Plasma Proteomic Profiling Reveals the Regulatory Factors of Milk Protein Synthesis in Holstein Cows
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Design
2.2. Plasma Sampling
2.3. Biochemical Index Measurement
2.4. Hormone Concentration Assay
2.5. Plasma Proteomic Analysis
2.6. Bovine Mammary Epithelial Cells Isolation and Culture
2.7. Cell Viability Assay
2.8. Western Blotting Assay
2.9. Statistical Analyses
3. Results
3.1. Plasma Biochemical Parameters and Hormones Concentrations
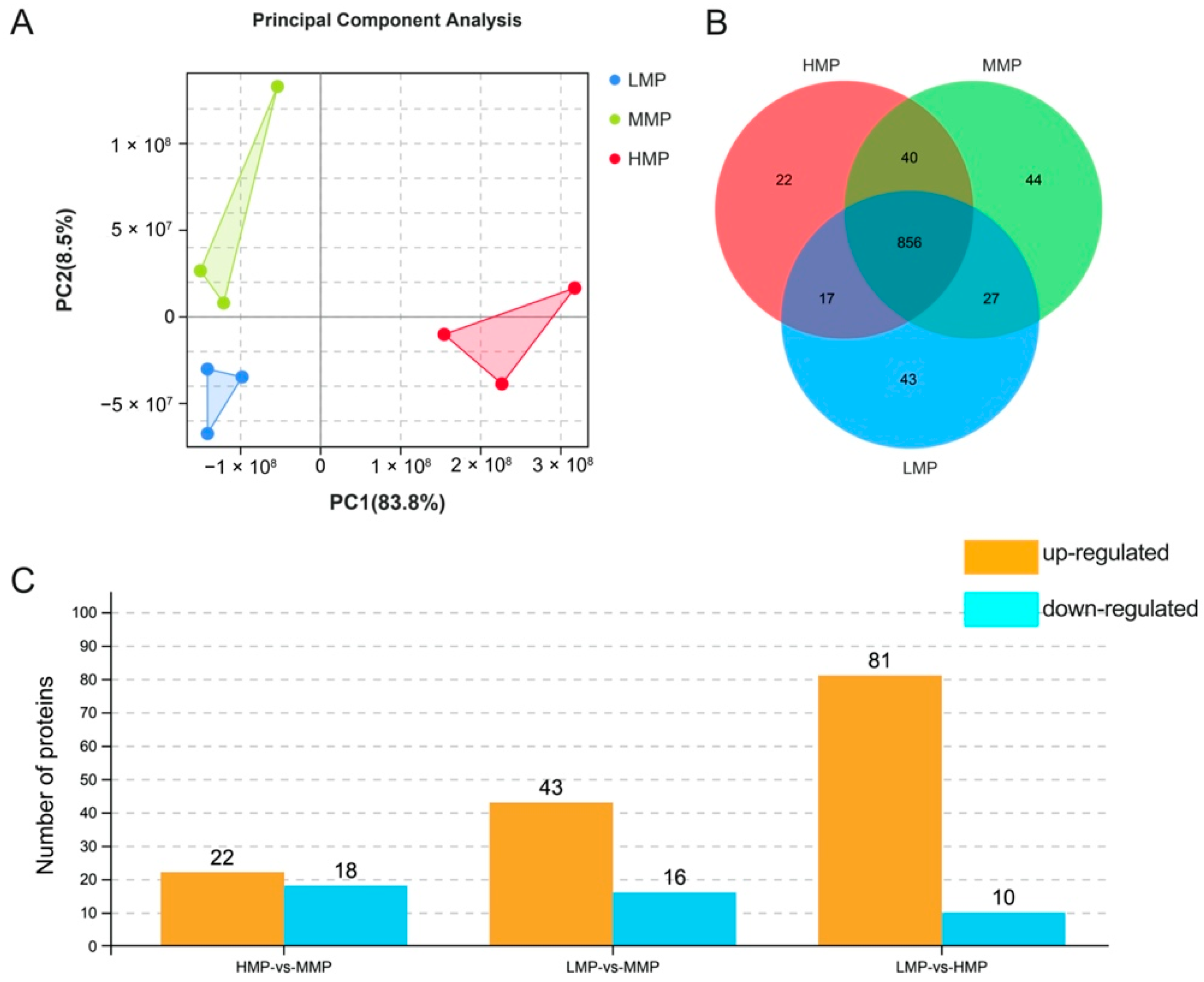
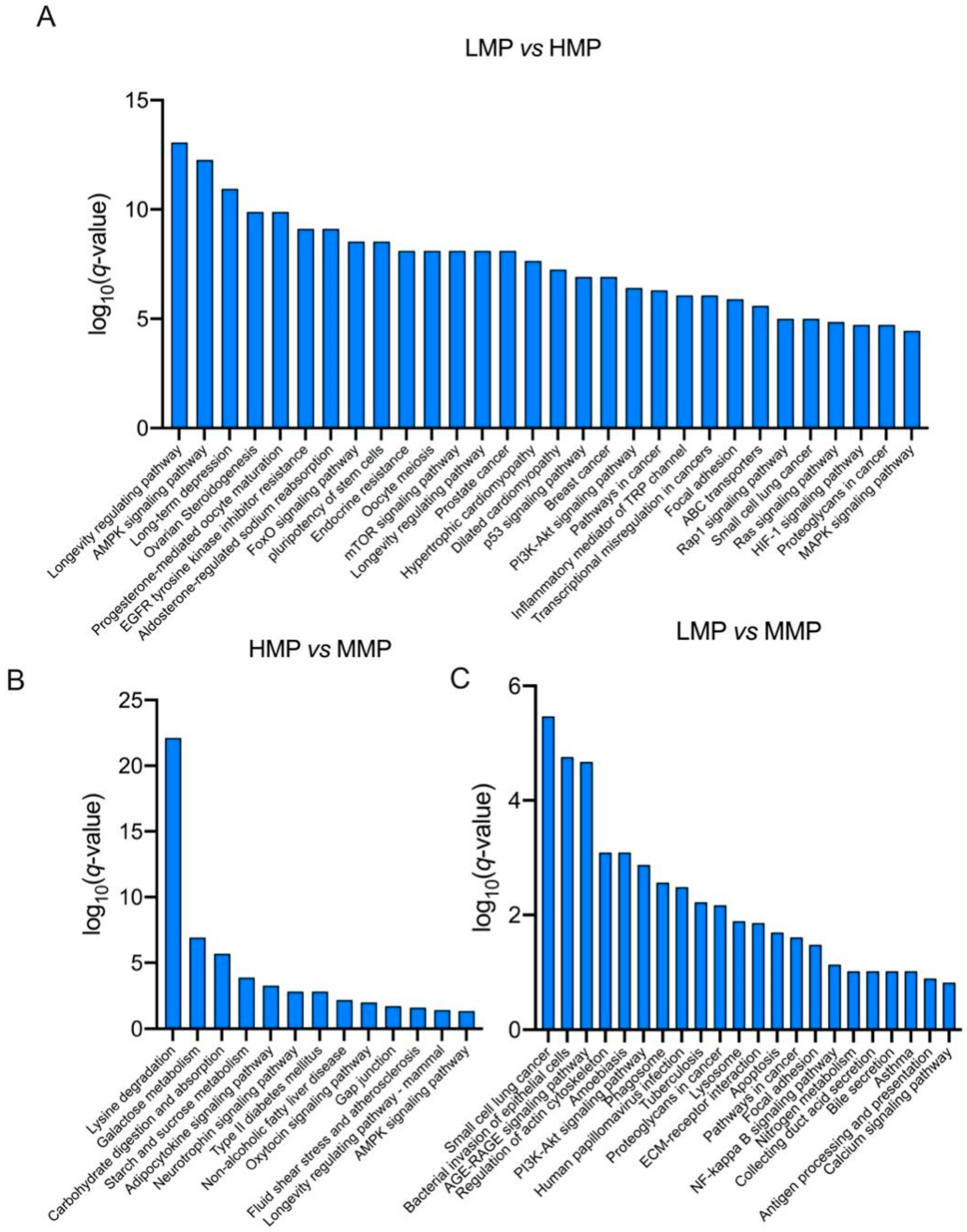
3.2. Plasma Proteome
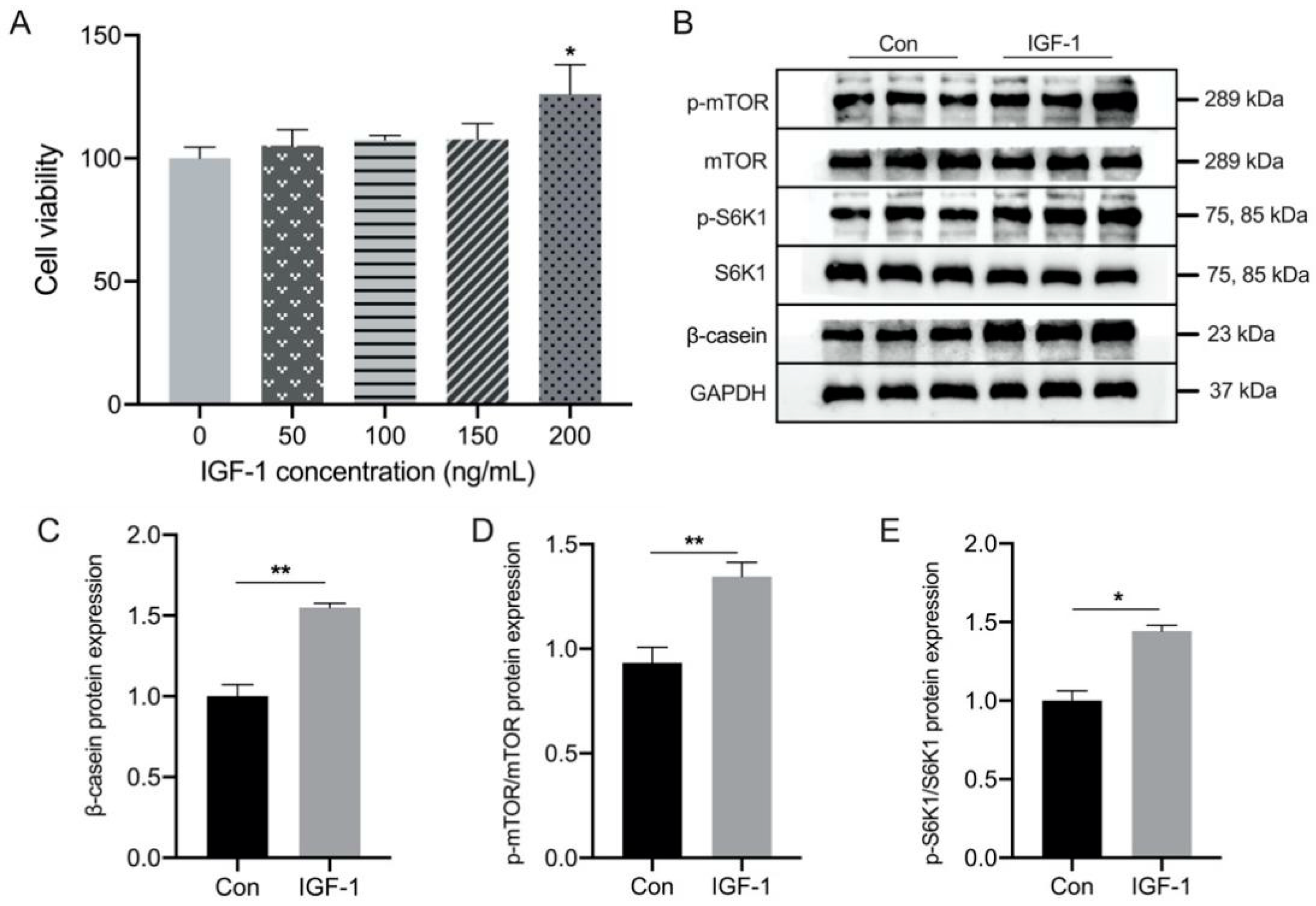
3.3. Effect of IGF-1 Treatment on the β-Casein and mTOR Signaling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, X.L.; Zeng, H.F.; Xu, J.; Zhai, Y.F.; Xia, H.B.; Xi, Y.M.; Han, Z.Y. Characteristics of ruminal microbiota and metabolome in Holstein cows differing in milk protein concentrations. J. Anim. Sci. 2022, skac253. [Google Scholar] [CrossRef] [PubMed]
- Toerien, C.A.; Trout, D.R.; Cant, J.P. Effect of nutrients on p70S6K activation in the bovine mammary gland. J. Anim. Feed. 2004, 13, 449–452. [Google Scholar] [CrossRef][Green Version]
- Xiao, C.B.; Zhang, L.; Cheng, H.C. Animal Anatomy and Histoembryology; China Agricultural University Press: Beijing, China, 2007. [Google Scholar]
- Emery, R.S.; Brown, L.D.; Bell, J.W. Correlation of Milk Fat with Dietary and Metabolic Factors in Cows Fed Restricted-Roughage Rations Supplemented with Magnesium Oxide or Sodium Bicarbonate1. J. Dairy Sci. 1965, 48, 1647–1651. [Google Scholar] [CrossRef]
- Cant, J.P.; DePeters, E.J.; Baldwin, R.L. Mammary Amino Acid Utilization in Dairy Cows Fed Fat and Its Relationship to Milk Protein Depression. J. Dairy Sci. 1993, 76, 762–774. [Google Scholar] [CrossRef]
- Burgos, S.A.; Dai, M.; Cant, J.P. Nutrient availability and lactogenic hormones regulate mammary protein synthesis through the mammalian target of rapamycin signaling pathway. J. Dairy Sci. 2010, 93, 153–161. [Google Scholar] [CrossRef]
- Burgos, S.A.; Cant, J.P. IGF-1 stimulates protein synthesis by enhanced signaling through mTORC1 in bovine mammary epithelial cells. Domest. Anim. Endocrinol. 2010, 38, 211–221. [Google Scholar] [CrossRef]
