Mechanistic Insight into the Enzymatic Inhibition of β-Amyrin against Mycobacterial Rv1636: In Silico and In Vitro Approaches
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence-Based Prediction of Rv1636 Protein: In Silico Studies
2.1.1. Retrieve Rv1636 Protein Sequence
2.1.2. Physiochemical Parameters
2.1.3. Prediction of the Protein Localization
2.1.4. Secondary (2D) Structure Prediction
2.1.5. Phylogenetic Studies of Rv1636
2.1.6. Interaction Analysis (PPI and PCI)
2.2. Structure-Based Prediction of Rv1636 Protein: In Silico Studies
2.2.1. Molecular Docking
2.2.2. Receptor–Ligand Interaction Profile
2.2.3. Pharmacokinetics Profile
2.2.4. Pharmacological Profile
2.2.5. Molecular Dynamics Simulation
2.3. Determination of Protein Expression and Purification of Rv1636 Protein: In Vitro Studies
2.3.1. Cloning of Rv1636 in pET28a Vector
2.3.2. Protein Expression and Purification
2.4. Biochemical and Biophysical Characterization of Rv1636 Protein: In Vitro Studies
2.4.1. Biochemical Characterization of Rv1636 to Determine Its ATPase Activity
2.4.2. Circular Dichroism (CD) to Determine the Secondary Structure of Rv1636
2.4.3. Isothermal Calorimetry (ITC) to Determine the Thermodynamic Properties of the Interaction of Rv1636 and β Amyrin
3. Results
3.1. Physiochemical Parameters
3.2. Subcellular Localization
3.3. Secondary Structure Prediction
3.4. Phylogenetic Analysis
3.5. PPI Interaction Studies of Rv1636
3.6. Molecular Docking
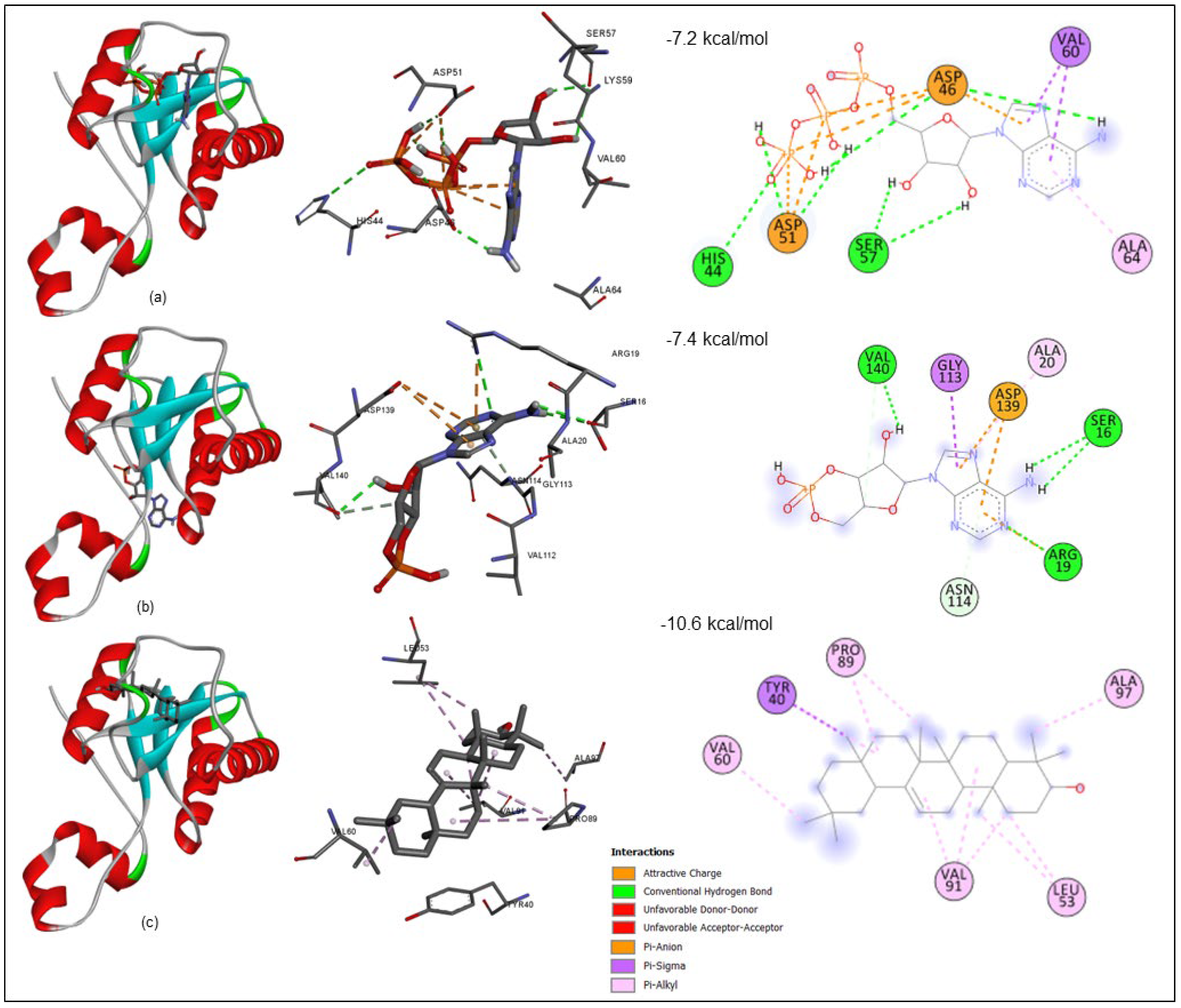
3.6.1. Receptor–Ligand Interaction Profile
3.6.2. Pharmacokinetics Profile
3.6.3. Pharmacological Profile
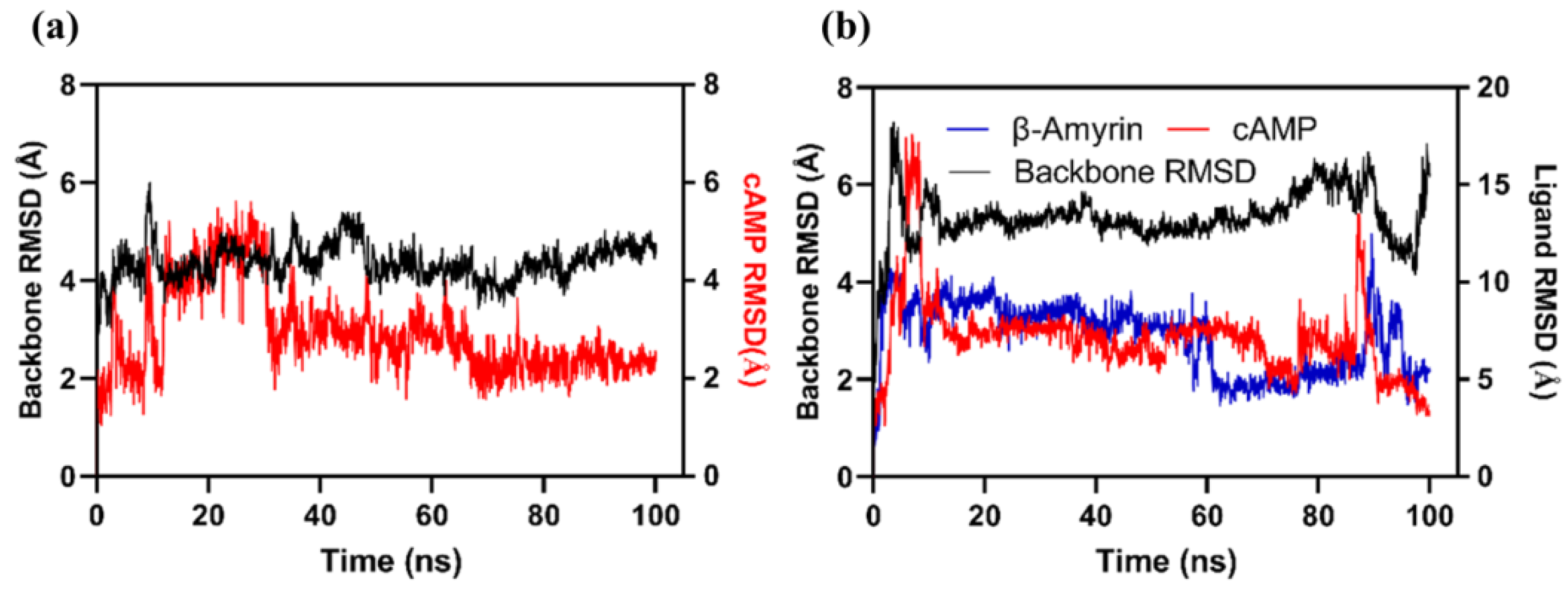
3.6.4. Molecular Dynamics Simulation
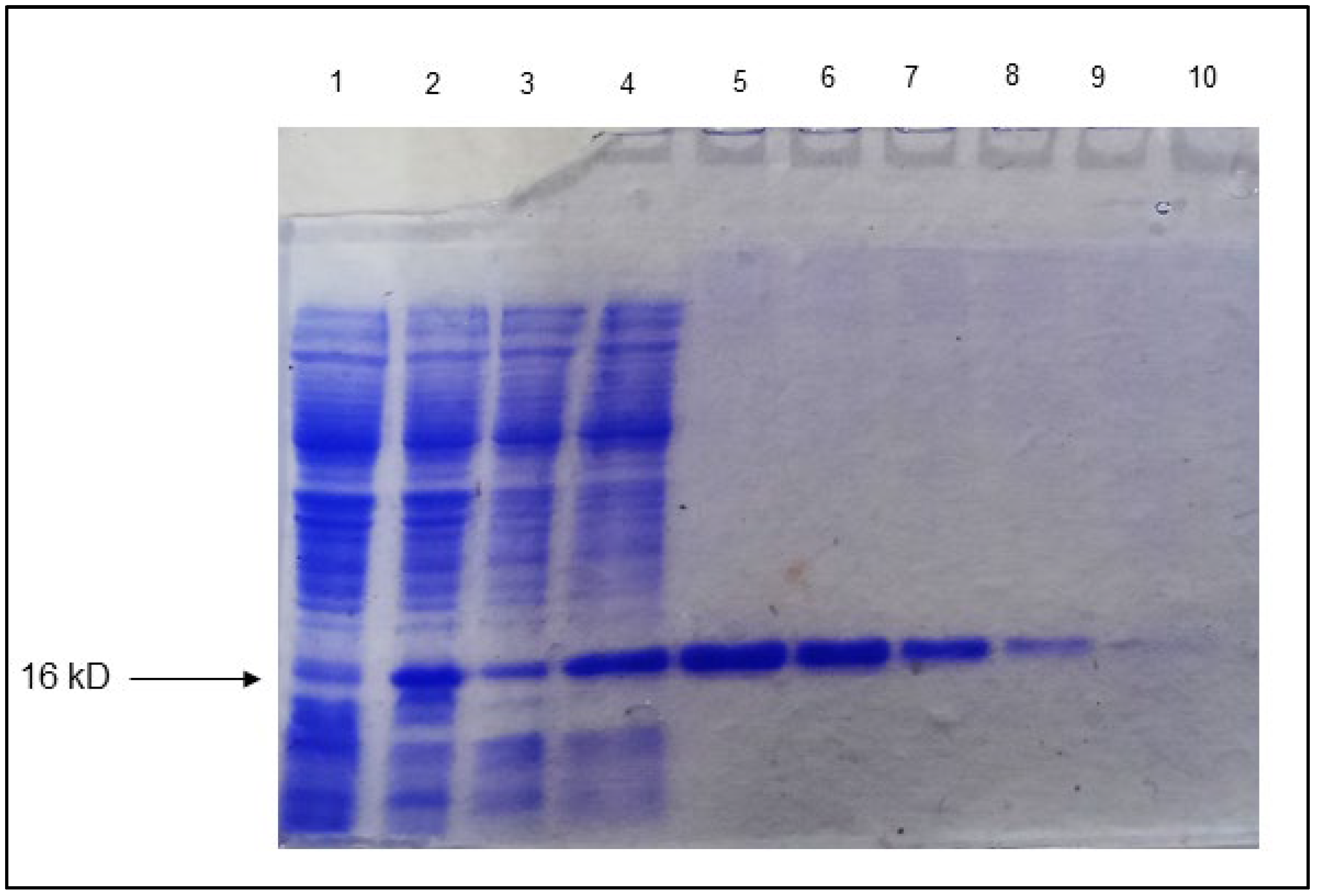
3.7. Purification of Rv1636
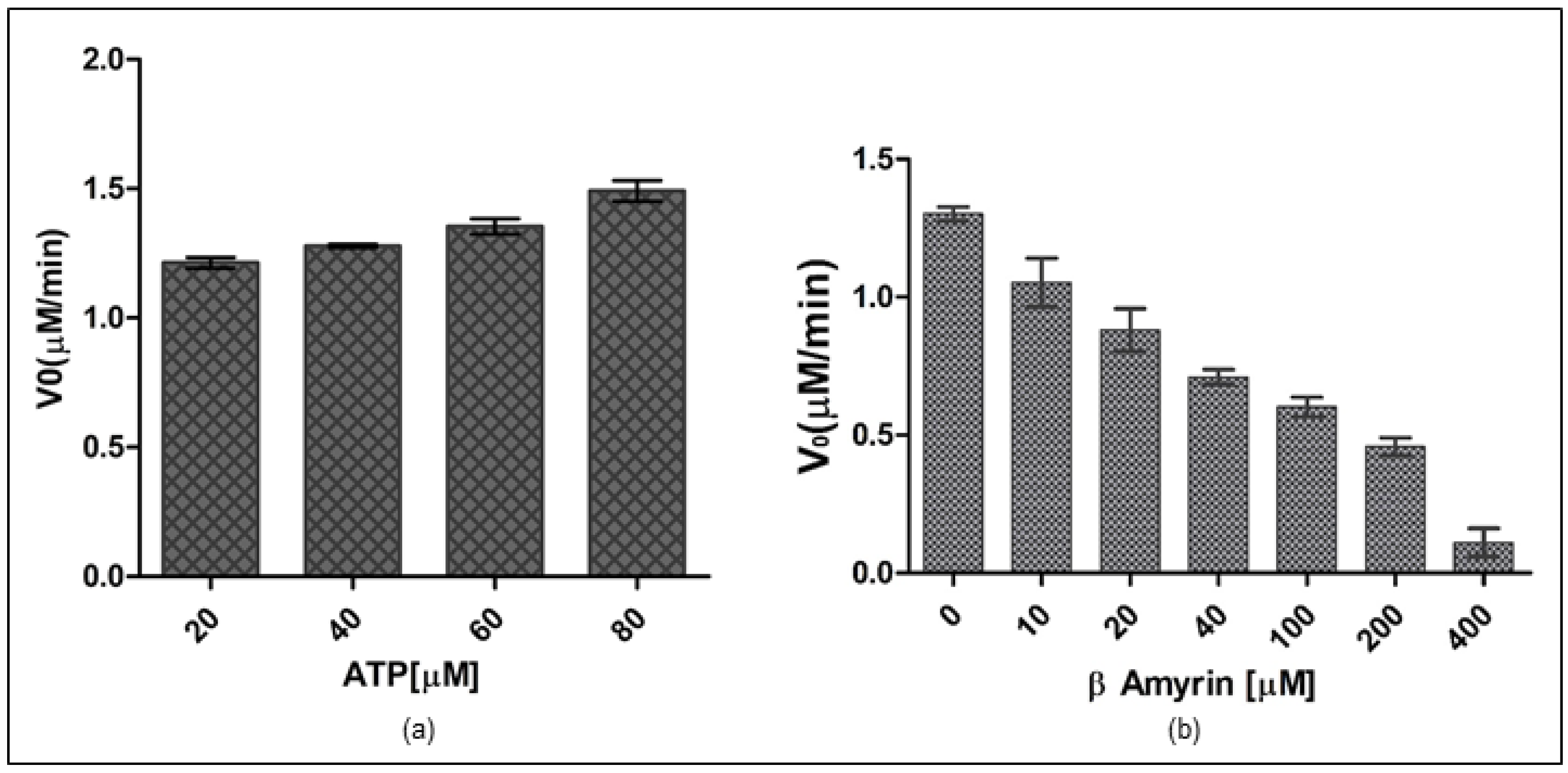
3.8. Rv1636 Contains Prominent ATPase Activity
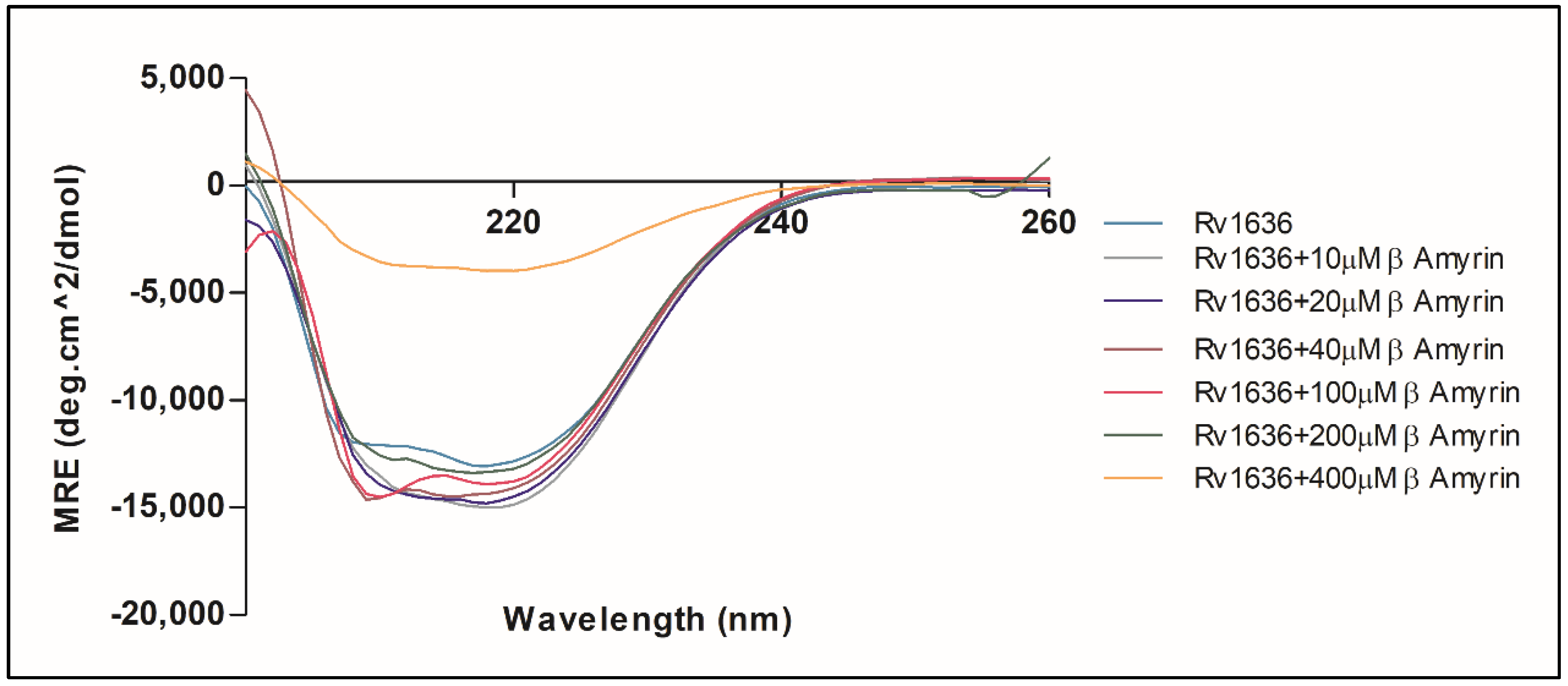
3.9. β-Amyrin Affects the Secondary Structure Pattern of Rv1636
3.10. The Thermodynamic Interaction of β-Amyrin and Rv1636
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, H.; Javid, B. Antibodies and tuberculosis: Finally coming of age? Nat. Rev. Immunol. 2018, 18, 591–596. [Google Scholar]
- Tanwar, P.; Shivangi, M.L. Cholesterol Metabolism: As a Promising Target Candidate for Tuberculosis Treatment by Nanomedicine. J. Nanomater. Mol. Nanotechnol. 2019, 8, 2. [Google Scholar]
- Vinod, V.; Vijayrajratnam, S.; Vasudevan, A.K.; Biswas, R. The cell surface adhesins of Mycobacterium tuberculosis. Microbiol. Res. 2019, 232, 126392. [Google Scholar] [CrossRef]
