Multiscale Analysis of Cellular Composition and Morphology in Intact Cerebral Organoids
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture of Cerebral Organoid
2.2. Lentiviral Transduction of Organoids
2.3. 3D Immunostaining
2.4. Resin Embedding
2.5. Confocal Microscopy
2.6. fMOST Imaging
2.7. Visualization and Reconstruction
3. Results
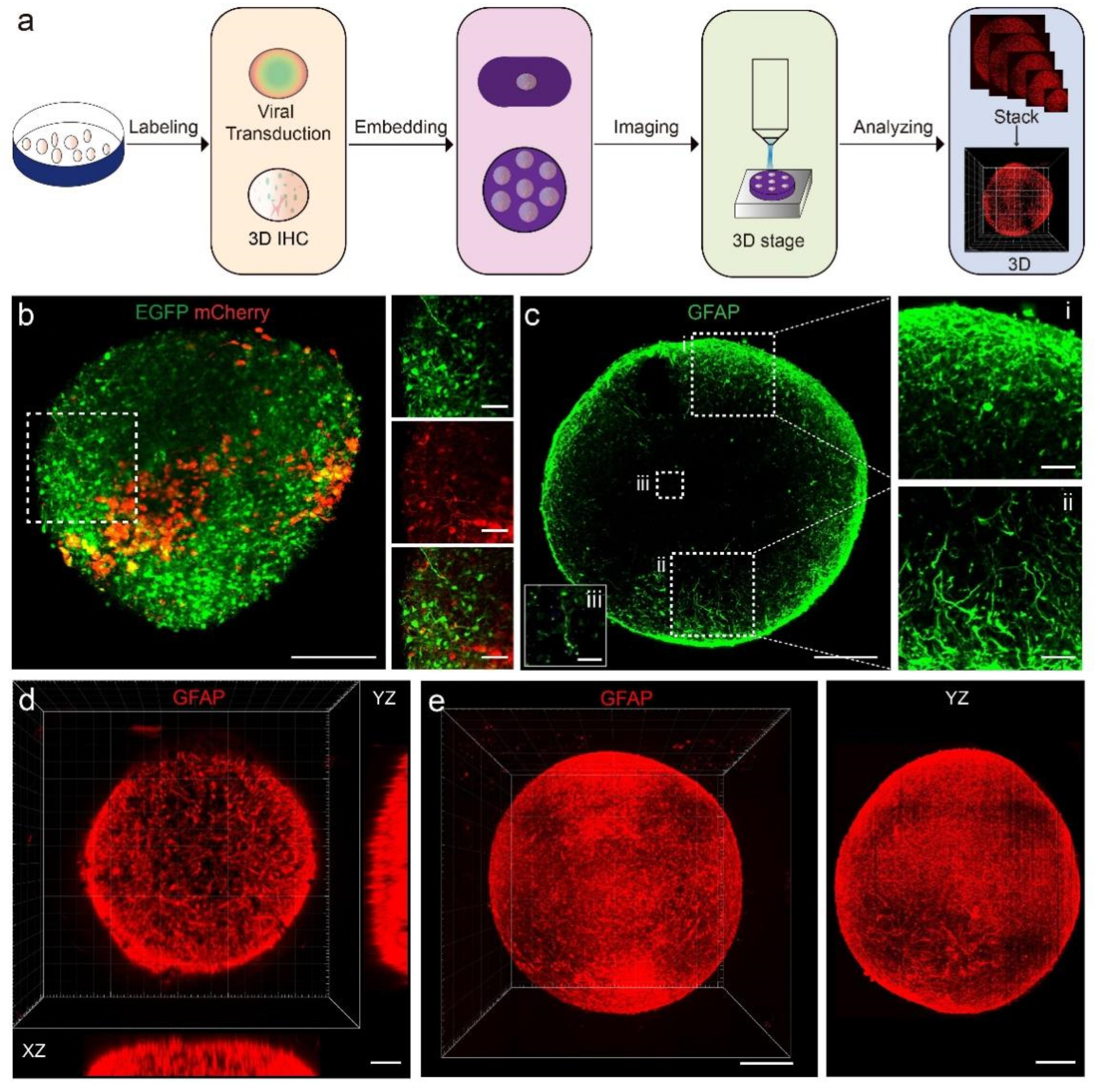
3.1. High-Resolution 3D Imaging Pipeline for Cerebral Organoids
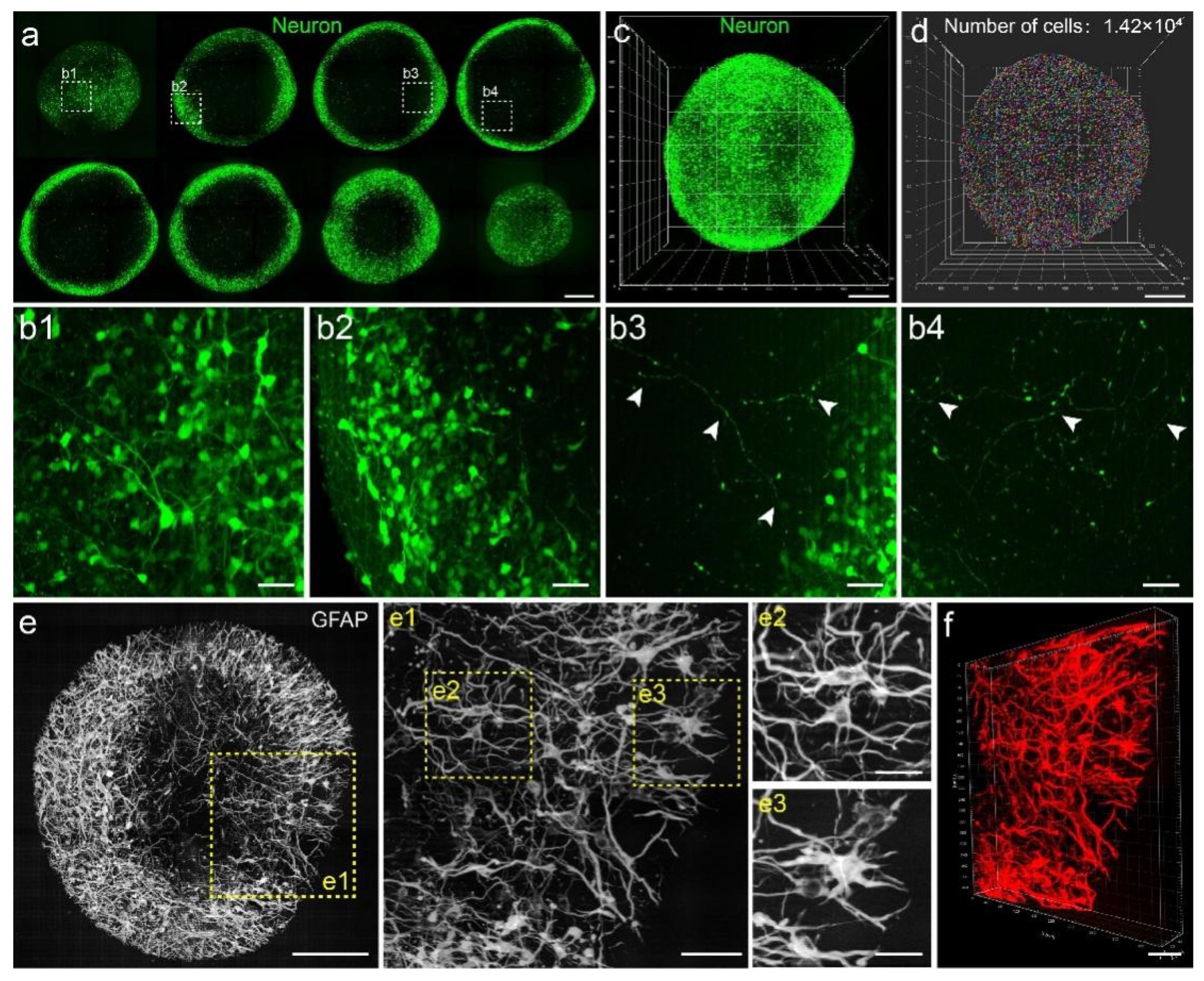
3.2. Visualization of Fine Structures of Neurons and GFAP+ Cells
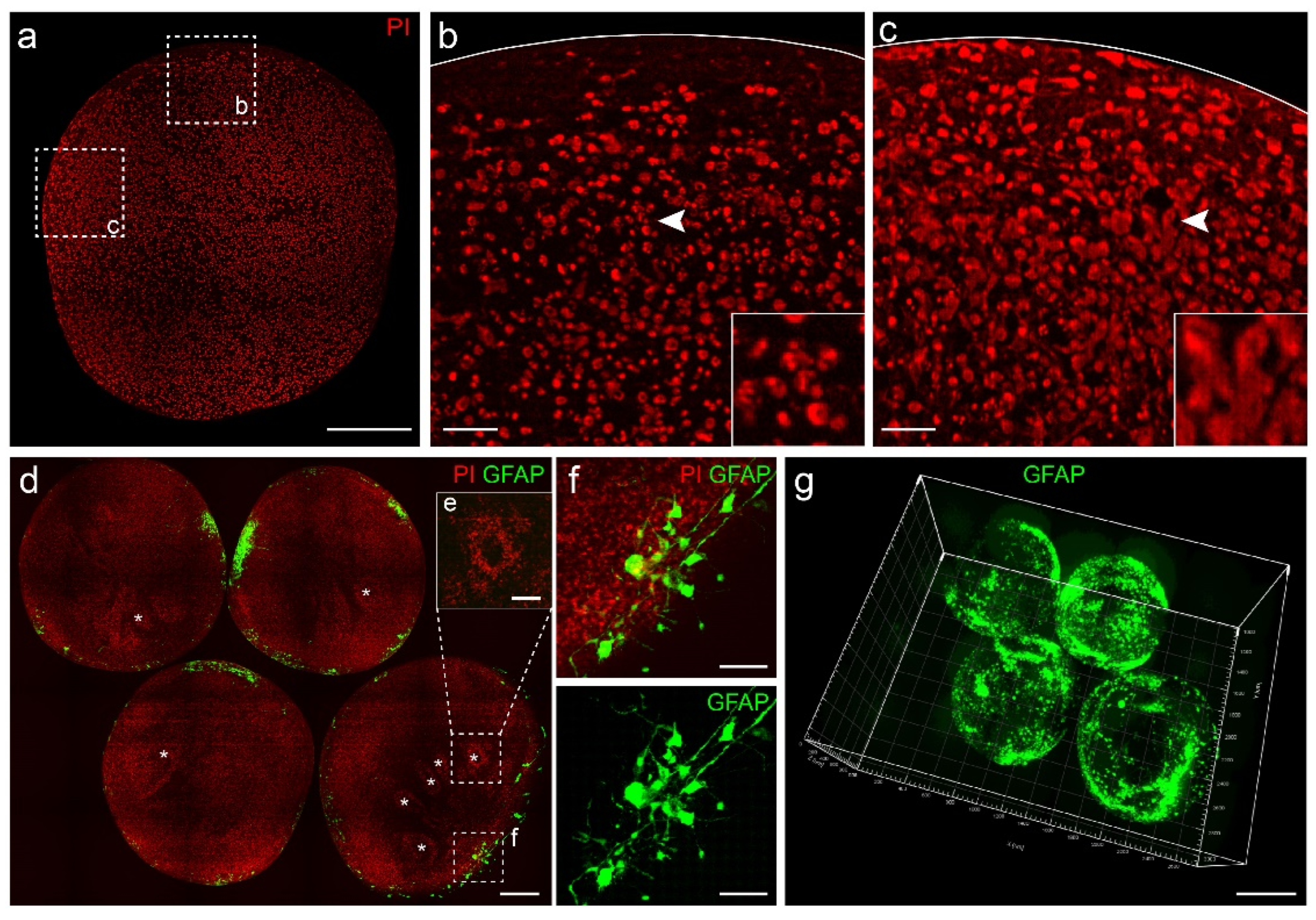
3.3. Propidium Iodide (PI) Staining Provided Cytoarchitecture Information
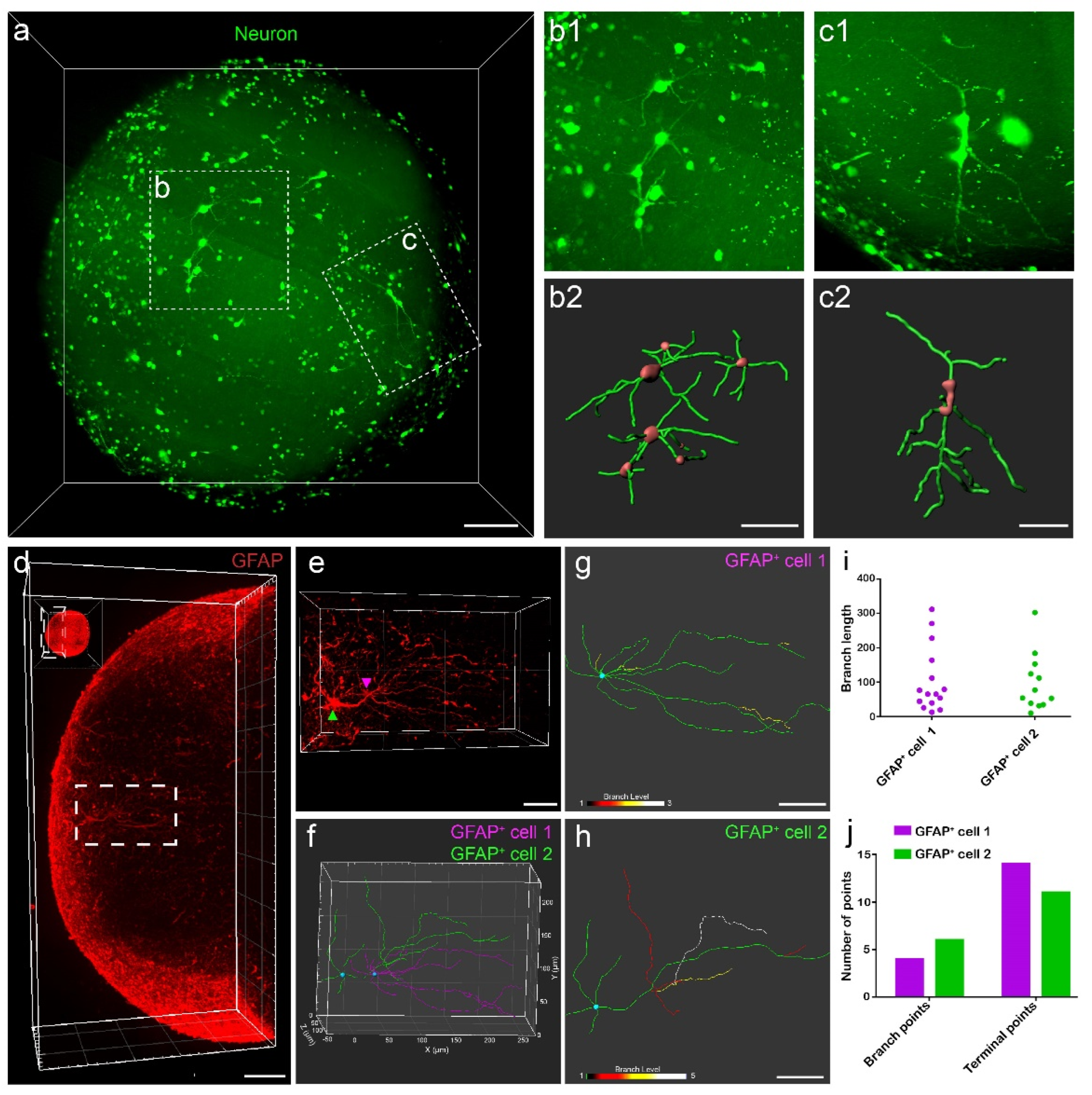
3.4. Single-Cell Reconstruction Using 3D Datasets of Organoids
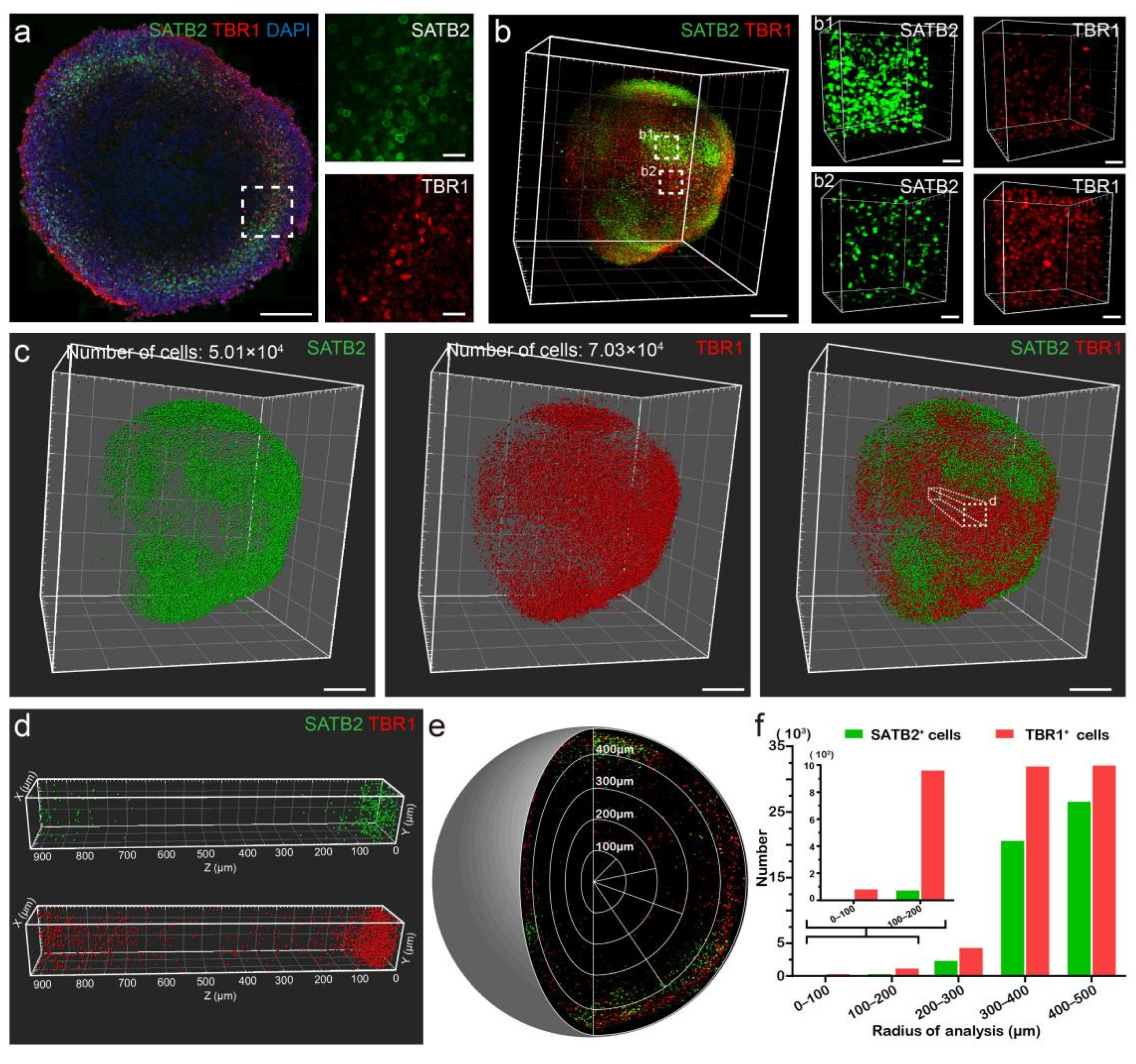
3.5. Spatial Localization of Cortical Layer Markers in Organoids
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kadoshima, T.; Sakaguchi, H.; Nakano, T.; Soen, M.; Ando, S.; Eiraku, M.; Sasai, Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 2013, 110, 20284–20289. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in Vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Czerniecki, S.M.; Cruz, N.M.; Harder, J.L.; Menon, R.; Annis, J.; Otto, E.A.; Gulieva, R.E.; Islas, L.V.; Kim, Y.K.; Tran, L.M.; et al. High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell 2018, 22, 929–940.e924. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, A.; Ozone, C.; Nakano, T.; Saito, K.; Eiraku, M.; Sasai, Y. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat. Commun. 2015, 6, 6286. [Google Scholar] [CrossRef]
- Assawachananont, J.; Mandai, M.; Okamoto, S.; Yamada, C.; Eiraku, M.; Yonemura, S.; Sasai, Y.; Takahashi, M. Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Rep. 2014, 2, 662–674. [Google Scholar] [CrossRef]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Chiaradia, I.; Lancaster, M.A. Brain organoids for the study of human neurobiology at the interface of in vitro and in Vivo. Nat. Neurosci. 2020, 23, 1496–1508. [Google Scholar] [CrossRef]
- Camp, J.G.; Badsha, F.; Florio, M.; Kanton, S.; Gerber, T.; Wilsch-Brauninger, M.; Lewitus, E.; Sykes, A.; Hevers, W.; Lancaster, M.; et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA 2015, 112, 15672–15677. [Google Scholar] [CrossRef]
- Qian, X.; Su, Y.; Adam, C.D.; Deutschmann, A.U.; Pather, S.R.; Goldberg, E.M.; Su, K.; Li, S.; Lu, L.; Jacob, F.; et al. Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell 2020, 26, 766–781.e769. [Google Scholar] [CrossRef]