- Sílvia, B.-B.; Ramon, G. Proteomics in obesity research. Proteom.-Clin. Appl. 2010, 3, 263–278. [Google Scholar]
- Herosimczyk, A.; Lepczyński, A.; Ogo, M.; Dratwa-Chaupnik, A.; Michaek, K.; Skrzypczak, W.F. Blood plasma protein and lipid profile changes in calves during the first week of life. Pol. J. Vet. Sci. 2013, 16, 425–434. [Google Scholar] [CrossRef]
- Marques, I.; Vasconcelos, F.R.; Alves, J.; Montenegro, A.R.; Rondina, D. Proteome of milk fat globule membrane and mammary gland tissue in goat fed different lipid supplementation. Small Rumin. Res. 2021, 199, 106378. [Google Scholar] [CrossRef]
- Koh, Y.Q.; Peiris, H.N.; Vaswani, K.; Almughlliq, F.B.; Meier, S.; Burke, C.R.; Roche, J.R.; Reed, C.B.; Arachchige, B.J.; Reed, S. Proteome profiling of exosomes derived from plasma of heifers with divergent genetic merit for fertility. J. Dairy Sci. 2018, 101, 6462–6473. [Google Scholar] [CrossRef]
- Yu, K.; Zhang, Y.; Chen, H.; Zhu, W. Hepatic Metabolomic and Transcriptomic Responses Induced by Cecal Infusion of Sodium Propionate in a Fistula Pig Model. J. Agric. Food Chem. 2019, 64, 13073–13081. [Google Scholar] [CrossRef] [PubMed]
- Muntel, J.; Xuan, Y.; Berger, S.T.; Reiter, L.; Bachur, R.; Kentsis, A.; Steen, H. Advancing Urinary Protein Biomarker Discovery by Data-Independent Acquisition on a Quadrupole-Orbitrap Mass Spectrometer. J. Proteome Res. 2015, 14, 4752–4762. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Xu, J.; Han, Z.Y. Enhancement of BCAT2-Mediated Valine Catabolism Stimulates-Casein Synthesis via the AMPK-mTOR Signaling Axis in Bovine Mammary Epithelial Cells. J. Agric. Food Chem. 2022, 70, 9898–9907. [Google Scholar] [CrossRef]
- Merhi, Z.; Bazzi, A.A.; Bonney, E.A.; Buyuk, E. Role of adiponectin in ovarian follicular development and ovarian reserve. Biomed. Rep. 2019, 10, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Shi, H.; Jin, Y.; Li, X.; Pan, J.; Lai, Y.; Lin, Y.; Jin, Y.; Gaurab, R.; Zhao, A. Adiponectin Deficiency Leads to Female Subfertility and Ovarian Dysfunctions in Mice. Endocrinology 2016, 157, 2015–2080. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wu, J.; Liu, D.; Shan, H.; Zhang, J. Anti-inflammatory effect of full-length adiponectin and proinflammatory effect of globular adiponectin in esophageal adenocarcinoma cells. Oncol. Res. 2013, 21, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Jeong, J.; Woo, J.; Lee, C.H.; Yoo, C.G. Globular Adiponectin Exerts a Pro-Inflammatory Effect via IκB/NF-κB Pathway Activation and Anti-Inflammatory Effect by IRAK-1 Downregulation. Mol. Cells 2018, 41, 762–770. [Google Scholar] [PubMed]
- Liu, Y.; Palanivel, R.; Rai, E.; Park, M.; Gabor, T.V.; Scheid, M.P.; Xu, A.; Sweeney, G. Adiponectin Stimulates Autophagy and Reduces Oxidative Stress to Enhance Insulin Sensitivity During High-Fat Diet Feeding in Mice. Diabetes 2015, 64, 36–48. [Google Scholar] [CrossRef]
- Sena, C.M.; Pereira, A.; Fernandes, R.; Letra, L.; Seia, R.M. Adiponectin improves endothelial function in mesenteric arteries of rats fed a high-fat diet: Role of perivascular adipose tissue. Br. J. Pharmacol. 2017, 174, 3514–3526. [Google Scholar] [CrossRef]
- Liu, Y.; Turdi, S.; Park, T.; Morris, N.J.; Deshaies, Y.; Xu, A.; Sweeney, G. Adiponectin Corrects High-Fat Diet-Induced Disturbances in Muscle Metabolomic Profile and Whole-Body Glucose Homeostasis. Diabetes 2013, 62, 743–752. [Google Scholar] [CrossRef]
- Mohammed, M.H.; Al-Thuwaini, T.M.; Al-Shuhaib, M. High association of a novel variant in the adiponectin gene with the litter size in Awassi ewes. J. Saudi Soc. Agric. Sci. 2022, 21, 296–301. [Google Scholar] [CrossRef]
- Waalkes, S.; Eggers, H.; Großhennig, A.; Hennenlotter, J.; Atschekzei, F.; Tränkenschuh, W.; Stenzl, A.; Schrader, A.J.; Merseburger, A.S.; Kuczyk, M.A. 738 fibronectin 1 Mrna expression correlates with risk of recurrence in renal cell cancer. Eur. Urol. Suppl. 2011, 10, 235. [Google Scholar] [CrossRef]
- Hof, G.; Vervoorn, M.D.; Lenaers, P.J.; Tamminga, S. Milk urea nitrogen as a tool to monitor the protein nutrition of dairy cows. J. Dairy Sci. 1997, 80, 3333–3340. [Google Scholar] [CrossRef]
- Blowey, R.W.; Wood, D.W.; Davis, J.R. A nutritional monitoring system for dairy herds based on blood glucose, urea and albumin levels. Vet. Record. 1973, 92, 691–696. [Google Scholar] [CrossRef]