- McNerney, R.; Maeurer, M.; Abubakar, I.; Marais, B.; Mchugh, T.D.; Ford, N.; Weyer, K.; Lawn, S.; Grobusch, M.P.; Memish, Z.; et al. Tuberculosis Diagnostics and Biomarkers: Needs, Challenges, Recent Advances, and Opportunities. J. Infect. Dis. 2012, 205, S147–S158. [Google Scholar] [CrossRef]
- Parasa, V.R.; Rahman, M.J.; Hoang, A.T.N.; Svensson, M.; Brighenti, S.; Lerm, M. Modeling Mycobacterium tuberculosis early granuloma formation in experimental human lung tissue. Dis. Models Mech. 2013, 7, 281–288. [Google Scholar] [CrossRef]
- Stallings, C.L.; Glickman, M.S. Is Mycobacterium tuberculosis stressed out? A critical assessment of the genetic evidence. Microbes Infect. 2010, 12, 1091–1101. [Google Scholar] [CrossRef]
- Simmons, J.; Stein, C.; Seshadri, C.; Campo, M.; Alter, G.; Fortune, S.; Schurr, E.; Wallis, R.S.; Churchyard, G.; Mayanja-Kizza, H.; et al. Immunological mechanisms of human resistance to persistent Mycobacterium tuberculosis infection. Nat. Rev. Immunol. 2018, 18, 575–589. [Google Scholar] [CrossRef]
- Li, H.; Dang, G.; Liu, H.; Wang, Z.; Cui, Z.; Song, N.; Chen, L.; Liu, S. Characterization of a novel Mycobacterium tuberculosis serine protease (Rv3194c) activity and pathogenicity. Tuberculosis 2019, 119, 101880. [Google Scholar] [CrossRef]
- Barry, C.E., 3rd; Boshoff, H.I.; Dartois, V.; Dick, T.; Ehrt, S.; Flynn, J.; Schnappinger, D.; Wilkinson, R.; Young, D. The spectrum of latent tuberculosis: Rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 2009, 7, 845–855. [Google Scholar] [CrossRef]
- Cressy, N.L. The Tubercle Bacillus in the Pulmonary Lesion of Man. Yale J. Biol. Med. 1955, 28, 72–73. [Google Scholar]
- Gupta, U.D.; Katoch, V.M. Understanding the phenomenon of persistence in mycobacterial infections. Indian J. Lepr. 1997, 69, 385–393. [Google Scholar]
- O’Toole, R.; Smeulders, M.J.; Blokpoel, M.C.; Kay, E.J.; Lougheed, K.; Williams, H.D. A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J. Bacteriol. 2003, 185, 1543–1554. [Google Scholar]
- Florczyk, M.A.; McCue, L.A.; Stack, R.F.; Hauer, C.R.; McDonough, K.A. Identification and Characterization of Mycobacterial Proteins Differentially Expressed understanding and Shaking Culture Conditions, Including Rv2623 from a Novel Class of Putative ATP-Binding Proteins. Infect. Immun. 2001, 69, 5777–5785. [Google Scholar] [CrossRef]
- Sherman, D.R.; Voskuil, M.; Schnappinger, D.; Liao, R.; Harrell, M.I.; Schoolnik, G.K. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. USA 2001, 98, 7534–7539. [Google Scholar] [CrossRef]
