COVID-19-Related Scientific Literature Exploration: Short Survey and Comparative Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Datasets
2.1. Unstructured Datasets
2.2. Structured Datasets
2.3. Hybrid
3. Exploratory Search Applications
3.1. Textual Exploratory Search
- Tasks: The tasks are related to textual data, and hence we suppose that we have a text database (or collection or corpus) as a string of N symbols drawn from an alphabet (i.e., all possible combinations of letters) . A vocabulary V is the set of unique words used in . is partitioned into n documents . A document d can be presented as in , including words from V. Queries are also strings (or sets of strings) composed of symbols drawn from . Symbols in may be letters, bytes or even words, and the documents may be articles, chromosomes or any other texts in which we need to search. In general, these tokens are extracted using tokenizers and further processed using lemmatization, stemming and other techniques that help normalize tokens. In the explored systems, we can identify the following tasks.
- −
- Document Retrieval (Indexing, Ranking): for this task, two sub-tasks can be identified [3].
- Document Listing: given query and a text that is partitioned into n documents, , the aim of this task is to return a list of the documents in which one or multiple tokens of Q appear at least once.
- Document Ranking: given a query , an integer , and a text that is partitioned into n documents , and returns the top-k documents ordered by a similarity measure .
- −
- Passage Retrieval (Indexing, Ranking): Given a query Q, and a set of documents D where each document is partitioned into passages, the aim of this task is to find relevant passages for the query [5]. Passage retrieval can also be used for sentence highlighting.
- −
- Question Answering: Given a Query made of m tokens and a passage made of k tokens, the aim of this task is to find an answer span in P [4].
- −
- Summarization: We will opt for the definition presented in [6]. Given a set of documents that we will call source documents, summarization aims to generate a text s (called summary) that is coherent and contains a significant amount of relevant information from the source text. Ref. [6] considered a good summary to have a compression rate (where is the word count in x, x can be a sentence or document or any grouping of words) of less than a third of the length of the original document.
- −
- Topic Modeling: The aim of topic modeling is to infer a set of K topics capturing a lower-dimensional representation suitable for summarization and prediction tasks [46]. According to [47], Given a text corpus with a vocabulary of size V and the predefined number of topics K, the major tasks of topic modeling can be defined as:
- Learning the word representation of topics : a topic in a given collection is defined as a multinomial distribution over the vocabulary V, i.e., .
- Learning the sparse topic representation of documents : the topic representation of a document d, , is defined as a multinomial distribution over K topics, i.e., .
In general, the task of topic modeling aims to find K salient topics from and to find the topic representation of each document . - −
- FAQ Matching: let F denote the set of question–answer pairs; given F and a user query Q, this task aims to rank the question–answer pairs in F. The top k QA pairs with high scores are returned to the user [48].
- −
- Recommendation: Given the set of all users and the set of all possible items that can be recommended . Let u be a utility function that measures the usefulness of item s to user c, i.e., , where is a totally ordered set (e.g., non-negative integers or real numbers within a certain range). The goal of this task is to choose the item(s) that maximize(s) the utility for each user [49].
- Feedback Loop: this characteristic is related to the use of user feedback data in any of the mentioned tasks.
- Representation Level for Text: In general, text can be represented in two distinct spaces: (a) bag-of-words space, (b) vector space. These representations can be shown on one or multiple levels of granularity of textual documents; that is, Document Level, Paragraph Level, Sentence Level and Word Level.
- Representation Levels for Graphs: Graphs can also be represented in a frequentist space or low-dimensional vectorial space. These representations can be shown on one or multiple levels of granularity of graphs; that is, Full Graph Level, Sub-graph Level, Node Level and Edge Level. Examples of graph representation in COVID-19 literature search engines are as follow:
- Novelty: a research paper is said to have novelty if the authors explored uncharted territories to solve old or new problems. Specifically, we characterize papers to have novelty if they contain new contributions to the design of models, learning objectives or data processing. We ignored the data aspect of this characterization because all the papers can be considered to be novel considering only data.
- Data Enrichment: Data enrichment refers, in general, to the process of adding more data to the already existing training data. Data enrichment methods can take two main forms, (a) data augmentation and (b) data supplementation. The former is characteristic of the set of methods that use the already existing data to generate more data, while the latter encapsulates methods that use external resources in order to supplement the available data. The latter is easy to accomplish as long as there are external resources. There are various data augmentations methods. For example, in CO-Search [39], in order to train a Siamese network, the authors generated negative (paragraph, reference) pairs based on positive pairs extracted from documents.
- Search Type:
- −
- Keyword: Keyword search refers to searching using queries composed of one specific word.
- −
- Regular Expression: In this type of search, the query takes the form of regular expressions that annotates textual patterns that we would like to retrieve. For example, ref. [50] used this search strategy to look for drugs with certain properties in a drug re-purposing database.
- −
- Open Questions: This type of search refers to using natural language queries with simple or complex structures.
- −
- Keyphrase Search: This type of search refers to using queries composed of one or multiple keywords, and the order is taken into consideration.
- KG Traversal: This refers to the use of knowledge graphs to search for entities or relationships that are relevant to achieving one or multiple tasks.
- Representation Combination (Rep.Comb.): This characteristic exists in one of two cases: (a) the combination of multiple levels of representation to achieve a task, or (b) the combination of KG and textual representation to achieve a task.
- Fast Prototyping and Deployment: Given the urgent nature of most of the applications, the researcher opted mainly for off-the-shelf technologies that are easy to work with. In addition, except for one application, all the other applications used existing models and algorithms, which can also be attributed to the urgency of the task.
- Textual Representation Methods: There are two categories of methods: (a) Bag-of-Words (BOW) models and (b) Vector Space Models (VSMs). The major difference is that VSMs capture more of the contextual elements of text than the BOW methods, but on the other hand, the VSMs are computationally more expensive during training and inference. Some works struck a balance by applying both categories of methods, e.g., [37,44,51,52], which is performed generally by using a multi-stage ranking scheme that applies the first ranking using BOW models, which is then followed by a re-ranking using a VSM of the output of the previous ranking. Some works compensate for the latency of neural language models [12] by pre-indexing documents offline.
- Granularity/Levels of Representations: We also noticed that the works used different levels of granularity, which depends on the intended tasks and the available computational resources. For example, to achieve the task of document retrieval, some works opted for simple document level representations [53], while other works either used more granular representations [12,32,37,40,50,54,55,56] or a mix of more granular representations with document level representations [16,24,38,39,44,51,52,57].
- Using KGs: Knowledge graphs were used in multiple works for different purposes. For example, ref. [38] used a KG (CKG [36]) embedding in tandem with textual representations for document recommendation, while [37] (CovEx KG [37]), [32] (Vapur KG [32]) and [44] (Citation KG [44]) traversed their respective KGs looking for similar entities to retrieve relevant papers. The authors of [56] (Blender-KG [34]) used a KG to extend queries and make the search more efficient.
- Query Transformation/Extension: Query transformation is also used in many applications to make the queries more expressive, which can help get more relevant results. For example, ref. [53] used an extensive database of medical terms to augment the queries made by novices to search an academic biomedical corpus.
- Multimedia (e.g., image, video, etc.) Grounding: Multimedia grounding is also used to couple textual data with relevant multimedia content. For example, ref. [54] used a self-supervised method to couple biomedical text with corresponding coordinates in a human atlas. This mapping was used to conduct two kinds of queries: (a) atlas-based document retrieval using textual queries (which contain mentions of body parts) and (b) atlas-based document retrieval using 3D atlas coordinates. In addition, ref. [34] associated figures that depict molecular structures in research papers with their chemical entities that exist in a KG by using the captions of the figures. This was done to augment the KG.