- Toerien, C.A.; Trout, D.R.; Cant, J.P. Nutritional Stimulation of Milk Protein Yield of Cows Is Associated with Changes in Phosphorylation of Mammary Eukaryotic Initiation Factor 2 and Ribosomal S6 Kinase 1. J. Nutr. 2010, 140, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.C.; Zhao, S.G.; Wang, S.S.; Luo, C.C.; Gao, H.N.; Zheng, N.; Wang, J.Q. d-Glucose and amino acid deficiency inhibits casein synthesis through JAK2/STAT5 and AMPK/mTOR signaling pathways in mammary epithelial cells of dairy cows. J. Dairy Sci. 2018, 101, 1737–1746. [Google Scholar] [CrossRef]
- Puppel, K.; Kuczyńska, B. Metabolic profiles of cow’s blood: A review. J. Sci. Food Agric. 2016, 96, 4321–4328. [Google Scholar] [CrossRef]
- Payne, A.H.; Hales, D.B. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr. Rev. 2004, 25, 947–970. [Google Scholar] [CrossRef]
- Sakowski, T.; Kuczyńska, B.; Puppel, K.; Metera, E.; Słoniewski, K.; Barszczewski, J. Relationships between physiological indicators in blood, and their yield, as well as chemical composition of milk obtained from organic dairy cows. J. Sci. Food Agric. 2012, 92, 2905–2912. [Google Scholar] [CrossRef]
- Xie, Y.; Wu, Z.; Wang, D.; Liu, J. Nitrogen partitioning and microbial protein synthesis in lactating dairy cows with different phenotypic residual feed intake. J. Anim. Sci. Biotechnol. 2019, 10, 54. [Google Scholar] [CrossRef]
- Jacquemet, N.; Prigge, E.C. Effect of prolactin infusion on lactation, glucose kinetics, and pancreatic hormones in lactating goats. J. Dairy Sci. 1990, 73, 3433–3438. [Google Scholar] [CrossRef]
- Hart, I.C. Effect of 2-bromo-α-ergocryptine on milk yield and the level of prolactin and growth hormone in the blood of the goat at milking. J. Endocrinol. 1973, 57, 179–180. [Google Scholar] [CrossRef] [PubMed]
- Lacasse, P.; Ollier, S.; Lollivier, V.; Boutinaud, M. New insights into the importance of prolactin in dairy ruminants. J. Dairy Sci. 2016, 99, 864–874. [Google Scholar] [CrossRef] [PubMed]
- Plaut, K.; Bauman, D.E.; Agergaard, N.; Akers, R.M. Effect of exogenous prolactin administration on lactational performance of dairy cows. Domest. Anim. Endocrinol. 1987, 4, 279–290. [Google Scholar] [CrossRef]
