Functional Characterization of Three GnRH Isoforms in Small Yellow Croaker Larimichthys polyactis Maintained in Captivity: Special Emphasis on Reproductive Dysfunction
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Fish and Rearing Condition
2.2. Tissue Collection for Gene Cloning
2.3. Tissue Collection for mRNA Expression Analysis from Different Experimental Conditions
2.3.1. Different Organ Tissues of Adult Small Yellow Croaker of Both Sexes
2.3.2. Different Brain Parts of Adult Small Yellow Croaker in Both Sexes
2.3.3. Brain and Pituitary at Different Gonadal Developmental Stages in Both Sexes
2.3.4. Brain of Small Yellow Croaker during Induced Spawning Event in Both Sexes
2.3.5. Pituitary of Small Yellow Croaker from In Vivo Induction of GnRH1 Peptide
2.3.6. Pituitary of Small Yellow Croaker from In Vitro Incubation with GnRH1 Peptide
2.3.7. Hypothalamus of Small Yellow Croaker from In Vitro Incubation with E2 and MT
2.4. RNA Extraction and cDNA Syntheses
2.5. Cloning and Sequencing of Three Full-Length GnRH Isoforms in Small Yellow Croaker
2.5.1. Cloning of Partial Sequences
2.5.2. Cloning of 5′- and 3′-RACE Sequences
2.6. In-Silico Analysis of Sequences of the Three Cloned GnRH Isoforms
2.7. Analysis of Amino Acid Sequence Alignment and Identity-Similarity Index
2.8. Prediction of 3D Protein Structure of Three GnRH Isoforms and Their GAP Regions
2.9. Phylogenetic Analysis
2.10. Synteny Analysis of Three GnRH Isoforms of Small Yellow Croaker
2.11. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.12. Statistical Analysis
3. Results
3.1. Cloning, Characterization, and In-Silico Analysis of Three GnRH Isoforms