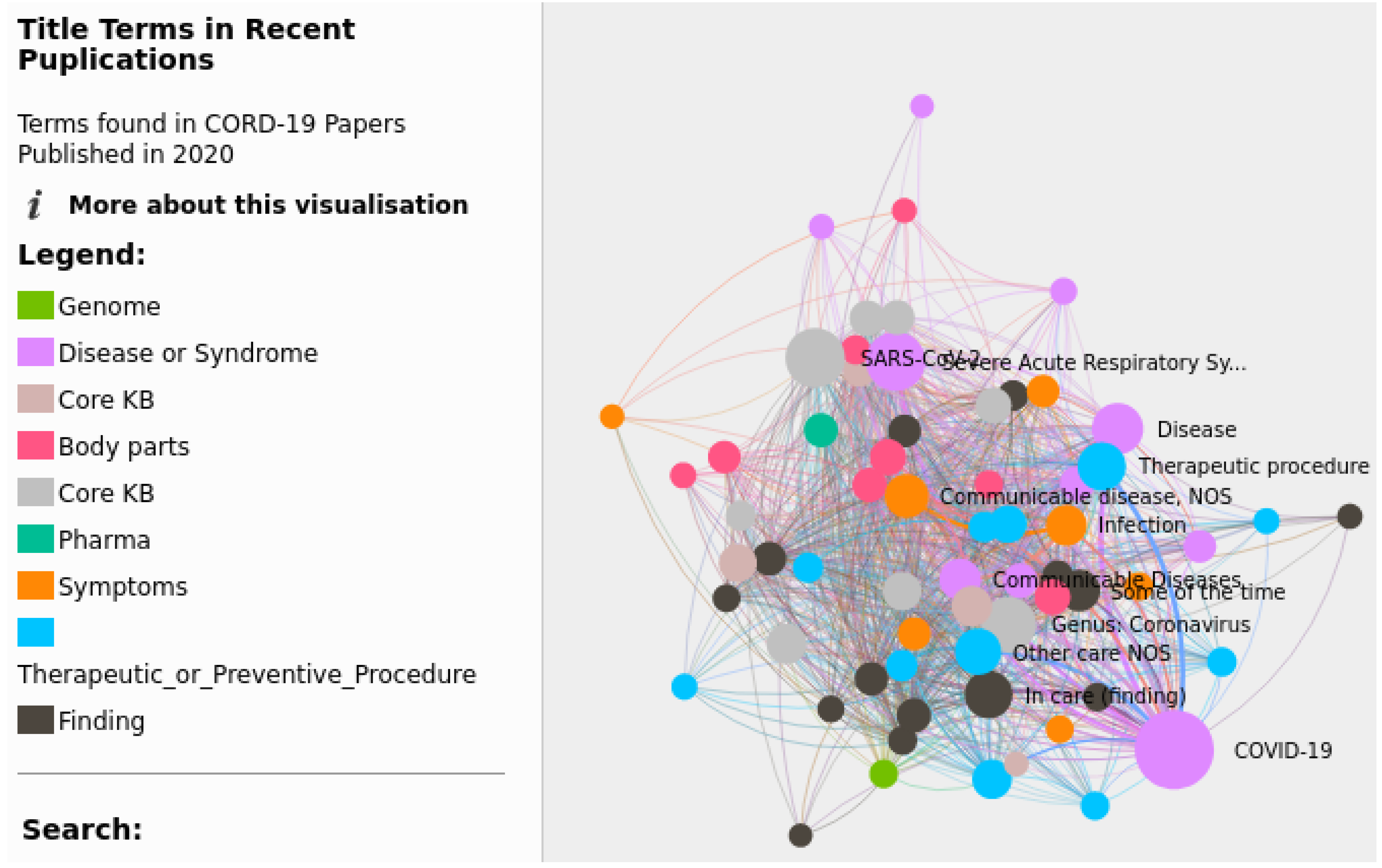
3.2. Visual Exploratory Search
4. Evaluation Methods
4.1. Human Evaluation
4.2. Automatic Evaluation
4.2.1. Intrinsic Evaluation
4.2.2. Extrinsic Evaluation
5. Discussion and Future Research Directions
- Fact Checking: Due to the rapid expansion of the COVID-19 literature and the existence of many contradictory claims concerning, for example, the incubation period of the virus and the optimal social distancing protocol stresses the importance of fact checking applications for COVID-19 claims. The authors of [65] created a claim verification application for the COVID-19 literature, which uses a passage and a claim as input and outputs if the claim is true or not given the passage. This type of application needs huge amounts of annotated data, which is particularly cumbersome in the case of COVID-19 since it needs skilled specialists to annotate it. Developing semi-supervised or unsupervised techniques would be useful.
- Data Bias: Some applications (e.g., [54]) can also benefit from reducing data bias, especially gender bias.
- Smart Querying: Some applications [56] use query functionalities that tend to be limited to simple word matching. This can be problematic in cases where the intent of the user is not evident in the query. This can be remedied by using embedding-based query matching, which uses contextual information for matching the queries to the results.
6. Conclusions
7. Limitations of This Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Q.; Allot, A.; Lu, Z. LitCOVID: An open database of COVID-19 literature. Nucleic Acids Res. 2021, 49, D1534–D1540. [Google Scholar] [CrossRef]
- Jo, T. Text mining. In Studies in Big Data; Springer International Publishing: Cham, Switzerland, 2019. [Google Scholar]
- Culpepper, J.S.; Navarro, G.; Puglisi, S.J.; Turpin, A. Top-k ranked document search in general text databases. In Proceedings of the European Symposium on Algorithms, Liverpool, UK, 6–8 September 2010; Springer: Berlin/Heidelberg, Germany, 2010; pp. 194–205. [Google Scholar]
- Liu, X.; Shen, Y.; Duh, K.; Gao, J. Stochastic answer networks for machine reading comprehension. arXiv 2017, arXiv:1712.03556. [Google Scholar]
- Ganesh, S.; Varma, V. Passage retrieval using answer type profiles in question answering. In Proceedings of the 23rd Pacific Asia Conference on Language, Information and Computation, Hong Kong, China, 3–5 December 2009; Volume 2, pp. 559–568. [Google Scholar]
- Torres-Moreno, J.M. Automatic text summarization; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Wang, L.L.; Lo, K.; Chandrasekhar, Y.; Reas, R.; Yang, J.; Eide, D.; Funk, K.; Kinney, R.; Liu, Z.; Merrill, W.; et al. CORD-19: The COVID-19 Open Research Dataset. arXiv 2020, arXiv:2004.10706v2. [Google Scholar]
- Voorhees, E.; Alam, T.; Bedrick, S.; Demner-Fushman, D.; Hersh, W.R.; Lo, K.; Roberts, K.; Soboroff, I.; Wang, L.L. TREC-COVID: Constructing a Pandemic Information Retrieval Test Collection. arXiv 2020, arXiv:2005.04474. [Google Scholar] [CrossRef]
- Wang, L.L.; Lo, K. Text mining approaches for dealing with the rapidly expanding literature on COVID-19. Brief. Bioinform. 2020, 22, 781–799. [Google Scholar] [CrossRef]
- Tsatsaronis, G.; Schroeder, M.; Paliouras, G.; Almirantis, Y.; Androutsopoulos, I.; Gaussier, E.; Gallinari, P.; Artieres, T.; Alvers, M.R.; Zschunke, M.; et al. BioASQ: A Challenge on Large-Scale Biomedical Semantic Indexing and Question Answering. In Proceedings of the AAAI Fall Symposium: Information Retrieval and Knowledge Discovery in Biomedical Text, Arlington, VA, USA, 2–4 November 2012. [Google Scholar]
- Tang, R.; Nogueira, R.; Zhang, E.; Gupta, N.; Cam, P.; Cho, K.; Lin, J. Rapidly Bootstrapping a Question Answering Dataset for COVID-19. arXiv 2020, arXiv:2004.11339. [Google Scholar]
- Lee, J.; Yi, S.S.; Jeong, M.; Sung, M.; Yoon, W.; Choi, Y.; Ko, M.; Kang, J. Answering questions on COVID-19 in real-time. arXiv 2020, arXiv:2006.15830. [Google Scholar]
- Möller, T.; Reina, A.; Jayakumar, R.; Pietsch, M. COVID-QA: A Question Answering Dataset for COVID-19. In Proceedings of the 1st Workshop on NLP for COVID-19 at ACL 2020, Seattle, WA, USA, 9–10 July 2020. [Google Scholar]
- Poliak, A.; Fleming, M.; Costello, C.; Murray, K.W.; Yarmohammadi, M.; Pandya, S.; Irani, D.; Agarwal, M.; Sharma, U.; Sun, S.; et al. Collecting verified COVID-19 question answer pairs. In Proceedings of the ACL 2020 Workshop NLP-COVID Submission, Seattle, WA, USA, 9–10 July 2020. [Google Scholar]
- Nguyen, T.; Rosenberg, M.; Song, X.; Gao, J.; Tiwary, S.; Majumder, R.; Deng, L. Ms Marco: A Human-Generated Machine Reading Comprehension Dataset. 2016. Available online: https://openreview.net/forum?id=rJ-Qj8-_ZH (accessed on 4 April 2022).
