; Lilienthal, I.; Kanno, T.; Lindqvist, A.; Brink, M.C.; Dantuma, N.P.; Katou, Y.; et al. The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement. PLoS Genet. 2014, 10, e1004680. [Google Scholar] [CrossRef] [PubMed]
- Hansen, P.; Hecht, J.; Ibrahim, D.M.; Krannich, A.; Truss, M.; Robinson, P.N. Saturation analysis of ChIP-seq data for reproducible identification of binding peaks. Genome Res. 2015, 25, 1391–1400. [Google Scholar] [CrossRef] [PubMed]
- Webster, D.E.; Barajas, B.; Bussat, R.T.; Yan, K.J.; Neela, P.H.; Flockhart, R.J.; Kovalski, J.; Zehnder, A.; Khavari, P.A. Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition. Genome Res. 2014, 24, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Cheli, Y.; Ohanna, M.; Ballotti, R.; Bertolotto, C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment. Cell Melanoma Res. 2009, 23, 27–40. [Google Scholar] [CrossRef]
- Vachtenheim, J.; Borovanský, J. “Transcription physiology” of pigment formation in melanocytes: Central role of MITF. Exp. Dermatol. 2010, 19, 617–627. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, H.; Zhao, H.; Liu, S.; Wang, T. Differences in MITF gene expression and histology between albino and normal sea cucumbers (Apostichopus japonicus Selenka). Chin. J. Oceanol. Limnol. 2012, 30, 80–91. [Google Scholar] [CrossRef]
- Wilbanks, E.G.; Larsen, D.J.; Neches, R.Y.; Yao, A.I.; Wu, C.; Kjolby, R.A.S.; Facciotti, M.T. A workflow for genome-wide mapping of archaeal transcription factors with ChIP-seq. Nucleic Acids Res. 2012, 40, e74. [Google Scholar] [CrossRef]
- Landt, S.G.; Marinov, G.K.; Kundaje, A.; Kheradpour, P.; Pauli, F.; Batzoglou, S.; Bernstein, B.E.; Bickel, P.; Brown, J.B.; Cayting, P.; et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012, 22, 1813–1831. [Google Scholar] [CrossRef]
- Pan, X.; Fan, Z.; Chen, L.; Liu, C.; Bai, F.; Wei, Y.; Tian, Z.; Dong, Y.; Shi, J.; Chen, H.; et al. PvrA is a novel regulator that contributes to Pseudomonas aeruginosa pathogenesis by controlling bacterial utilization of long chain fatty acids. Nucleic Acids Res. 2020, 48, 5967–5985. [Google Scholar] [CrossRef]
- Salmon-Divon, M.; Dvinge, H.; Tammoja, K.; Bertone, P. PeakAnalyzer: Genome-wide annotation of chromatin binding and modification loci. BMC Bioinform. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, L.; Yuan, J.; Sun, Y.; Gao, Y.; Zhang, L.; Li, S.; Dai, H.; Hamel, J.-F.; Liu, C.; et al. The sea cucumber genome provides insights into morphological evolution and visceral regeneration. PLoS Biol. 2017, 15, e2003790. [Google Scholar] [CrossRef]
- Goding, C.R. Mitf from neural crest to melanoma: Signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000, 14, 1712–1728. [Google Scholar] [CrossRef]
- Artavanis-Tsakonas, S.; Rand, M.D.; Lake, R.J. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef]
- Moriyama, M.; Osawa, M.; Mak, S.-S.; Ohtsuka, T.; Yamamoto, N.; Siu-Shan, M.; Delmas, V.; Kageyama, R.; Beermann, F.; LaRue, L.; et al. Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. J. Cell Biol. 2006, 173, 333–339. [Google Scholar] [CrossRef]
- Nickoloff, B.J.; Hendrix, M.J.C.; Pollock, P.M.; Trent, J.M.; Miele, L.; Qin, J.-Z. Notch and NOXA-related pathways in melanoma cells. J. Investig. Dermatol. Symp. Proc. 2005, 10, 95–104. [Google Scholar] [CrossRef]
- Yue, X.; Nie, Q.; Xiao, G.; Liu, B. Transcriptome Analysis of Shell Color-Related Genes in the Clam Meretrix meretrix. Mar. Biotechnol. 2015, 17, 364–374. [Google Scholar] [CrossRef]