- Renner, H.; Grabos, M.; Becker, K.J.; Kagermeier, T.E.; Wu, J.; Otto, M.; Peischard, S.; Zeuschner, D.; TsyTsyura, Y.; Disse, P.; et al. A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. Elife 2020, 9, e52904. [Google Scholar] [CrossRef]
- Bershteyn, M.; Nowakowski, T.J.; Pollen, A.A.; Di Lullo, E.; Nene, A.; Wynshaw-Boris, A.; Kriegstein, A.R. Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell 2017, 20, 435–449.e434. [Google Scholar] [CrossRef]
- Mora-Bermudez, F.; Badsha, F.; Kanton, S.; Camp, J.G.; Vernot, B.; Kohler, K.; Voigt, B.; Okita, K.; Maricic, T.; He, Z.S.; et al. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife 2016, 5, e18683. [Google Scholar] [CrossRef]
- Knight, G.T.; Lundin, B.F.; Iyer, N.; Ashton, L.M.T.; Sethares, W.A.; Willett, R.M.; Ashton, R.S. Engineering induction of singular neural rosette emergence within hPSC-derived tissues. Elife 2018, 7, e37549. [Google Scholar] [CrossRef]
- Albanese, A.; Swaney, J.M.; Yun, D.H.; Evans, N.B.; Antonucci, J.M.; Velasco, S.; Sohn, C.H.; Arlotta, P.; Gehrke, L.; Chung, K. Multiscale 3D phenotyping of human cerebral organoids. Sci. Rep. 2020, 10, 21487. [Google Scholar] [CrossRef]
- Masselink, W.; Reumann, D.; Murawala, P.; Pasierbek, P.; Taniguchi, Y.; Bonnay, F.; Meixner, K.; Knoblich, J.A.; Tanaka, E.M. Broad applicability of a streamlined ethyl cinnamate-based clearing procedure. Development 2019, 146, dev166884. [Google Scholar] [CrossRef]