- Voskuil, M.I.; Schnappinger, D.; Visconti, K.C.; Harrell, M.I.; Dolganov, G.M.; Sherman, D.R.; Schoolnik, G.K. Inhibition of Respiration by Nitric Oxide Induces a Mycobacterium tuberculosis Dormancy Program. J. Exp. Med. 2003, 198, 705–713. [Google Scholar] [CrossRef]
- Park, H.-D.; Guinn, K.M.; Harrell, M.I.; Liao, R.; Voskuil, M.I.; Tompa, M.; Schoolnik, G.K.; Sherman, D.R. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 2003, 48, 833–843. [Google Scholar] [CrossRef]
- Kvint, K.; Nachin, L.; Diez, A.; Nyström, T. The bacterial universal stress protein: Function and regulation. Curr. Opin. Microbiol. 2003, 6, 140–145. [Google Scholar] [CrossRef]
- Nyström, T.; Neidhardt, F.C. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Eschericha coli. Mol. Microbiol. 1992, 6, 3187–3198. [Google Scholar]
- Nyström, T.; Neidhardt, F.C. Isolation and properties of a mutant of Escherichia coli with an insertional inactivation of the uspA gene, which encodes a universal stress protein. J. Bacteriol. 1993, 175, 3949–3956. [Google Scholar] [CrossRef][Green Version]
- Nyström, T.; Neidhardt, F.C. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 1994, 11, 537–544. [Google Scholar] [CrossRef]
- Nachin, L.; Nannmark, U.; Nyström, T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 2005, 187, 6265–6272. [Google Scholar]
- Liu, W.-T.; Karavolos, M.H.; Bulmer, D.M.; Allaoui, A.; Hormaeche, R.D.C.E.; Lee, J.J.; Khan, C.A. Role of the universal stress protein UspA of Salmonella in growth arrest, stress and virulence. Microb. Pathog. 2007, 42, 2–10. [Google Scholar] [CrossRef]
- Boes, N.; Schreiber, K.; Härtig, E.; Jaensch, L.; Schobert, M. The Pseudomonas aeruginosa Universal Stress Protein PA4352 Is Essential for Surviving Anaerobic Energy Stress. J. Bacteriol. 2006, 188, 6529–6538. [Google Scholar] [CrossRef]
- Schreiber, K.; Boes, N.; Eschbach, M.; Jaensch, L.; Wehland, J.; Bjarnsholt, T.; Givskov, M.; Hentzer, M.; Schobert, M. Anaerobic Survival of Pseudomonas aeruginosa by Pyruvate Fermentation Requires an Usp-Type Stress Protein. J. Bacteriol. 2006, 188, 659–668. [Google Scholar] [CrossRef]
- Zarembinski, T.I.; Hung, L.W.; Mueller-Dieckmann, H.J.; Kim, K.K.; Yokota, H.; Kim, R.; Kim, S.H. Structure-based assignment of the biochemical function of a hypothetical protein: A test case of structural ge-nomics. Proc. Natl. Acad. Sci. USA 1998, 95, 15189–15193. [Google Scholar]