- Wu, Z.; Guan, K.-L. Hippo Signaling in Embryogenesis and Development. Trends Biochem. Sci. 2020, 46, 51–63. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
| Sample | Raw Reads | Clean Reads | Bases | Q20 | Q30 | Error Rate | GC Content |
|---|---|---|---|---|---|---|---|
| IP_G | 50,834,380 | 48,211,710 | 6,965,268,667 | 98.32% | 95.18% | 0.01% | 44.48% |
| IP_P | 40,186,312 | 38,936,494 | 5,602,175,868 | 98.47% | 95.86% | 0.01% | 39.89% |
| IP_W | 42,857,474 | 41,156,574 | 6,008,796,738 | 98.55% | 95.87% | 0.01% | 45.03% |
| In_G | 58,644,000 | 56,691,046 | 8,203,781,133 | 98.18% | 94.98% | 0.01% | 37.86% |
| In_P | 44,763,546 | 43,446,150 | 6,325,150,652 | 98.49% | 95.92% | 0.01% | 37.81% |
| In_W | 58,268,370 | 55,632,032 | 8,109,868,722 | 98.51% | 95.96% | 0.01% | 37.62% |
| Sample | Total Reads | Mapped Reads | Map Rate | Paired | Single |
|---|---|---|---|---|---|
| IP_G | 48,321,829 | 24,458,808 | 50.62% | 22,989,908 | 706,691 |
| IP_P | 39,062,377 | 27,048,644 | 69.24% | 25,214,094 | 799,505 |
| IP_W | 41,246,498 | 20,189,447 | 48.95% | 19,071,040 | 543,201 |
| In_G | 56,932,965 | 46,878,697 | 82.34% | 43,263,590 | 1,590,244 |
| In_P | 43,646,150 | 36,173,208 | 82.88% | 33,337,656 | 1,173,866 |
| In_W | 55,946,442 | 46,076,330 | 82.36% | 42,772,052 | 1,344,408 |
| Sample | Peak Number | Peak Total Length | Peak Average Length | Average Tag |
|---|---|---|---|---|
| IP_G | 984 | 349,932 | 355 | 922 |
| IP_P | 732 | 282,705 | 386 | 974 |
| IP_W | 1191 | 412,510 | 346 | 706 |
| Rank | Motif | Log (p Value) | Name |
|---|---|---|---|
| 1 |  | −11.73 | IRF:BATF(IRF:bZIP)/pDC-Irf8-ChIP-Seq(GSE66899)/Homer |
| 2 |  | −9.47 | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer |
| 3 |  | −9.08 | LXRE(NR),DR4/RAW-LXRb.biotin-ChIP-Seq(GSE21512)/Homer |
| 4 |  | −7.66 | RORgt(NR)/EL4-RORgt.Flag-ChIP-Seq(GSE56019)/Homer |
| 5 |  | −7.28 | CRE(bZIP)/Promoter/Homer |
| Rank | Motif | Log (p Value) | Name |
|---|---|---|---|
| 1 |  | −13.47 | HRE(HSF)/Striatum-HSF1-ChIP-Seq(GSE38000)/Homer |
| 2 |  | −10.92 | THRa(NR)/C17.2-THRa-ChIP-Seq(GSE38347)/Homer |
| 3 |  | −10.24 | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer |
| 4 |  | −9.73 | GEI-11(Myb?)/cElegans-L4-GEI11-ChIP-Seq(modEncode)/Homer |
| 5 |  | −8.97 | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, L.; Liu, S.; Zhang, L.; Yang, H.; Sun, L. MITF Contributes to the Body Color Differentiation of Sea Cucumbers Apostichopus japonicus through Expression Differences and Regulation of Downstream Genes. Biology 2023, 12, 1. https://doi.org/10.3390/biology12010001
Xing L, Liu S, Zhang L, Yang H, Sun L. MITF Contributes to the Body Color Differentiation of Sea Cucumbers Apostichopus japonicus through Expression Differences and Regulation of Downstream Genes. Biology. 2023; 12(1):1. https://doi.org/10.3390/biology12010001
Chicago/Turabian StyleXing, Lili, Shilin Liu, Libin Zhang, Hongsheng Yang, and Lina Sun. 2023. "MITF Contributes to the Body Color Differentiation of Sea Cucumbers Apostichopus japonicus through Expression Differences and Regulation of Downstream Genes" Biology 12, no. 1: 1. https://doi.org/10.3390/biology12010001
APA StyleXing, L., Liu, S., Zhang, L., Yang, H., & Sun, L. (2023). MITF Contributes to the Body Color Differentiation of Sea Cucumbers Apostichopus japonicus through Expression Differences and Regulation of Downstream Genes. Biology, 12(1), 1. https://doi.org/10.3390/biology12010001