- Nurnberg, E.; Vitacolonna, M.; Klicks, J.; von Molitor, E.; Cesetti, T.; Keller, F.; Bruch, R.; Ertongur-Fauth, T.; Riedel, K.; Scholz, P.; et al. Routine Optical Clearing of 3D-Cell Cultures: Simplicity Forward. Front. Mol. Biosci. 2020, 7, 20. [Google Scholar] [CrossRef]
- Cora, V.; Haderspeck, J.; Antkowiak, L.; Mattheus, U.; Neckel, P.H.; Mack, A.F.; Bolz, S.; Ueffing, M.; Pashkovskaia, N.; Achberger, K.; et al. A Cleared View on Retinal Organoids. Cells 2019, 8, 391. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Alieva, M.; Wellens, L.M.; Ariese, H.C.R.; Jamieson, P.R.; Vonk, A.M.; Amatngalim, G.D.; Hu, H.; Oost, K.C.; Snippert, H.J.G.; et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 2019, 14, 1756–1771. [Google Scholar] [CrossRef]
- Li, Y.; Muffat, J.; Omer, A.; Bosch, I.; Lancaster, M.A.; Sur, M.; Gehrke, L.; Knoblich, J.A.; Jaenisch, R. Induction of Expansion and Folding in Human Cerebral Organoids. Cell Stem Cell 2017, 20, 385–396.e383. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, M.A.; Corsini, N.S.; Wolfinger, S.; Gustafson, E.H.; Phillips, A.W.; Burkard, T.R.; Otani, T.; Livesey, F.J.; Knoblich, J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017, 35, 659–666. [Google Scholar] [CrossRef]