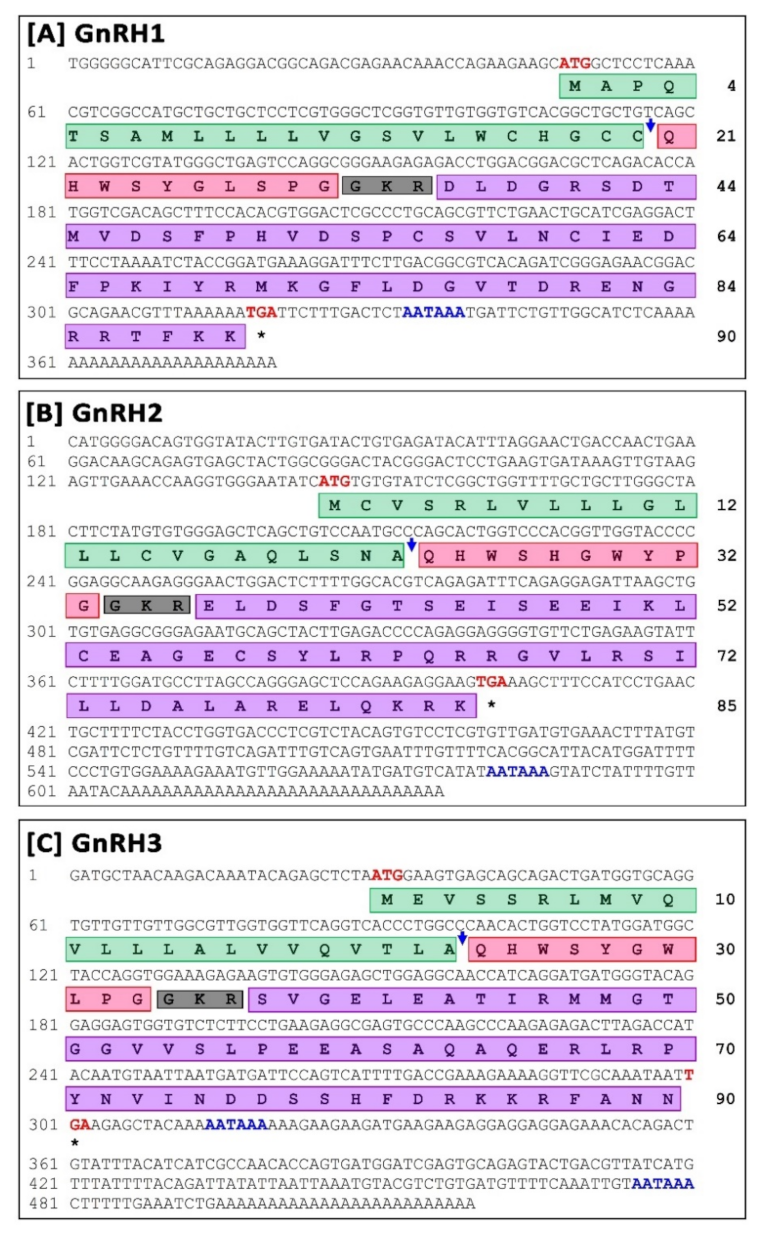
3.1.1. Molecular Characterization and In-Silico Analysis of sbGnRH (GnRH1) Isoform
3.1.2. Molecular Characterization and In-Silico Analysis of cGnRH-II (GnRH2) Isoform
3.1.3. Molecular Characterization and In-Silico Analysis of sGnRH (GnRH3) Isoform
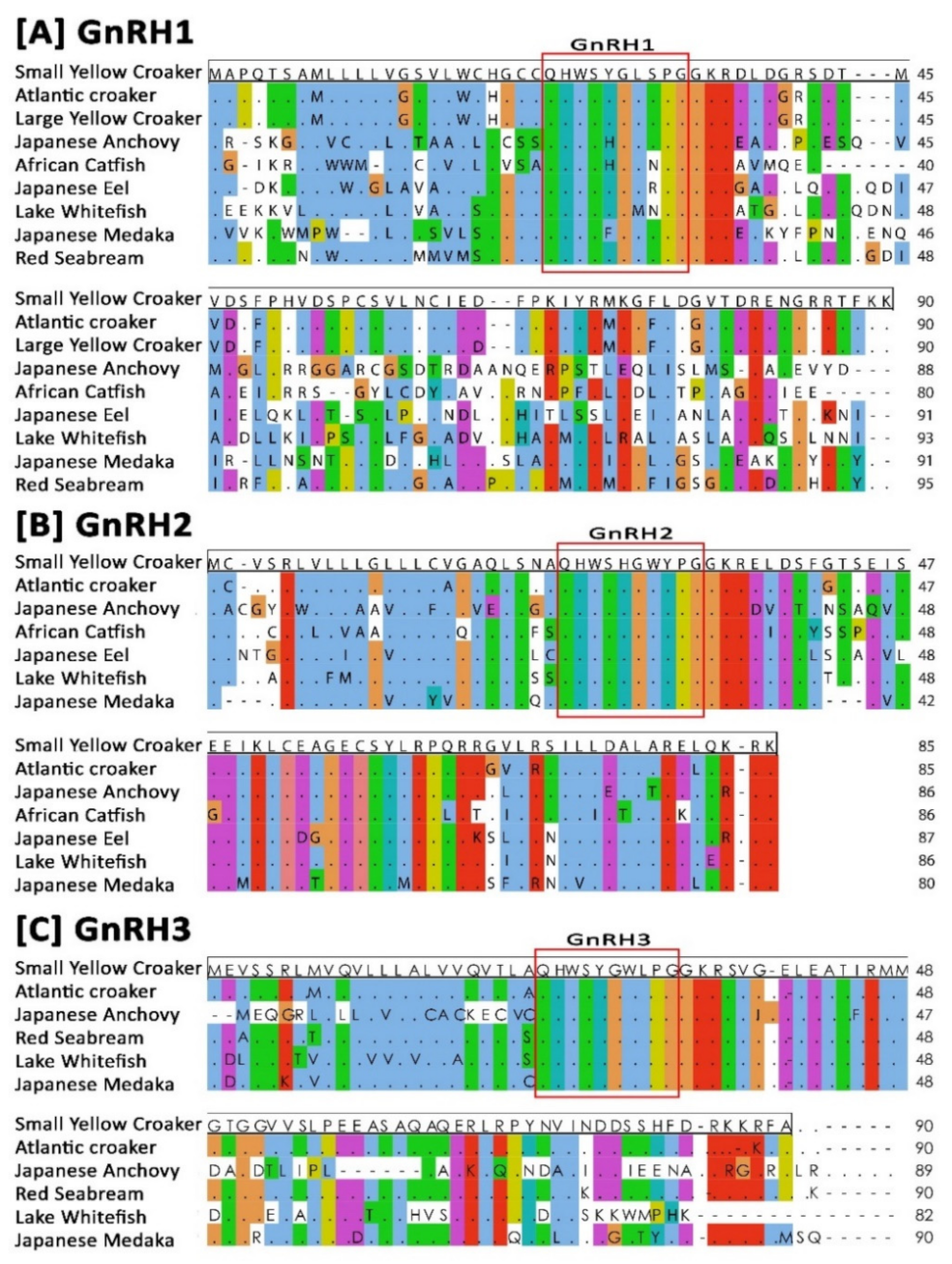
3.2. Sequence Alignment and Identity–Similarity Index
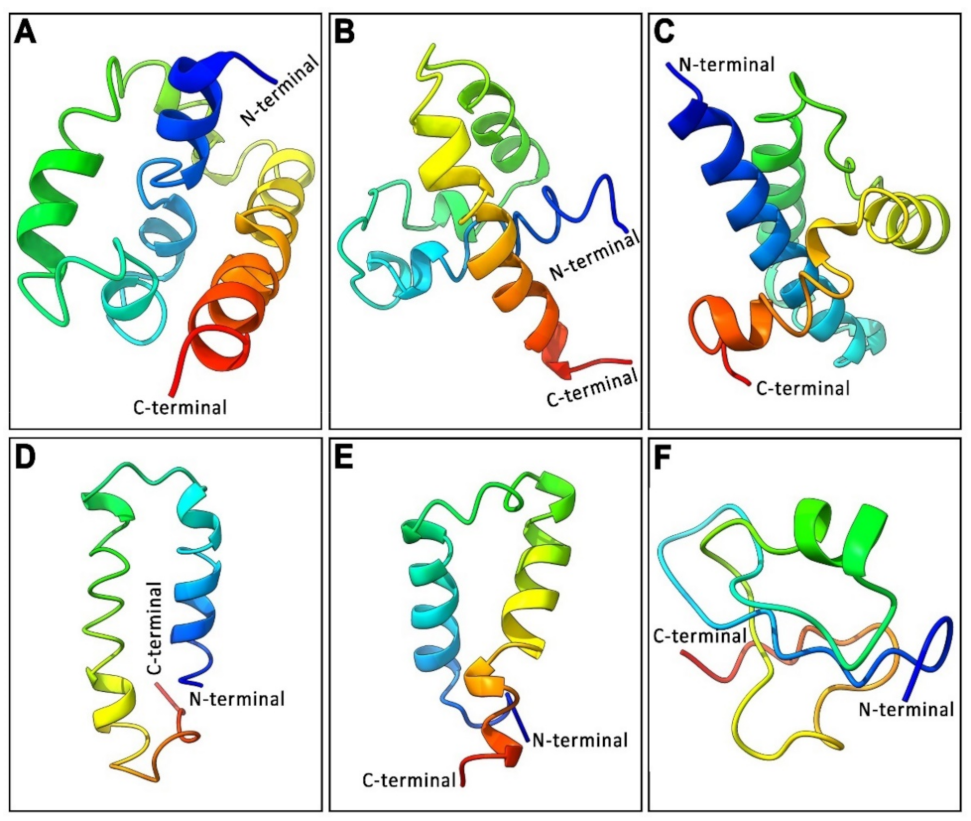
3.3. Analysis of 3D Protein Structure of Three GnRH Isoforms and Their GAP Region