- O’Toole, R.; Williams, H. Universal stress proteins and Mycobacterium tuberculosis. Res. Microbiol. 2003, 154, 387–392. [Google Scholar] [CrossRef]
- Hingley-Wilson, S.; Lougheed, K.; Ferguson, K.; Leiva, S.; Williams, H. Individual Mycobacterium tuberculosis universal stress protein homologues are dispensable in vitro. Tuberculosis 2010, 90, 236–244. [Google Scholar] [CrossRef]
- Schnappinger, D.; Ehrt, S.; Voskuil, M.I.; Liu, Y.; Mangan, J.A.; Monahan, I.M.; Dolganov, G.; Efron, B.; Butcher, P.D.; Nathan, C.; et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: Insights into the phagosomal environment. J. Exp. Med. 2003, 198, 693–704. [Google Scholar]
- Banerjee, A.; Adolph, R.S.; Gopalakrishnapai, J.; Kleinboelting, S.; Emmerich, C.; Steegborn, C.; Visweswariah, S.S. A Universal Stress Protein (USP) in Mycobacteria Binds cAMP. J. Biol. Chem. 2015, 290, 12731–12743. [Google Scholar] [CrossRef]
- McDonough, K.A.; Rodriguez, A. The myriad roles of cyclic AMP in microbial pathogens: From signal to sword. Nat. Rev. Genet. 2011, 10, 27–38. [Google Scholar] [CrossRef]
- Shenoy, A.R.; Visweswariah, S.S. New messages from old messengers: cAMP and mycobacteria. Trends Microbiol. 2006, 14, 543–550. [Google Scholar] [CrossRef]
- Boshoff, H.I.; Reed, M.B.; Barry, C.E., III; Mizrahi, V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuber-culosis. Cell 2003, 113, 183–193. [Google Scholar]
- Borland, G.; Smith, B.O.; Yarwood, S.J. EPAC proteins transduce diverse cellular actions of cAMP. J. Cereb. Blood Flow Metab. 2009, 158, 70–86. [Google Scholar] [CrossRef]
- Harman, J.G. Allosteric regulation of the cAMP receptor protein. Biochim. Et Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 2001, 1547, 1–17. [Google Scholar]
- Kapopoulou, A.; Lew, J.M.; Cole, S.T. The MycoBrowser portal: A comprehensive and manually annotated resource for myco-bacterial genomes. Tuberculosis 2011, 91, 8–13. [Google Scholar]
- Beg, M.A.; Hejazi, I.I.; Thakur, S.C.; Athar, F. Domain-wise differentiation of Mycobacterium tuberculosis H37Rv hypothetical proteins: A roadmap to discover bacterial survival potentials. Biotechnol. Appl. Biochem. 2022, 69, 296–312. [Google Scholar]
- Garg, V.K.; Avashthi, H.; Tiwari, A.; Jain, P.A.; Ramkete, P.W.; Kayastha, A.M.; Singh, V.K. MFPPI–multi FASTA ProtParam interface. Bioinformation 2016, 12, 74. [Google Scholar]
- Rashid, M.; Saha, S.; Raghava, G.P. Support Vector Machine-based method for predicting subcellular localization of mycobacterial proteins using evolutionary information and motifs. BMC Bioinform. 2007, 8, 337. [Google Scholar] [CrossRef]