- MacAvaney, S.; Cohan, A.; Goharian, N. SLEDGE-Z: A Zero-Shot Baseline for COVID-19 Literature Search. arXiv 2020, arXiv:2010.05987. [Google Scholar]
- Kwiatkowski, T.; Palomaki, J.; Redfield, O.; Collins, M.; Parikh, A.; Alberti, C.; Epstein, D.; Polosukhin, I.; Devlin, J.; Lee, K.; et al. Natural questions: A benchmark for question answering research. Trans. Assoc. Comput. Linguist. 2019, 7, 453–466. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Zhang, J.; Lopyrev, K.; Liang, P. Squad: 100,000+ questions for machine comprehension of text. arXiv 2016, arXiv:1606.05250. [Google Scholar]
- Arora, A.; Shrivastava, A.; Mohit, M.; Lecanda, L.S.M.; Aly, A. Cross-lingual Transfer Learning for Intent Detection of COVID-19 Utterances. In Proceedings of the EMNLP 2020 Workshop NLP-COVID Submission, Virtual, 19 November 2020. [Google Scholar]
- Choi, E.; He, H.; Iyyer, M.; Yatskar, M.; Yih, W.t.; Choi, Y.; Liang, P.; Zettlemoyer, L. Quac: Question answering in context. arXiv 2018, arXiv:1808.07036. [Google Scholar]
- Dang, H.T. Overview of DUC 2005. In Proceedings of the Document Understanding Conference, Vancouver, BC, Canada, 9–10 October 2005; Volume 2005, pp. 1–12. [Google Scholar]
- Hoa, T. Overview of DUC 2006. In Proceedings of the Document Understanding Conference, Brooklyn, NY, USA, 8–9 June 2006. [Google Scholar]
- Nema, P.; Khapra, M.; Laha, A.; Ravindran, B. Diversity driven attention model for query-based abstractive summarization. arXiv 2017, arXiv:1704.08300. [Google Scholar]
- Dan, S.; Xu, Y.; Yu, T.; Siddique, F.B.; Barezi, E.; Fung, P. CAiRE-COVID: A Question Answering and Query-focused Multi-Document Summarization System for COVID-19 Scholarly Information Management. arXiv 2020, arXiv:2005.03975. [Google Scholar]
- Kim, J.D.; Ohta, T.; Tateisi, Y.; Tsujii, J. GENIA corpus—A semantically annotated corpus for bio-textmining. Bioinformatics 2003, 19, i180–i182. [Google Scholar] [CrossRef]
- Kim, J.D.; Ohta, T.; Tsuruoka, Y.; Tateisi, Y.; Collier, N. Introduction to the bio-entity recognition task at JNLPBA. In Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and Its Applications, Geneva, Switzerland, 28–29 August 2004; pp. 70–75. [Google Scholar]
- Krallinger, M.; Rabal, O.; Leitner, F.; Vazquez, M.; Salgado, D.; Lu, Z.; Leaman, R.; Lu, Y.; Ji, D.; Lowe, D.M.; et al. The CHEMDNER corpus of chemicals and drugs and its annotation principles. J. Cheminform. 2015, 7, 1–17. [Google Scholar] [CrossRef]
- Doğan, R.I.; Leaman, R.; Lu, Z. NCBI disease corpus: A resource for disease name recognition and concept normalization. J. Biomed. Inform. 2014, 47, 1–10. [Google Scholar] [CrossRef]
- Kringelum, J.; Kjaerulff, S.K.; Brunak, S.; Lund, O.; Oprea, T.I.; Taboureau, O. ChemProt-3.0: A global chemical biology diseases mapping. Database 2016, 2016, bav123. [Google Scholar] [CrossRef]
- Li, J.; Sun, Y.; Johnson, R.J.; Sciaky, D.; Wei, C.H.; Leaman, R.; Davis, A.P.; Mattingly, C.J.; Wiegers, T.C.; Lu, Z. BioCreative V CDR task corpus: A resource for chemical disease relation extraction. Database 2016, 2016, baw068. [Google Scholar] [CrossRef]
- Basu, S.; Chakraborty, S.; Hassan, A.; Siddique, S.; Anand, A. ERLKG: Entity Representation Learning and Knowledge Graph based association analysis of COVID-19 through mining of unstructured biomedical corpora. In Proceedings of the First Workshop on Scholarly Document Processing, Online, 19 November 2020; pp. 127–137. [Google Scholar]
- Köksal, A.; Dönmez, H.; Özçelik, R.; Ozkirimli, E.; Özgür, A. Vapur: A Search Engine to Find Related Protein–Compound Pairs in COVID-19 Literature. arXiv 2020, arXiv:2009.02526. [Google Scholar]
- Amini, A.; Hope, T.; Wadden, D.; van Zuylen, M.; Horvitz, E.; Schwartz, R.; Hajishirzi, H. Extracting a knowledge base of mechanisms from COVID-19 papers. arXiv 2020, arXiv:2010.03824. [Google Scholar]
- Wang, Q.; Li, M.; Wang, X.; Parulian, N.; Han, G.; Ma, J.; Tu, J.; Lin, Y.; Zhang, H.; Liu, W.; et al. COVID-19 literature knowledge graph construction and drug repurposing report generation. arXiv 2020, arXiv:2007.00576. [Google Scholar]