- Xu, J.C.; Fan, J.; Wang, X.; Eacker, S.M.; Kam, T.I.; Chen, L.; Yin, X.; Zhu, J.; Chi, Z.; Jiang, H.; et al. Cultured networks of excitatory projection neurons and inhibitory interneurons for studying human cortical neurotoxicity. Sci. Transl. Med. 2016, 8, 333ra48. [Google Scholar] [CrossRef]
- Renier, N.; Wu, Z.; Simon, D.J.; Yang, J.; Ariel, P.; Tessier-Lavigne, M. iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 2014, 159, 896–910. [Google Scholar] [CrossRef]
- Ren, M.; Tian, J.J.; Zhao, P.L.; Luo, J.L.; Feng, Z.; Gong, H.; Li, X.N. Simultaneous Acquisition of Multicolor Information From Neural Circuits in Resin-Embedded Samples. Front. Neurosci. 2018, 12, 885. [Google Scholar] [CrossRef]
- Gong, H.; Xu, D.L.; Yuan, J.; Li, X.N.; Guo, C.D.; Peng, J.; Li, Y.X.; Schwarz, L.A.; Li, A.A.; Hu, B.H.; et al. High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat. Commun. 2016, 7, 12142. [Google Scholar] [CrossRef]
- Jiang, Y.; Gong, H.S.; Jiang, S.H.; She, C.W.; Cao, Y. Multi-walled carbon nanotubes decrease neuronal NO synthase in 3D brain organoids. Sci. Total Environ. 2020, 748, 141384. [Google Scholar] [CrossRef]
- Birey, F.; Andersen, J.; Makinson, C.D.; Islam, S.; Wei, W.; Huber, N.; Fan, H.C.; Metzler, K.R.C.; Panagiotakos, G.; Thom, N.; et al. Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545, 54–59. [Google Scholar] [CrossRef]