- Yu, C.-S.; Cheng, C.-W.; Su, W.-C.; Chang, S.-C.; Huang, S.-W.; Hwang, J.-K.; Lu, C.-H. CELLO2GO: A Web Server for Protein Subcellular Localization Prediction with Functional Gene Ontology Annotation. PLoS ONE 2014, 9, e99368. [Google Scholar] [CrossRef]
- Goldberg, T.; Hecht, M.; Hamp, T.; Karl, T.; Yachdav, G.; Ahmed, N.; Altermann, U.; Angerer, P.; Ansorge, S.; Balasz, K.; et al. LocTree3 prediction of localization. Nucleic Acids Res. 2014, 42, W350–W355. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Bryson, K.; Jones, D.T.J.B. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Rost, B.; Yachdav, G.; Liu, J. The PredictProtein server. Nucleic Acids Res. 2004, 32, W321–W326. [Google Scholar] [CrossRef]
- Lavigne, R.; Seto, D.; Mahadevan, P.; Ackermann, H.-W.; Kropinski, A.M. Unifying classical and molecular taxonomic classification: Analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 2008, 159, 406–414. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Beg, A.; Shivangi; Thakur, S.C.; Meena, L.S. Structural Prediction and Mutational Analysis of Rv3906c Gene of Mycobacterium tuberculosis H37Rv to Determine Its Essentiality in Survival. Adv. Bioinform. 2018, 2018, 6152014. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar]
- Szklarczyk, D.; Santos, A.; Von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar]
- Mohammad, T.; Mathur, Y.; Hassan, I. InstaDock: A single-click graphical user interface for molecular docking-based virtual high-throughput screening. Brief. Bioinform. 2020, 22, bbaa279. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided-Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Chakraborti, S.; Chakraborty, M.; Bose, A.; Srinivasan, N.; Visweswariah, S.S. Identification of Potential Binders of Mtb Universal Stress Protein (Rv1636) Through an in silico Approach and Insights Into Compound Selection for Experimental Validation. Front. Mol. Biosci. 2021, 8, 599221. [Google Scholar] [CrossRef]
- Sharma, K.; Singh, P.; Beg, A.; Dohare, R.; Athar, F.; Syed, M.A. Revealing new therapeutic opportunities in hypertension through network-driven integrative genetic analysis and drug target prediction approach. Gene 2021, 801, 145856. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef]
- Beg, M.; Athar, F. Pharmacokinetic and molecular docking studies of Achyranthes aspera phytocompounds to exploring potential anti-tuberculosis activity. J. Bacteriol. Mycol. Open Access 2020, 8, 18–27. [Google Scholar]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef]