- Cernile, G.; Heritage, T.; Sebire, N.J.; Gordon, B.; Schwering, T.; Kazemlou, S.; Borecki, Y. Network graph representation of COVID-19 scientific publications to aid knowledge discovery. BMJ Health Care Inform. 2020, 28, e100254. [Google Scholar] [CrossRef] [PubMed]
- Wise, C.; Ioannidis, V.N.; Calvo, M.R.; Song, X.; Price, G.; Kulkarni, N.; Brand, R.; Bhatia, P.; Karypis, G. COVID-19 knowledge graph: Accelerating information retrieval and discovery for scientific literature. arXiv 2020, arXiv:2007.12731. [Google Scholar]
- Rahdari, B.; Brusilovsky, P.; Thaker, K.; Chau, H.K. CovEx: An Exploratory Search System for COVID-19 Scientific Literature; University of Pittsburgh: Pittsburgh, PA, USA, 2020. [Google Scholar]
- Bhatia, P.; Arumae, K.; Pourdamghani, N.; Deshpande, S.; Snively, B.; Mona, M.; Wise, C.; Price, G.; Ramaswamy, S.; Kass-Hout, T. AWS CORD19-search: A scientific literature search engine for COVID-19. arXiv 2020, arXiv:2007.09186. [Google Scholar]
- Esteva, A.; Kale, A.; Paulus, R.; Hashimoto, K.; Yin, W.; Radev, D.; Socher, R. Co-search: COVID-19 information retrieval with semantic search, question answering, and abstractive summarization. arXiv 2020, arXiv:2006.09595. [Google Scholar]
- Otegi, A.; Campos, J.A.; Azkune, G.; Soroa, A.; Agirre, E. Automatic Evaluation vs. User Preference in Neural Textual Question Answering over COVID-19 Scientific Literature. In Proceedings of the 1st Workshop on NLP for COVID-19 (Part 2) at EMNLP 2020, Seattle, WA, USA, 9–10 July 2020. [Google Scholar]
- Gangi Reddy, R.; Iyer, B.; Arafat Sultan, M.; Zhang, R.; Sil, A.; Castelli, V.; Florian, R.; Roukos, S. End-to-End QA on COVID-19: Domain Adaptation with Synthetic Training. arXiv 2020, arXiv:2012.01414. [Google Scholar]
- Lee, S.; Sedoc, J. Using the Poly-encoder for a COVID-19 Question Answering System. In Proceedings of the 1st Workshop on NLP for COVID-19 (Part 2) at EMNLP 2020, Seattle, WA, USA, 9–10 July 2020. [Google Scholar]
- Tu, J.; Verhagen, M.; Cochran, B.; Pustejovsky, J. Exploration and discovery of the COVID-19 literature through semantic visualization. arXiv 2020, arXiv:2007.01800. [Google Scholar]
- Das, D.; Katyal, Y.; Verma, J.; Dubey, S.; Singh, A.; Agarwal, K.; Bhaduri, S.; Ranjan, R. Information retrieval and extraction on COVID-19 clinical articles using graph community detection and bio-bert embeddings. In Proceedings of the 1st Workshop on NLP for COVID-19 at ACL 2020, Seattle, WA, USA, 9–10 July 2020. [Google Scholar]
- Ciampaglia, G.L.; Shiralkar, P.; Rocha, L.M.; Bollen, J.; Menczer, F.; Flammini, A. Computational fact checking from knowledge networks. PLoS ONE 2015, 10, e0128193. [Google Scholar]
- Virtanen, S.; Girolami, M. Precision-Recall Balanced Topic Modelling. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; pp. 6750–6759. [Google Scholar]
- Qiang, J.; Qian, Z.; Li, Y.; Yuan, Y.; Wu, X. Short text topic modeling techniques, applications, and performance: A survey. IEEE Trans. Knowl. Data Eng. 2020, 34, 1427–1445. [Google Scholar] [CrossRef]
- Damani, S.; Narahari, K.N.; Chatterjee, A.; Gupta, M.; Agrawal, P. Optimized Transformer Models for FAQ Answering. In Proceedings of the Pacific-Asia Conference on Knowledge Discovery and Data Mining, Singapore, 11–14 May 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 235–248. [Google Scholar]
- Adomavicius, G.; Tuzhilin, A. Toward the next generation of recommender systems: A survey of the state-of-the-art and possible extensions. IEEE Trans. Knowl. Data Eng. 2005, 17, 734–749. [Google Scholar] [CrossRef]
- Tworowski, D.; Gorohovski, A.; Mukherjee, S.; Carmi, G.; Levy, E.; Detroja, R.; Mukherjee, S.B.; Frenkel-Morgenstern, M. COVID-19 Drug Repository: Text-mining the literature in search of putative COVID-19 therapeutics. Nucleic Acids Res. 2020, 49, D1113–D1121. [Google Scholar] [CrossRef]
- Zhang, E.; Gupta, N.; Tang, R.; Han, X.; Pradeep, R.; Lu, K.; Zhang, Y.; Nogueira, R.; Cho, K.; Fang, H.; et al. COVIDex: Neural ranking models and keyword search infrastructure for the COVID-19 open research dataset. arXiv 2020, arXiv:2007.07846. [Google Scholar]