- Qian, Z.; Su, H.; Wang, F.; Cao, Z.; Li, S. Effects of energy density in close-up diets and postpartum supplementation of extruded full-fat soybean on lactation performance and metabolic and hormonal status of dairy cows. J. Dairy Sci. 2015, 98, 7115–7130. [Google Scholar]
- Bionaz, M.; Loor, J.J. Gene Networks Driving Bovine Mammary Protein Synthesis During the Lactation Cycle. Bioinform. Biol. Insights 2011, 2011, 83–98. [Google Scholar] [CrossRef]
- Menzies, K.K.; Lefèvre, C.; Macmillan, K.L.; Nicholas, K.R. Insulin regulates milk protein synthesis at multiple levels in the bovine mammary gland. Funct. Integr. Genom. 2009, 9, 197–217. [Google Scholar] [CrossRef]
- Sciascia, Q.; Pacheco, D.; Mccoard, S.A. Increased milk protein synthesis in response to exogenous growth hormone is associated with changes in mechanistic (mammalian) target of rapamycin (mTOR)C1-dependent and independent cell signaling. J. Dairy Sci. 2013, 96, 2327–2338. [Google Scholar] [CrossRef]
- Capuco, A.V.; Wood, D.L.; Baldwin, R. Mammary cell number, proliferation, and apoptosis during a bovine lactation: Relation to milk production and effect of bST. J. Dairy Sci. 2001, 84, 2177–2187. [Google Scholar] [CrossRef]
- Chaiyabutr, N.; Boonsanit, D.; Chanpongsang, S. Effects of Cooling and Exogenous Bovine Somatotropin on Hematological and Biochemical Parameters at Different Stages of Lactation of Crossbred Holstein Friesian Cow in the Tropics. Asian Australas. J. Anim. Sci. 2011, 24, 230–238. [Google Scholar] [CrossRef]
- Prosser, C.G.; Fleet, I.R.; Corps, A.N. Increased secretion of insulin-like growth factor I into milk of cows treated with recombinantly derived bovine growth hormone. J. Dairy Res. 1989, 56, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, A.A.; Nones, K.; Roy, N.C.; Mcnabb, W.C.; Mackenzie, D.S.; Pacheco, D.; Mccoard, S. Initiation and elongation steps of mRNA translation are involved in the increase in milk protein yield caused by growth hormone administration during lactation. J. Dairy Sci. 2009, 92, 1889–1899. [Google Scholar] [CrossRef] [PubMed]


| Parameter | Group | p-Value 1 | ||
|---|---|---|---|---|
| LMP | MMP | HMP | ||
| Aspartate aminotransferase, IU/L | 109.89 ± 7.12 a | 86.22 ± 5.78 b | 90.22 ± 4.63 b | 0.02 |