- Studio, B.D. Discovery Studio Modeling Environment; Dassault Systèmes San Diego: San Diego, CA, USA, 2016. [Google Scholar]
- Hejazi, I.I.; Beg, M.A.; Imam, A.; Athar, F.; Islam, A. Glossary of phytoconstituents: Can these be repurposed against SARS CoV-2? A quick in silico screening of various phytoconstituents from plant Glycyrrhiza glabra with SARS CoV-2 main protease. Food Chem. Toxicol. 2021, 150, 112057. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Filimonov, D.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.; Druzhilovskii, D.S.; Pogodin, P.V.; Poroikov, V.V. Prediction of the Biological Activity Spectra of Organic Compounds Using the Pass Online Web Resource. Chem. Heterocycl. Compd. 2014, 50, 444–457. [Google Scholar] [CrossRef]
- Shivangi; Ekka, M.K.; Meena, L.S. Essential biochemical, biophysical and computational inputs on efficient functioning of Mycobacterium tuberculosis H37Rv FtsY. Int. J. Biol. Macromol. 2021, 171, 59–73. [Google Scholar] [CrossRef]
- Yan, J.X. Prokaryotic expression, purification of Mycobacterium tuberculosis PZA-resistant protein. Gene Ther. Mol. Biol. 2013, 15, 185–192. [Google Scholar]
- Ali, N.; Amir, M.; Hassan, I.; Ahmad, F.; Islam, A. Purification, modeling and structural insights of calmodulin-binding receptor like cytoplasmic kinase 2 from Oroxylum Indicum. Int. J. Biol. Macromol. 2018, 123, 704–712. [Google Scholar] [CrossRef]
- Van Veldhoven, P.P.; Mannaerts, G.P. Inorganic and organic phosphate measurements in the nanomolar range. Anal. Biochem. 1987, 161, 45–48. [Google Scholar] [CrossRef]
- Agarwal, N.; Pareek, M.; Thakur, P.; Pathak, V. Functional characterization of EngAMS, a P-loop GTPase of Mycobacterium smegmatis. PLoS ONE 2012, 7, e34571. [Google Scholar]
- Yang, J.T.; Wu, C.-S.C.; Martinez, H.M. [11] Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986, 130, 208–269. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Duff, M.R., Jr.; Grubbs, J.; Howell, E.E. Isothermal titration calorimetry for measuring macromolecule-ligand affinity. J. Vis. Exp. 2011, e2796. [Google Scholar]
- Anwar, S.; Shamsi, A.; Shahbaaz, M.; Queen, A.; Khan, P.; Hasan, G.M.; Islam, A.; Alajmi, M.F.; Hussain, A.; Ahmad, F.; et al. Rosmarinic Acid Exhibits Anticancer Effects via MARK4 Inhibition. Sci. Rep. 2020, 10, 14336. [Google Scholar] [CrossRef]