- Farokhnejad, M.; Pranesh, R.R.; Vargas-Solar, G.; Mehr, D.A. S_COVID: An Engine to Explore COVID-19 Scientific Literature. In Proceedings of the 24th International Conference on Extending Database Technology (EDBT), Nicosia, Cyprus, 23–26 March 2021. [Google Scholar]
- He, D.; Wang, Z.; Thaker, K.; Zou, N. Translation and expansion: Enabling laypeople access to the COVID-19 academic collection. Data Inf. Manag. 2017, 4, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Grujicic, D.; Radevski, G.; Tuytelaars, T.; Blaschko, M.B. Self-supervised context-aware COVID-19 document exploration through atlas grounding. In Proceedings of the ACL 2020 Workshop NLP-COVID Submission, Seattle, WA, USA, 9–10 July 2020. [Google Scholar]
- Tabib, H.T.; Shlain, M.; Sadde, S.; Lahav, D.; Eyal, M.; Cohen, Y.; Goldberg, Y. Interactive extractive search over biomedical corpora. In Proceedings of the 19th SIGBioMed Workshop on Biomedical Language Processing, Online, 9 July 2020; pp. 28–37. [Google Scholar]
- Wang, X.; Guan, Y.; Liu, W.; Chauhan, A.; Jiang, E.; Li, Q.; Liem, D.; Sigdel, D.; Caufield, J.; Ping, P.; et al. Evidenceminer: Textual evidence discovery for life sciences. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics: System Demonstrations, Online, 5–10 July 2020; pp. 56–62. [Google Scholar]
- Zhang, E.; Gupta, N.; Nogueira, R.; Cho, K.; Lin, J. Rapidly deploying a neural search engine for the COVID-19 open research dataset: Preliminary thoughts and lessons learned. arXiv 2020, arXiv:2004.05125. [Google Scholar]
- Le Bras, P.; Gharavi, A.; Robb, D.A.; Vidal, A.F.; Padilla, S.; Chantler, M.J. Visualising COVID-19 Research. arXiv 2020, arXiv:2005.06380. [Google Scholar]
- Hope, T.; Portenoy, J.; Vasan, K.; Borchardt, J.; Horvitz, E.; Weld, D.S.; Hearst, M.A.; West, J. SciSight: Combining faceted navigation and research group detection for COVID-19 exploratory scientific search. arXiv 2020, arXiv:2005.12668. [Google Scholar]
- Wolinski, F. Visualization of Diseases at Risk in the COVID-19 Literature. arXiv 2020, arXiv:2005.00848. [Google Scholar]
- Lin, C.Y. Rouge: A package for automatic evaluation of summaries. In Proceedings of the Text Summarization Branches Out, Barcelona, Spain, 25–26 July 2004; pp. 74–81. [Google Scholar]
- Chen, D.; Fisch, A.; Weston, J.; Bordes, A. Reading wikipedia to answer open-domain questions. arXiv 2017, arXiv:1704.00051. [Google Scholar]
- Karpukhin, V.; Oğuz, B.; Min, S.; Wu, L.; Edunov, S.; Chen, D.; Yih, W.T. Dense Passage Retrieval for Open-Domain Question Answering. arXiv 2020, arXiv:2004.04906. [Google Scholar]
- Zhu, M. Recall, Precision and Average Precision; Department of Statistics and Actuarial Science, University of Waterloo: Waterloo, ON, USA, 2004; Volume 2, p. 6. [Google Scholar]
- Wadden, D.; Lo, K.; Wang, L.L.; Lin, S.; van Zuylen, M.; Cohan, A.; Hajishirzi, H. Fact or Fiction: Verifying Scientific Claims. arXiv 2020, arXiv:2004.14974. [Google Scholar]
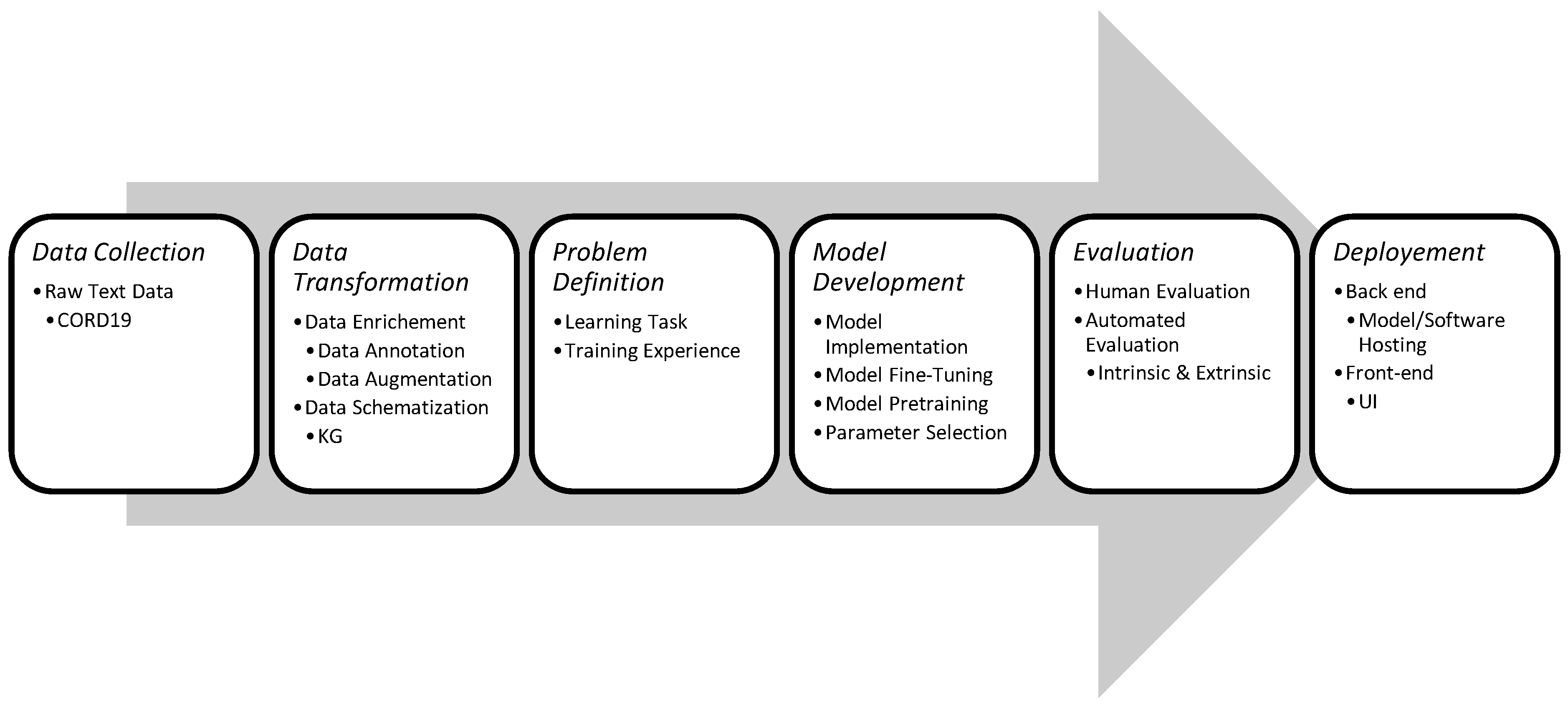
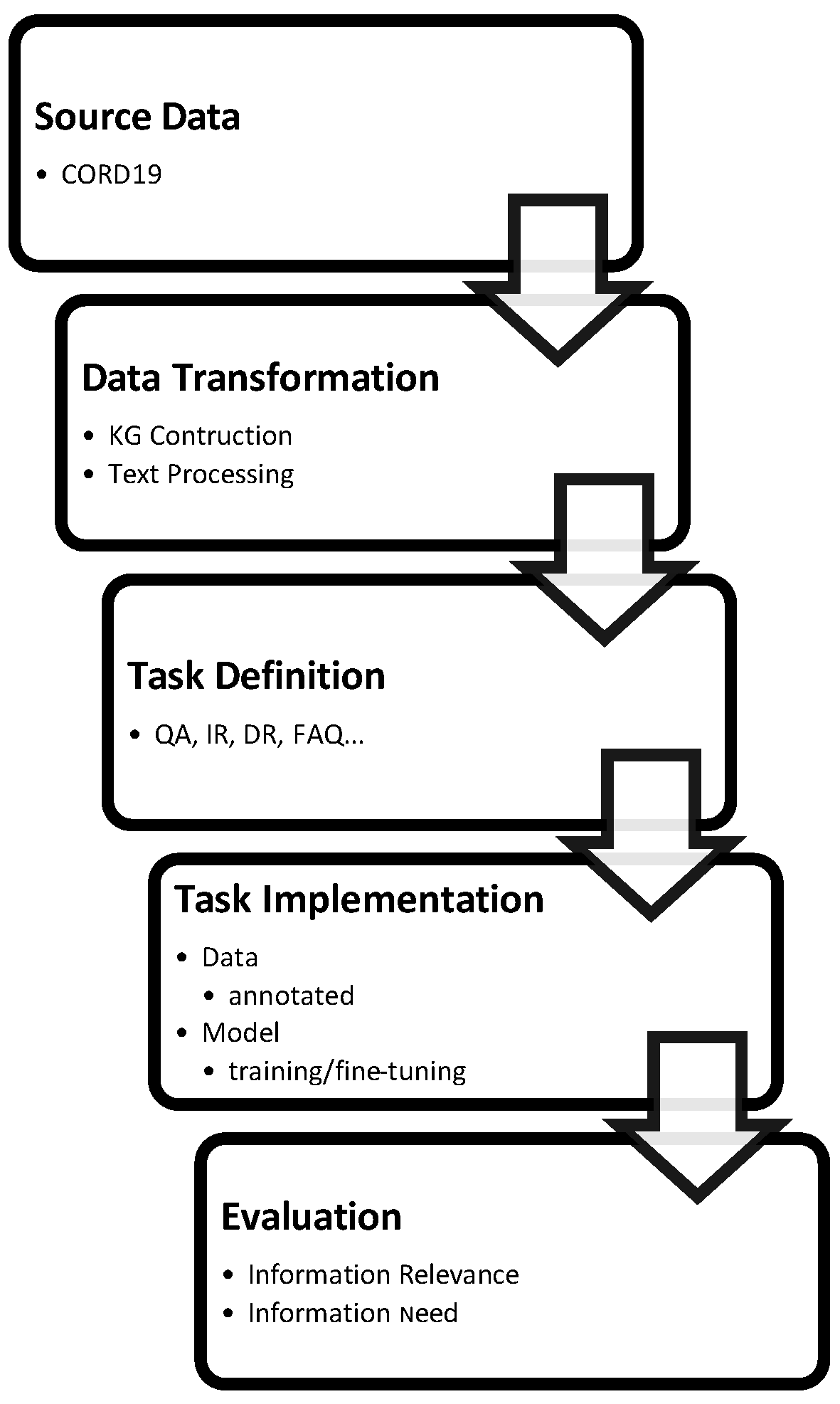
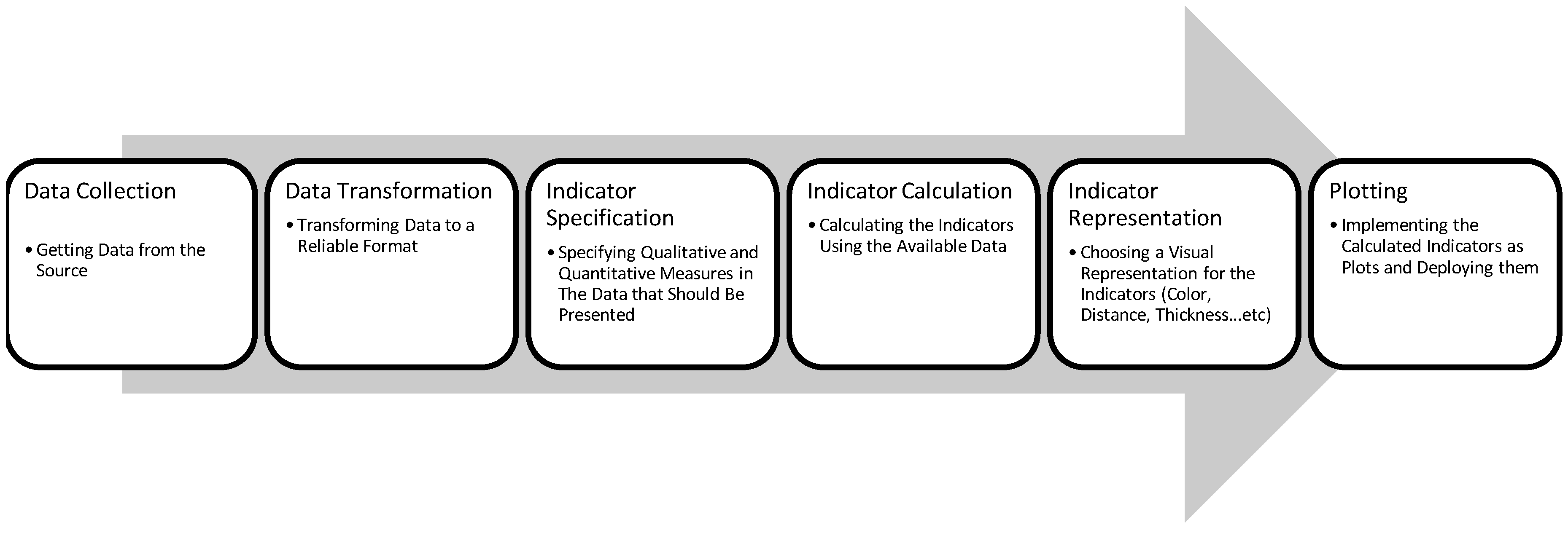
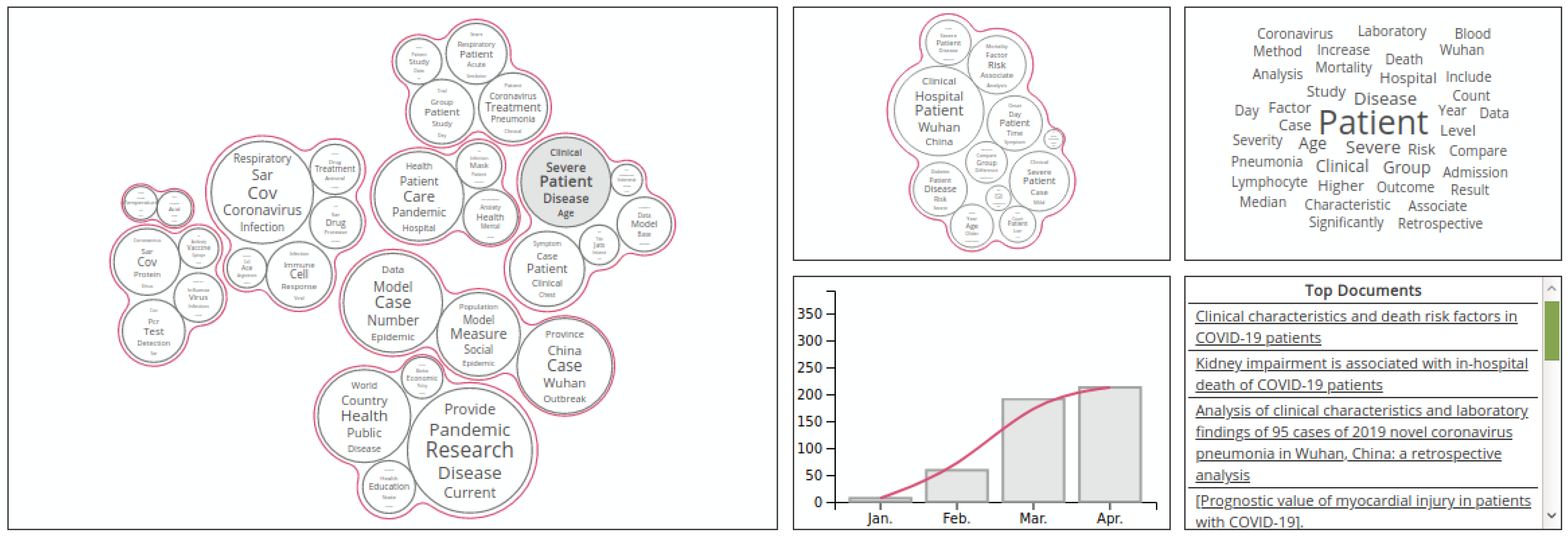
| Dataset | Application Refs | Tasks | Statistics | URL |
|---|---|---|---|---|
| TREC-COVID [8] | [16,38,39,40] | DR | The TREC-COVID dataset has many versions which correspond to TREC-COVID challenges. For example, round three contains a total of 16,677 unique journal articles in CORD-19 with a relevance annotation. | https://www.kaggle.com/c/trec-COVID-information-retrieval/data (accessed on 4 April 2022) |
| COVIDQA * [11] | [24,38,41] | QA | The dataset contains 147 question–article–answer triples with 27 unique questions and 104 unique articles. | https://github.com/castorini/pygaggle/tree/master/data (accessed on 4 April 2022) |
| COVID-19 Questions * [12] | [12,41] | QA | The dataset contains 111 question–answer pairs with 53 interrogative and 58 keyword-style queries. | https://drive.google.com/file/d/1z7jW0fovgTfTScCanZvrvrUax1HAMEFV/view?usp=sharing (accessed on 4 April 2022) |
| COVID-QA * [13] | [40,41] | QA | The dataset consists of 2019 question–article–answer triples. | https://github.com/deepset-ai/COVID-QA (accessed on 4 April 2022) |