| Alanine aminotransferase, IU/L | 36.30 ± 3.33 a | 28.40 ± 2.42 b | 38.00 ± 2.00 a | 0.04 |
| Total protein, mmol/L | 69.34 ± 1.62 ab | 65.10 ± 2.45 b | 72.29 ± 2.80 a | 0.11 |
| Albumin, g/L | 29.15 ± 0.56 | 28.54 ± 0.86 | 28.36 ± 0.78 | 0.73 |
| Globulin, g/L | 40.19 ± 1.11 ab | 36.55 ± 1.90 b | 43.93 ± 2.17 a | 0.02 |
| Total bilirubin, μmol/L | 0.99 ± 0.11 | 1.65 ± 0.90 | 0.86 ± 0.20 | 0.57 |
| Total bile acid, μmol/L | 31.50 ± 7.88 | 24.68 ± 4.52 | 31.96 ± 3.86 | 0.61 |
| Lactate dehydrogenase, g/L | 955.50 ± 36.31 | 902.20 ± 50.71 | 939.44 ± 39.69 | 0.66 |
| α-Hydroxybutyric acid, g/L | 970.70 ± 43.15 | 978.20 ± 69.51 | 968.00 ± 48.34 | 0.99 |
| Urea, mmol/L | 5.04 ± 0.26 | 4.15 ± 0.29 | 4.36 ± 0.52 | 0.20 |
| Creatinine, umol/L | 61.05 ± 3.13 | 60.72 ± 5.77 | 65.49 ± 2.63 | 0.68 |
| Glucose, mmol/L | 3.16 ± 0.06 b | 3.27 ± 0.11 ab | 3.50 ±0.11 a | 0.06 |
| Total cholesterol, mmol/L | 5.10 ±0.34 a | 4.29 ± 0.36 ab | 3.80 ± 0.26 b | 0.03 |
| Triglyceride, mmol/L | 0.14 ± 0.01 | 0.18 ± 0.04 | 0.16 ± 0.01 | 0.52 |
| High-density lipoprotein, mmol/L | 1.70 ± 0.06 a | 1.35 ± 0.08 b | 1.43 ± 0.07 b | <0.01 |
| Low-density lipoprotein, mmol/L | 1.94 ± 0.16 a | 1.61 ± 0.14 ab | 1.47 ± 0.11 b | 0.07 |
| ID | Protein | Group | |||||
|---|---|---|---|---|---|---|---|
| MMP vs. HMP | MMP vs. LMP | LMP vs. HMP | |||||
| log2FC 1 | p-Value 2 | log2FC | p-Value | log2FC | p-Value | ||
| NP_783630.2 | CGN1 | 1.090 | 0.003 | 0.189 | 0.031 | 0.900 | 0.000 |
| NP_001029390.1 | FETUB | −1.120 | 0.000 | −1.563 | 0.000 | 0.444 | 0.000 |
| XP_015327629.1 | ADIPOQ | 0.663 | 0.001 | −0.454 | 0.000 | 1.117 | 0.000 |
| NP_803450.2 | TF | 0.273 | 0.000 | −0.349 | 0.693 | 0.622 | 0.000 |
| NP_777246.1 | SERPING1 | 0.207 | 0.077 | −0.493 | 0.000 | 0.700 | 0.000 |
| NP_001015590.2 | ITIH4 | 0.456 | 0.078 | −0.262 | 0.000 | 0.717 | 0.000 |
| NP_001137569.1 | CRP | 0.007 | 0.127 | 1.181 | 0.000 | −1.174 | 0.000 |
| NP_001157250.1 | FN1 | 0.354 | 0.602 | −0.650 | 0.000 | 1.004 | 0.000 |
| NP_001159957.1 | C4A | 0.320 | 0.513 | −0.343 | 0.000 | 0.663 | 0.000 |
| NP_001094702.1 | CPN2 | 0.147 | 0.085 | −0.673 | 0.001 | 0.820 | 0.000 |
| NP_001193699.1 | MAP2K5 | 0.627 | 0.002 | −0.262 | 0.291 | 0.890 | 0.000 |