- Pamies, D.; Block, K.; Lau, P.; Gribaldo, L.; Pardo, C.A.; Barreras, P.; Smirnova, L.; Wiersma, D.; Zhao, L.; Harris, G.; et al. Rotenone exerts developmental neurotoxicity in a human brain spheroid model. Toxicol Appl. Pharm. 2018, 354, 101–114. [Google Scholar] [CrossRef]
- Kobolak, J.; Teglasi, A.; Bellak, T.; Janstova, Z.; Molnar, K.; Zana, M.; Bock, I.; Laszlo, L.; Dinnyes, A. Human Induced Pluripotent Stem Cell-Derived 3D-Neurospheres are Suitable for Neurotoxicity Screening. Cells 2020, 9, 1122. [Google Scholar] [CrossRef]
- Renner, M.; Lancaster, M.A.; Bian, S.; Choi, H.; Ku, T.; Peer, A.; Chung, K.; Knoblich, J.A. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017, 36, 1316–1329. [Google Scholar] [CrossRef]
- Bagley, J.A.; Reumann, D.; Bian, S.; Levi-Strauss, J.; Knoblich, J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods 2017, 14, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Tanaka, Y.; Patterson, B.; Kang, Y.J.; Govindaiah, G.; Roselaar, N.; Cakir, B.; Kim, K.Y.; Lombroso, A.P.; Hwang, S.M.; et al. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 2017, 21, 383–398.e387. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.F.; Tanaka, Y.; Cakir, B.; Patterson, B.; Kim, K.Y.; Sun, P.N.; Kang, Y.J.; Zhong, M.; Liu, X.R.; Patra, P.; et al. hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell 2019, 24, 487–497.e7. [Google Scholar] [CrossRef] [PubMed]