| InfoBot Dataset * [14] | [42] | QA, FAQ | 2200 COVID-19-related Frequently asked Question–Answer pairs. | https://covid-19-infobot.org/data/ (accessed on 4 April 2022) |
| MS-MARCO [15] | [16] | QA | 1,000,000 training instances. | https://microsoft.github.io/msmarco/ (accessed on 4 April 2022) |
| Med-MARCO [16] | [16] | QA | 79K of the original MS-MARCO questions (9.7%). | https://github.com/Georgetown-IR-Lab/covid-neural-ir/blob/master/med-msmarco-train.txt (accessed on 4 April 2022) |
| Natural Questions [17] | [12] | QA | The public release consists of 307,373 training examples with single annotations; 7830 examples with 5-way annotations for development data; and a further 7842 examples with 5-way annotated sequestered as test data. | https://ai.google.com/research/NaturalQuestions/ (accessed on 4 April 2022) |
| SQuAD [18] | [12] | QA | The dataset contains 107,785 question–answer pairs on 536 articles. | https://rajpurkar.github.io/SQuAD-explorer/ (accessed on 4 April 2022) |
| BioASQ [10] | [12] | QA, DR | 500 questions with their relevant documents, text span answers and perfect answers. | http://www.bioasq.org/news/golden-datasets-2nd-edition-bioasq-challenge-are-now-available (accessed on 4 April 2022) |
| M-CID [19] | [19] | QA | The dataset is composed of 6871 natural language utterances across 16 COVID-19-specific intents and 4 languages: English, Spanish, French and German. | https://fb.me/covid_mcid_dataset (accessed on 4 April 2022) |
| QuAC [20] | [40] | QA | 14K information-seeking QA dialogs, and 100K questions in total. | http://quac.ai/ (accessed on 4 April 2022) |
| GENIA [25] | [37] | NER | 2000 abstracts taken from the MEDLINE database; contains more than 400,000 words and almost 100,000 annotations. | http://www.geniaproject.org/genia-corpus/term-corpus (accessed on 4 April 2022) |
| DUC 2005, 2006 [21,22] | [24] | SMZ | The dataset is composed of 50 topics. | https://www-nlpir.nist.gov/projects/duc/data.html (accessed on 4 April 2022) |
| Debatepedia [23] | [24] | SMZ | It consists of 10,859 training examples, 1357 testing and 1357 validation samples. The average number of words in summary, documents and query is 11.16, 66.4 and 10, respectively. | https://github.com/PrekshaNema25/DiverstiyBasedAttentionMechanism (accessed on 4 April 2022) |
| JNLPBA [26] | [31] | NER | This dataset contains a subset of the GENIA dataset V3.02. This subset is composed of 2404 abstracts. The articles were chosen to contain the MeSH terms “human”, “blood cells” and “transcription factors”, and their publication year ranges from 1990 to 1999. | http://www.geniaproject.org/shared-tasks/bionlp-jnlpba-shared-task-2004 (accessed on 4 April 2022) |
| CHEMDNER [27] | [31] | NER | 10,000 PubMed abstracts that contain a total of 84,355 chemical entities. | https://biocreative.bioinformatics.udel.edu/resources/biocreative-iv/chemdner-corpus/ (accessed on 4 April 2022) |
| NCBI Disease Corpus [28] | [31] | NER | 793 PubMed abstracts that were annotated. A total of 6892 disease mentions, which are mapped to 790 unique disease concepts that were extracted. | https://github.com/spyysalo/ncbi-disease (accessed on 4 April 2022) |
| CHEMPROT [29] | [31] | NER, RE | 2500 PubMed abstracts, from which 32,000 chemical entities and 31,000 protein entities were extracted. In addition, 10,000 chemical-protein relationships were extracted. | http://www.biocreative.org/accounts/login/?next=/resources/corpora/chemprot-corpus-biocreative-vi/ (accessed on 4 April 2022) |
| BC5CDR [30] | [31] | NER, RE | 1500 PubMed articles with 4409 annotated chemicals, 5818 diseases and 3116 chemical-disease interactions. | https://github.com/shreyashub/BioFLAIR/tree/master/data/ner (accessed on 4 April 2022) |
| COV19_729 * [31] | [31] | NER | The dataset is composed of 729 examples. Each example is a triple comprising an entity, the class that that entity belongs to (i.e., disease, protein, chemical), and a physician’s rating of how related those entities are to COVID-19. | https://github.com/sayantanbasu05/ERKLG (accessed on 4 April 2022) |
| Entities | Properties | Description | ID |
|---|---|---|---|