| XP_005202014.2 | ICA | 0.005 | 0.390 | −0.587 | 0.002 | 0.593 | 0.000 |
| NP_001033643.1 | ADPRH | 0.416 | 0.043 | −0.312 | 0.073 | 0.728 | 0.002 |
| NP_001095368.1 | ITIH3 | −1.075 | 0.000 | −0.533 | 0.010 | −0.542 | 0.002 |
| NP_001106748.1 | KNG2 | −0.572 | 0.030 | −0.982 | 0.005 | 0.409 | 0.003 |
| XP_002699259.1 | MSANTD2 | 1.229 | 0.342 | −0.425 | 0.664 | 1.654 | 0.003 |
| NP_777253.1 | GLYCAM1 | 0.794 | 0.055 | 0.095 | 0.744 | 0.699 | 0.004 |
| XP_005228030.1 | ATRX | 1.079 | 0.300 | 0.264 | 0.757 | 0.815 | 0.004 |
| NP_001073103.1 | CRISP3 | 0.571 | 0.352 | −0.569 | 0.006 | 1.140 | 0.005 |
| NP_001069646.1 | CFP | 0.257 | 0.241 | −0.507 | 0.430 | 0.764 | 0.010 |
| NP_001094532.1 | PF4 | −0.230 | 0.872 | −0.832 | 0.067 | 0.602 | 0.010 |
| XP_024855781.1 | Gucy1b2 | 0.558 | 0.137 | −0.357 | 0.401 | 0.915 | 0.012 |
| NP_001039988.1 | HAVCR1 | −0.026 | 0.168 | 0.576 | 0.088 | −0.602 | 0.014 |
| NP_001179392.1 | GOLM1 | 1.777 | 0.184 | 0.285 | 0.779 | 1.493 | 0.016 |
| NP_001107218.1 | ABCA3 | 0.459 | 0.012 | −0.300 | 0.242 | 0.759 | 0.020 |
| NP_001033763.1 | LBP | 0.334 | 0.969 | −0.687 | 0.008 | 1.021 | 0.022 |
| XP_024838406.1 | ITIH4 | 0.703 | 0.184 | −0.324 | 0.693 | 1.026 | 0.023 |
| NP_001033174.1 | RPP38 | −0.428 | 0.190 | 0.544 | 0.195 | −0.972 | 0.024 |
| XP_002689991.2 | SUSD1 | −0.431 | 0.581 | 0.432 | 0.256 | −0.864 | 0.026 |
| NP_001071296.1 | IGF1 | −0.148 | 0.529 | −0.814 | 0.135 | 0.666 | 0.028 |
| NP_001069769.1 | APOD | 0.467 | 0.078 | 0.785 | 0.023 | −0.318 | 0.029 |
| NP_776392.1 | TTR | 0.788 | 0.000 | 0.093 | 0.019 | 0.695 | 0.030 |
| NP_001035678.1 | SYCP3 | 0.332 | 0.152 | −0.280 | 0.425 | 0.613 | 0.034 |
| XP_005216584.2 | SERPING1 | 1.627 | 0.056 | 0.393 | 0.037 | 1.234 | 0.045 |
| XP_005217472.2 | CFHR5 | 1.445 | 0.818 | 0.811 | 0.338 | 0.633 | 0.050 |
| NP_001029638.1 | APCS | 1.150 | 0.000 | 0.756 | 0.392 | 0.394 | 0.056 |
| XP_005217494.2 | FGA | −0.363 | 0.050 | −1.018 | 0.022 | 0.655 | 0.074 |
| XP_010815084.2 | C6 | 0.627 | 0.003 | 0.837 | 0.025 | −0.210 | 0.117 |
| NP_001180035.1 | Gripap1 | −0.354 | 0.473 | 1.698 | 0.040 | −2.052 | 0.170 |
| NP_001070299.1 | COL3A1 | 1.742 | 0.040 | −0.752 | 0.671 | 2.494 | 0.291 |