- Adesnik, H.; Abdeladim, L. Probing neural codes with two-photon holographic optogenetics. Nat. Neurosci. 2021, 24, 1356–1366. [Google Scholar] [CrossRef]
- Ghatak, S.; Dolatabadi, N.; Trudler, D.; Zhang, X.T.; Wu, Y.; Mohata, M.; Ambasudhan, R.; Talantova, M.; Lipton, S.A. Mechanisms of hyperexcitability in Alzheimer’s disease hiPSC-derived neurons and cerebral organoids vs isogenic controls. eLife 2019, 8, e50333. [Google Scholar] [CrossRef]
- Lin, Y.T.; Seo, J.; Gao, F.; Feldman, H.M.; Wen, H.L.; Penney, J.; Cam, H.P.; Gjoneska, E.; Raja, W.K.; Cheng, J.; et al. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron 2018, 98, 1294. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, L.; Yu, H.; Yin, F.; Wang, Y.; Liu, H.; Jiang, L.; Qin, J. In situ generation of human brain organoids on a micropillar array. Lab Chip 2017, 17, 2941–2950. [Google Scholar] [CrossRef]
- Govindan, S.; Batti, L.; Osterop, S.F.; Stoppini, L.; Roux, A. Mass Generation, Neuron Labeling, and 3D Imaging of Minibrains. Front. Bioeng. Biotechnol. 2021, 8, 582650. [Google Scholar] [CrossRef]
| Antibody | Dilution | Cat. No. | Company |
|---|---|---|---|
| Anti-SATB2, mouse | 1:200 | ab51502 | Abcam, Cambridge, UK |
| Anti-TBR1, rabbit | 1:200 | ab31940 | Abcam, Cambridge, UK |
| Anti-GFAP, rabbit | 1:200 | Z0334 | Dako, Glostrup, Denmark |
| Anti-NeuN, rabbit | 1:200 | ab177487 | Abcam, Cambridge, UK |