| Paper | title, publication date, journal, Digital Object Identifier (DOI), link | Representation of research paper entities. | E1 |
| Author | identifier, first names, middle names, last names | Representation of the paper authors. | E2 |
| Affiliation | identifier, name, country, city | Representation of a research structure where an author belongs. | E3 |
| Concept | concept identifier, textual value, concept type (gene, disease, topic, chemical, etc.) | Representation of a domain specific concept. | E4 |
| Source Entity | Dest. Entity | Relation | Description | ID |
|---|---|---|---|---|
| Paper | Paper | cites | This relation connects paper entities with paper references indicating a citation relation. | R1 |
| Author | Author | co-author | This relation connects an author entity with another author entity indicating a co-authorship relation. | R2 |
| Concept | Concept | relate concepts | This relationship links two concepts with any general relationship that might link them. | R3 |
| Paper | Author | authored by | This relation connects paper entities with author entities and indicates an authorship relation. | R4 |
| Paper | Concept | associated concept | This relation connects paper entities with concept entities. | R5 |
| Author | Affiliation | affiliated with | This relation connects author entities with institution entities. | R6 |
| Author | Concept | research area | This relation connects author entities with concept entities indicating a research area of the author. | R7 |
| KG | Usage | Ent. | Rel. |
|---|---|---|---|
| CKG [36] | Article recommendations, citation-based navigation, and search result ranking. | E1, E2, E3, E4 | R1, R4, R6, R5 |
| CovEx KG [37] | Document Retrieval. | E1, E2, E4 | R1, R4, R5, R7 |
| ERLKG [31] | Link prediction. | E4 | R3 |
| COVID-KG [34] (Blender-KG [43]) | QA, Semantic Visualization, Drug Re-purposing. | E4 | R3 |
| COFIE KG [33] | KG search over relations and entities using a query. | E4 | R3 |
| Network Visualization KG [35] | Data Visualization. | E4 | R3 |
| Vapur KG [32] | Query extension. | E4 | R3 |
| Citation KG [44] | Document Ranking. | E1 | R1 |
| System | CO-Search [39] | AWS CORD-19 Search (ACS) [38] | COVID-19 Drug Repository [50] | CovEx [37] | COVIDex [51,57] | Vapur [32] | COVIDASK [12] | [40] | CAiRE-COVID [24] | [41] | CORD19-Explorer [54] | SLEDGE-Z [16] | S_COVID [52] | [44] | SLIC [53] | SPIKE [55] | EVIDENCEMINER [56] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Uses Raw Text (Uses KG) | ✓(❙) | ✓(✓) | ✓(❙) | ✓(✓) | ✓(❙) | ✓(✓) | ✓(❙) | ✓(❙) | ✓(❙) | ✓(❙) | ✓(❙) | ✓(❙) | ✓(❙) | ✓(✓) | ✓(❙) | ✓(❙) | ✓(✓) | |
| Publicly Available | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ❙ | ✓ | ❙ | ✓ | ❙ | ✓ | ❙ | ❙ | ✓ | ✓ | |
| Feedback Loop | ❙ | ❙ | ❙ | ✓ | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | |
| Multistage Ranking | ❙ | ❙ | ❙ | ✓ | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ✓ | ✓ | ❙ | ❙ | ❙ | |
| KG Traversal | ❙ | ✓ | ❙ | ❙ | ❙ | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ✓ | ❙ | ❙ | ✓ | |
| Text Representations Levels (KG Representation Level) | Document (KG) | ✓(❙) | ✓(❙) | ❙(❙) | ❙(❙) | ✓(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ✓(❙) | ❙(❙) | ❙(❙) | ✓(❙) | ✓(❙) | ✓(❙) | ✓(❙) | ❙(❙) | ❙(❙) |
| Paragraph (Sub-graph) | ✓(❙) | ✓(✓) | ❙(❙) | ❙(❙) | ✓(❙) | ❙(❙) | ❙(❙) | ✓(❙) | ✓(❙) | ✓(❙) | ❙(❙) | ✓(❙) | ❙(❙) | ❙(✓) | ❙(❙) | ✓(❙) | ❙(❙) | |
| Sentence (Edge) | ✓(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ✓(❙) | ❙(❙) | ✓(❙) | ✓(❙) | ✓(❙) | ❙(❙) | ✓(✓) | ✓(❙) | ❙(❙) | ✓(❙) | ✓(❙) | |
| Word (Node) | ❙(❙) | ❙(❙) | ❙(❙) | ✓(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ✓(❙) | ❙(❙) | ❙(❙) | ✓(❙) | ✓(❙) | |
| n-gram (Node Property) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ✓(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙)) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | |
| Keyphrase (Edge Property) | ❙(❙) | ❙(❙) | ✓(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ❙(❙) | ✓(❙) | ✓(❙) | |
| Rep.Comb. | Inter-Level | ✓ | ❙ | ❙ | ❙ | ✓ | ❙ | ❙ | ❙ | ✓ | ❙ | ❙ | ✓ | ✓ | ✓ | N❙ | ✓ | ❙ |
| Text & KG | ❙ | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ✓ | ❙ | ❙ | ❙ | |