- Beg, M.A.; Shivangi; Afzal, O.; Akhtar, M.S.; Altamimi, A.; Hussain, A.; Imam, M.A.; Ahmad, M.N.; Chopra, S.; Athar, F. Potential Efficacy of β-Amyrin Targeting Mycobacterial Universal Stress Protein by In Vitro and In Silico Approach. Molecules 2022, 27, 4581. [Google Scholar] [CrossRef]




| Protein | AA | MW | pI | EC | Instability Index | Aliphatic Index | GRAVY | |
|---|---|---|---|---|---|---|---|---|
| Rv1636 | 146 | ~15.31 | 5.51 | 5960 | 25.53 | Stable | 106.30 | 0.005 |
| S. No. | Webserver | Predicted Localization |
|---|---|---|
| 1 | TBpred AACB-based SVM/ Score | Cytoplasmic protein/3.225639 |
| 2 | TBpred DCB-based SVM/ Score | Cytoplasmic protein/0.34192113 |
| 3 | CELLO2GO/ Score | Cytoplasmic/4.833 |
| 4 | LocTree3/ Score | Cytoplasmic/99% |
| 5 | PSORTbv.3.0/ Score | Cytoplasmic /2.50 |
| Protein Name | Webservers | Methods | Alpha Helix | Extended Strand | Random Coil |
|---|---|---|---|---|---|
| Rv1636 | SOPMA | Self-optimized prediction (SOPM) | 67 (45.89%) | 28 (19.18%) | 43 (29.45%) |
| PSIPRED | Artificial neural network machine learning (NNML) | 65 (44.52%) | 31 (21.23%) | 50 (34.25%) | |
| Jpred4 | JNet algorithm | 54 (36.98%) | 34 (23.29) | 58 (39.72) | |
| Predict Protein | Deep learning embeddings | 69 (47.26%) | 25 (17.12%) | 52 (35.62%) |
| Predicted Functional Partners | Name | Score |
|---|---|---|
| Protein–Proteins Interaction (PPI) | ||
| TB16.3 | Conserved protein tb16.3 | 0.864 |
| uspA | Probable uspA, sugar-transport integral membrane protein | 0.764 |
| TB31.7 | Universal stress protein family protein tb31.7 | 0.754 |
| Rv1360 | Probable oxidoreductase | 0.748 |
| TB9.4 | conserved protein tb18.6 | 0.735 |
| TB18.6 | Conserved protein tb18.6 | 0.730 |
| mpt63 | Immunogenic protein mpt63 | 0.651 |
| kdpD | Probable sensor protein kdpD | 0.625 |
| ssb | Single-strand binding protein (ssb) | 0.578 |
| ufaA1 | Tuberculostearic acid methyltransferase ufaa1 | 0.577 |
| Protein–Compounds Interaction (PCI) | ||
| SeMet | SeMet (196.1 g/mol) | 0.846 |
| carboxy | Formic acid | 0.839 |
| chloride | Acriflavine is a topical antiseptic | 0.825 |
| nitrate | Nitric acid | 0.674 |
| MgATP | MgATP (507.2 g/mol) | 0.661 |
| Fe | Sodium nitroprusside is an inorganic compound | 0.639 |
| AMPPCP | AMPPCP (505.2 g/mol) | 0.611 |
| manganese | A trace element | 0.600 |
| 1,3 bisphospho | 1,3-bisphosphoglycerate | 0.596 |
| hydrogen | a hydron is the general name for a cationic form | 0.596 |
| Name of the Ligand | BE | LE | pKi | Torsional Energy |
|---|---|---|---|---|