| NP_001093176.1 | ECM1 | 0.083 | 0.933 | −0.785 | 0.047 | 0.867 | 0.299 |
| NP_001029392.1 | CCL4 | −0.859 | 0.291 | −0.606 | 0.047 | −0.254 | 0.379 |
| NP_777124.1 | CHIA | −0.418 | 0.023 | −0.855 | 0.017 | 0.437 | 0.496 |
| NP_001073741.2 | OSMR | −0.395 | 0.198 | −0.737 | 0.000 | 0.342 | 0.533 |
| NP_848667.1 | CA2 | −1.350 | 0.346 | −1.767 | 0.039 | 0.417 | 0.563 |
| NP_776532.1 | MBL | −0.217 | 0.581 | −0.692 | 0.016 | 0.475 | 0.605 |
| NP_001029784.1 | HPX | −0.345 | 0.125 | −0.744 | 0.041 | 0.399 | 0.641 |
| NP_776359.1 | LUM | −0.324 | 0.079 | −0.585 | 0.007 | 0.261 | 0.649 |
| XP_024847454.1 | FBLN1 | −0.626 | 0.008 | −0.963 | 0.016 | 0.337 | 0.668 |
| XP_003587429.1 | Wfdc18 | −0.513 | 0.488 | −0.637 | 0.049 | 0.124 | 0.679 |
| XP_005205978.1 | MGAM | 0.896 | 0.000 | 0.552 | 0.011 | 0.344 | 0.692 |
| NP_001028787.1 | CTSS | −0.246 | 0.302 | −0.779 | 0.000 | 0.532 | 0.702 |
| XP_024837895.1 | SERPINA3−7 | −0.690 | 0.254 | −0.817 | 0.004 | 0.126 | 0.770 |
| NP_001002237.1 | CL43 | 0.575 | 0.048 | −0.666 | 0.016 | 1.241 | 0.778 |
| XP_010808587.1 | Itsn2 | −0.489 | 0.051 | −0.665 | 0.019 | 0.175 | 0.808 |
| XP_024847033.1 | KMT2C | −0.830 | 0.008 | −0.777 | 0.062 | −0.053 | 0.817 |
| XP_024833616.1 | IGLL5 | −0.394 | 0.001 | −1.325 | 0.000 | 0.931 | 0.930 |
| NP_001192115.1 | IGHA2 | −0.648 | 0.219 | −1.576 | 0.022 | 0.928 | 0.967 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Xu, J.; Han, Z. Plasma Proteomic Profiling Reveals the Regulatory Factors of Milk Protein Synthesis in Holstein Cows. Biology 2022, 11, 1239. https://doi.org/10.3390/biology11081239
Wang X, Xu J, Han Z. Plasma Proteomic Profiling Reveals the Regulatory Factors of Milk Protein Synthesis in Holstein Cows. Biology. 2022; 11(8):1239. https://doi.org/10.3390/biology11081239
Chicago/Turabian StyleWang, Xinling, Jie Xu, and Zhaoyu Han. 2022. "Plasma Proteomic Profiling Reveals the Regulatory Factors of Milk Protein Synthesis in Holstein Cows" Biology 11, no. 8: 1239. https://doi.org/10.3390/biology11081239
APA StyleWang, X., Xu, J., & Han, Z. (2022). Plasma Proteomic Profiling Reveals the Regulatory Factors of Milk Protein Synthesis in Holstein Cows. Biology, 11(8), 1239. https://doi.org/10.3390/biology11081239