| Tasks | Document Retrieval (Indexing, Ranking) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ❙ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Passage Retrieval (Indexing, Ranking) | ✓ | ✓ | ❙ | ❙ | ✓ | ❙ | ✓ | ✓ | ✓ | ✓ | ❙ | ❙ | ✓ | ✓ | ❙ | ✓ | ✓ | |
| Question Answering | ✓ | ✓ | ❙ | ❙ | ✓ | ❙ | ✓ | ✓ | ✓ | ✓ | ❙ | ❙ | ❙ | ✓ | ❙ | ❙ | ❙ | |
| Summarization | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ✓ | ❙ | ❙ | ❙ | ❙ | ✓ | ❙ | ❙ | ❙ | |
| Topic Modeling | ❙ | ✓ | ❙ | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ✓ | ✓ | ❙ | ❙ | ❙ | |
| Recommendation | ❙ | ✓ | ❙ | ✓ | ❙ | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | |
| FAQ Matching | ❙ | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | |
| Search Type | Keyword | ❙ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ❙ | ✓ | ❙ | ✓ | ❙ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Open Questions | ✓ | ✓ | ❙ | ❙ | ❙ | ❙ | ✓ | ✓ | ✓ | ✓ | ❙ | ✓ | ✓ | ✓ | ✓ | ❙ | ✓ | |
| Keyphrases | ❙ | ❙ | ❙ | ✓ | ❙ | ❙ | ✓ | ❙ | ✓ | ❙ | ✓ | ❙ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Regular Expression | ❙ | ❙ | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ✓ | ❙ | |
| Novelty | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | |
| Data Enrichment | From External Resources | ❙ | ✓ | ✓ | ❙ | ✓ | ✓ | ✓ | ❙ | ❙ | ✓ | ✓ | ✓ | ❙ | ❙ | ✓ | ❙ | ✓ |
| From Internal Resources | ✓ | ✓ | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | ✓ | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | ❙ | |
| System | Vidar-19 [60] | TopicMaps [58] | Network Visualisations [35] | SciSight [59] | Semviz [43] | EvidenceMiner [56] | |
|---|---|---|---|---|---|---|---|
| Available Charts | Pie Chart | ❙ | ❙ | ❙ | ❙ | ❙ | ✓ |
| Histogram | ✓ | ✓ | ❙ | ✓ | ❙ | ❙ | |
| Data Tables | ❙ | ✓ | ❙ | ❙ | ✓ | ❙ | |
| Heat Map | ❙ | ❙ | ❙ | ❙ | ✓ | ❙ | |
| Tile Chart | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | |
| Word Cloud | ❙ | ✓ | ❙ | ✓ | ✓ | ❙ | |
| Stacked Barplot | ✓ | ❙ | ❙ | ❙ | ❙ | ❙ | |
| Bar Plot | ❙ | ❙ | ❙ | ❙ | ✓ | ✓ | |
| Bubble Maps | ❙ | ✓ | ❙ | ❙ | ❙ | ❙ | |
| Network/Graph | ❙ | ❙ | ✓ | ✓ | ❙ | ❙ | |
| Chord Diagram | ❙ | ❙ | ❙ | ✓ | ❙ | ❙ | |
| Indicators | Frequency | ✓ | ✓ | ✓ | ✓ | ❙ | ❙ |
| Count | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Other Indicators | ❙ | ✓ | ❙ | ❙ | ❙ | ❙ | |
| Related Tasks | IE | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Topic Modeling | ❙ | ✓ | ❙ | ✓ | ❙ | ❙ | |
| NER | ❙ | ❙ | ✓ | ✓ | ✓ | ❙ | |
| Network Analysis | ❙ | ❙ | ✓ | ✓ | ❙ | ❙ | |
| Data Source | Raw Text | ✓ | ✓ | ❙ | ✓ | ✓ | ✓ |
| KG | ❙ | ❙ | ✓ | ✓ | ✓ | ✓ | |
| Reactivity | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Public Availability | ✓ https://fran6wol.eu.pythonanywhere.com/ | ✓ http://strategicfutures.org/TopicMaps/COVID-19/dimensions.html | ✓ https://nlp.inspirata.com/NetworkVisualisations/TitleNetwork/, https://nlp.inspirata.com/NetworkVisualisations/TreatmentNetwork/, https://nlp.inspirata.com/NetworkVisualisations/LungNetwork/, https://nlp.inspirata.com/NetworkVisualisations/CardioNetwork/ | ✓ https://scisight.apps.allenai.org/ | ✓ https://www.semviz.org/ | ✓ https://evidenceminer.firebaseapp.com/analytics?kw=CORONAVIRUS&corpus=COVID-19 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adil, B.; Lhazmir, S.; Ghogho, M.; Benbrahim, H. COVID-19-Related Scientific Literature Exploration: Short Survey and Comparative Study. Biology 2022, 11, 1221. https://doi.org/10.3390/biology11081221
Adil B, Lhazmir S, Ghogho M, Benbrahim H. COVID-19-Related Scientific Literature Exploration: Short Survey and Comparative Study. Biology. 2022; 11(8):1221. https://doi.org/10.3390/biology11081221
Chicago/Turabian StyleAdil, Bahaj, Safae Lhazmir, Mounir Ghogho, and Houda Benbrahim. 2022. "COVID-19-Related Scientific Literature Exploration: Short Survey and Comparative Study" Biology 11, no. 8: 1221. https://doi.org/10.3390/biology11081221
APA StyleAdil, B., Lhazmir, S., Ghogho, M., & Benbrahim, H. (2022). COVID-19-Related Scientific Literature Exploration: Short Survey and Comparative Study. Biology, 11(8), 1221. https://doi.org/10.3390/biology11081221