| cAMP | −7.6 | 0.271 | 5.57 | 1.2452 |
| ATP | −7.2 | 0.141 | 5.28 | 4.6695 |
| β-Amyrin | −10.6 | 0.312 | 7.77 | 0.3113 |
| Absorption | Distribution | Metabolism | Excretion | Toxicity | |
|---|---|---|---|---|---|
| Intestinal absorption | Water Solubility | BBB/CNS permeation | CYP2D6/ CYP1A2/ CYP2C19 inhibitor | OCT2 substrate | AMES/ROAT/Carcinogenicity/Eye irritation and corrosion |
| 93.733% | Poor | No | No | No | No |
| Pa | Pi | Biological Activity |
|---|---|---|
| 0.977 | 0.001 | Insulin promoter |
| 0.976 | 0.002 | Caspase 3 stimulant |
| 0.944 | 0.001 | Transcription factor stimulant |
| 0.944 | 0.001 | Transcription factor NF kappa B stimulant |
| 0.939 | 0.004 | Mucomembranous protector |
| 0.926 | 0.002 | Hepatoprotectant |
| 0.923 | 0.004 | Apoptosis agonist |
| 0.916 | 0.005 | Antineoplastic |
| 0.913 | 0.002 | Oxidoreductase inhibitor |
| 0.909 | 0.002 | Membrane integrity antagonist |
| 0.903 | 0.002 | Chemopreventive |
| Ka (Association Constant) M−1 | ∆H (Enthalpy Change) cal/mol | ∆S (cal/mol/deg) |
|---|---|---|
| Ka1 = 5.61 × 104 ± 1.10 × 105 | ∆H1 = −6.58 × 105 ± 1.007 × 106 | ∆S1 = −2.18 × 103 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beg, M.A.; Sadaf; Shamsi, A.; Sahoo, S.; Yousuf, M.; Najm, M.Z.; Almutawif, Y.A.; Islam, A.; Aloliqi, A.A.; Athar, F. Mechanistic Insight into the Enzymatic Inhibition of β-Amyrin against Mycobacterial Rv1636: In Silico and In Vitro Approaches. Biology 2022, 11, 1214. https://doi.org/10.3390/biology11081214
Beg MA, Sadaf, Shamsi A, Sahoo S, Yousuf M, Najm MZ, Almutawif YA, Islam A, Aloliqi AA, Athar F. Mechanistic Insight into the Enzymatic Inhibition of β-Amyrin against Mycobacterial Rv1636: In Silico and In Vitro Approaches. Biology. 2022; 11(8):1214. https://doi.org/10.3390/biology11081214
Chicago/Turabian StyleBeg, Md Amjad, Sadaf, Anas Shamsi, Sibasis Sahoo, Mohd Yousuf, Mohammad Zeeshan Najm, Yahya Ahmad Almutawif, Asimul Islam, Abdulaziz A. Aloliqi, and Fareeda Athar. 2022. "Mechanistic Insight into the Enzymatic Inhibition of β-Amyrin against Mycobacterial Rv1636: In Silico and In Vitro Approaches" Biology 11, no. 8: 1214. https://doi.org/10.3390/biology11081214
APA StyleBeg, M. A., Sadaf, Shamsi, A., Sahoo, S., Yousuf, M., Najm, M. Z., Almutawif, Y. A., Islam, A., Aloliqi, A. A., & Athar, F. (2022). Mechanistic Insight into the Enzymatic Inhibition of β-Amyrin against Mycobacterial Rv1636: In Silico and In Vitro Approaches. Biology, 11(8), 1214. https://doi.org/10.3390/biology11081214