| Alexa Fluor 488 donkey anti-mouse lgG (H + L) | 1:500 | A21202 | Invitrogen, Waltham, MA, USA |
| Alexa Fluor 594 donkey anti-rabbit lgG (H + L) | 1:500 | A21207 | Invitrogen, Waltham, MA, USA |
| Alexa Fluor 488 donkey anti-rabbit lgG (H + L) | 1:500 | A21206 | Invitrogen, Waltham, MA, USA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, H.; Chen, J.; Deng, Z.; Sun, T.; Luo, Q.; Gong, H.; Li, X.; Long, B. Multiscale Analysis of Cellular Composition and Morphology in Intact Cerebral Organoids. Biology 2022, 11, 1270. https://doi.org/10.3390/biology11091270
Ma H, Chen J, Deng Z, Sun T, Luo Q, Gong H, Li X, Long B. Multiscale Analysis of Cellular Composition and Morphology in Intact Cerebral Organoids. Biology. 2022; 11(9):1270. https://doi.org/10.3390/biology11091270
Chicago/Turabian StyleMa, Haihua, Juan Chen, Zhiyu Deng, Tingting Sun, Qingming Luo, Hui Gong, Xiangning Li, and Ben Long. 2022. "Multiscale Analysis of Cellular Composition and Morphology in Intact Cerebral Organoids" Biology 11, no. 9: 1270. https://doi.org/10.3390/biology11091270
APA StyleMa, H., Chen, J., Deng, Z., Sun, T., Luo, Q., Gong, H., Li, X., & Long, B. (2022). Multiscale Analysis of Cellular Composition and Morphology in Intact Cerebral Organoids. Biology, 11(9), 1270. https://doi.org/10.3390/biology11091270