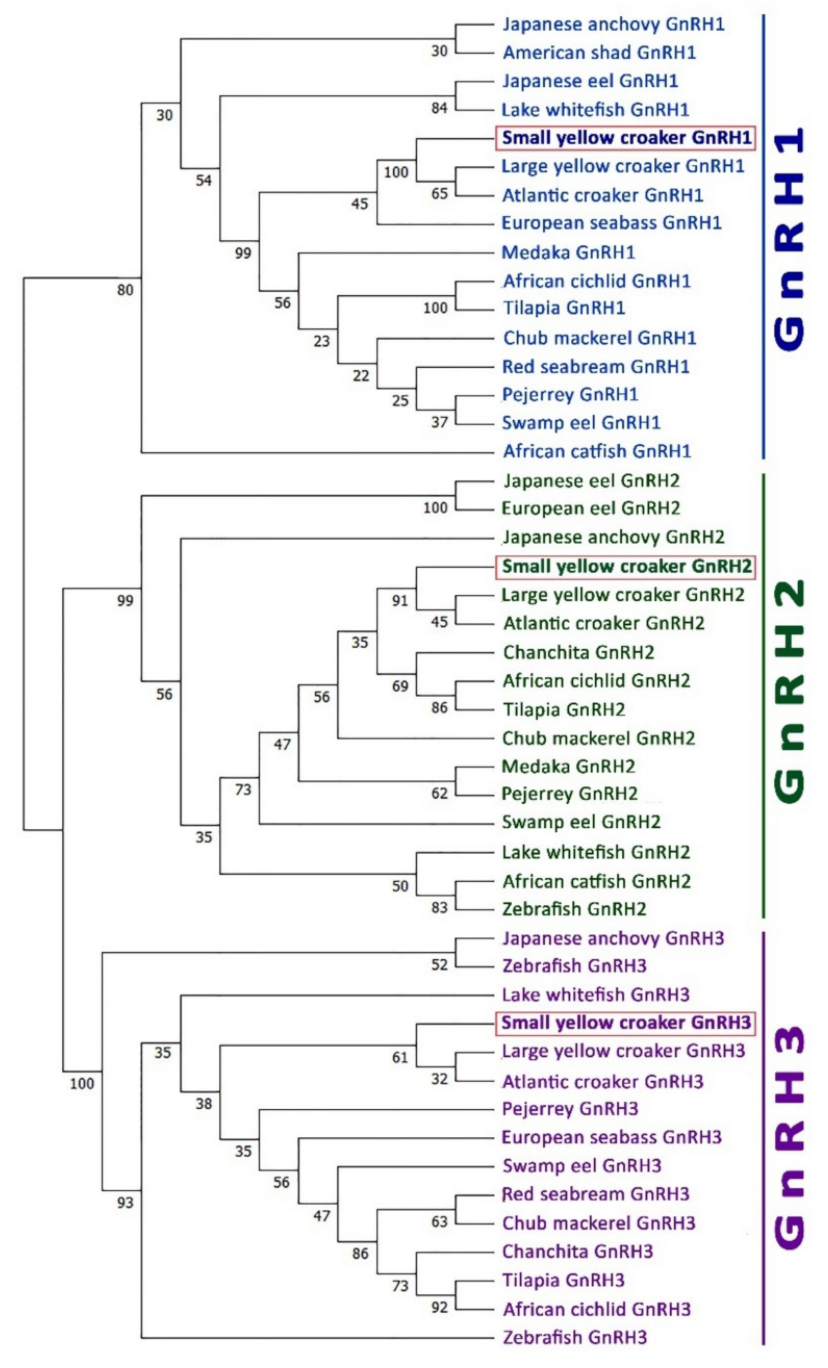
3.4. Phylogenetic Analysis
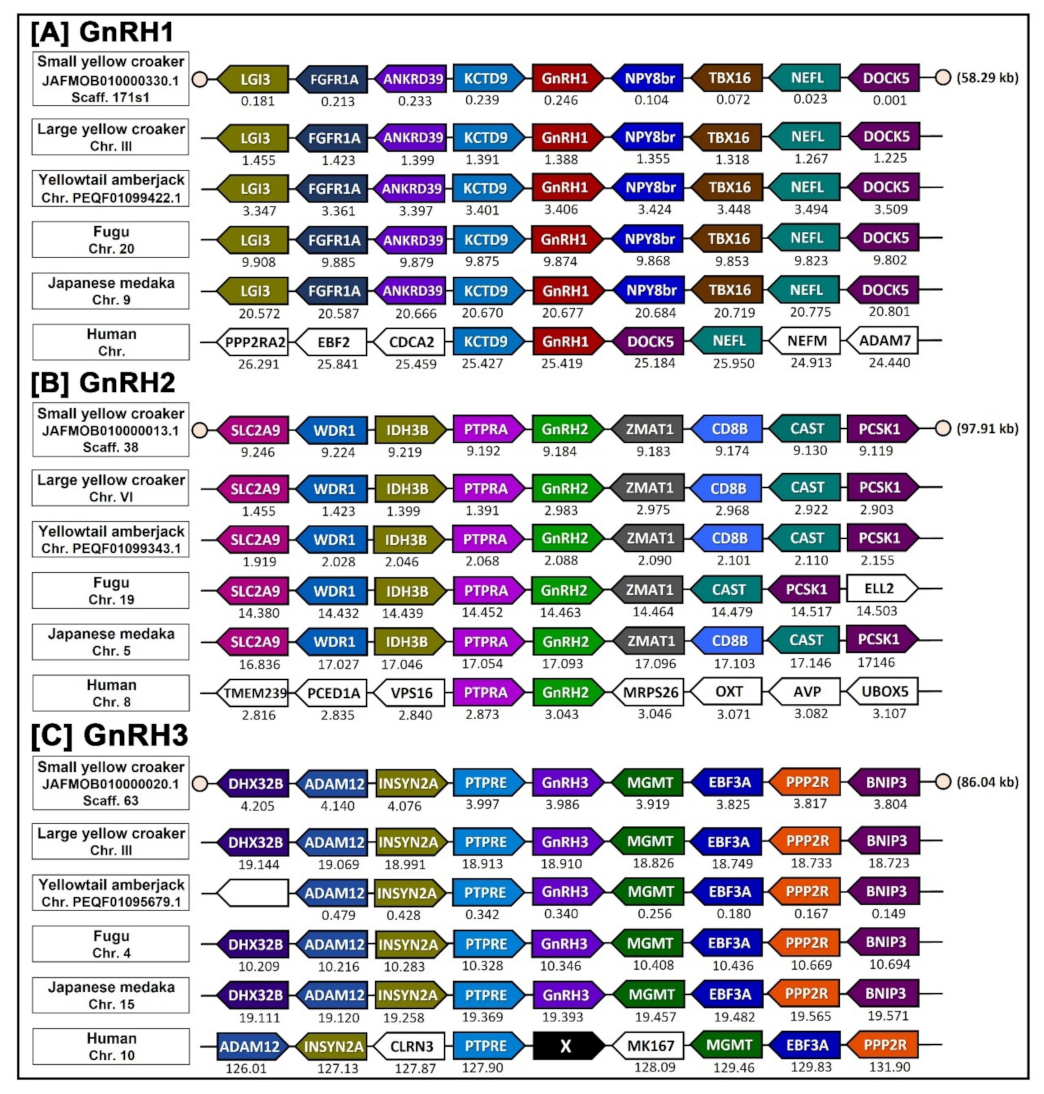
3.5. Synteny Analysis
3.6. Changes in mRNA Expression Levels of Three GnRH Isoforms in Different Organs of Small Yellow Croaker in Both Sexes
3.7. Changes in mRNA Expression Levels of Three GnRH Isoforms in Different Brain Parts of Small Yellow Croaker in Both Sexes
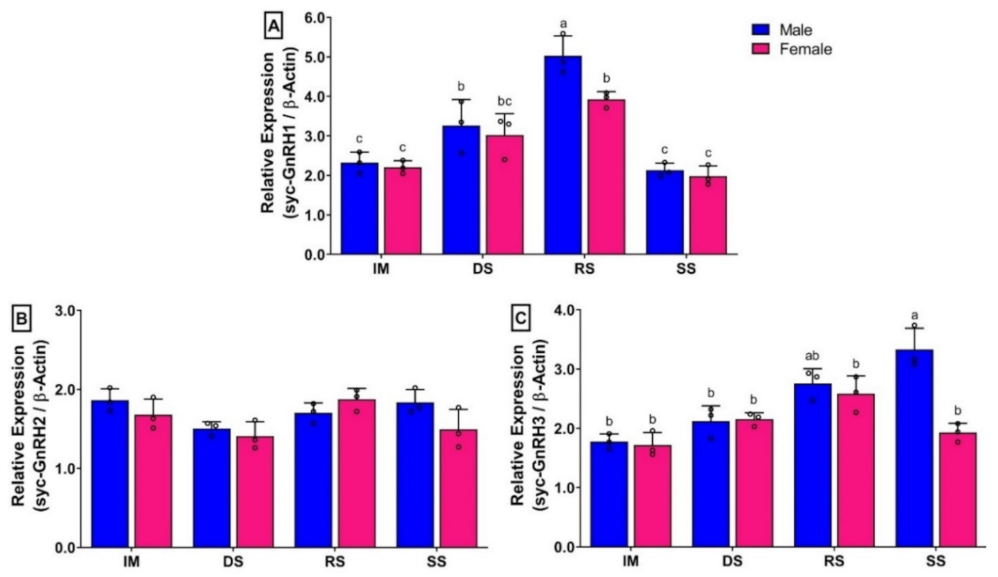
3.8. Changes in mRNA Expression Levels of Three GnRH Isoforms in Brain of Small Yellow Croaker during Gonadal Developmental Stages
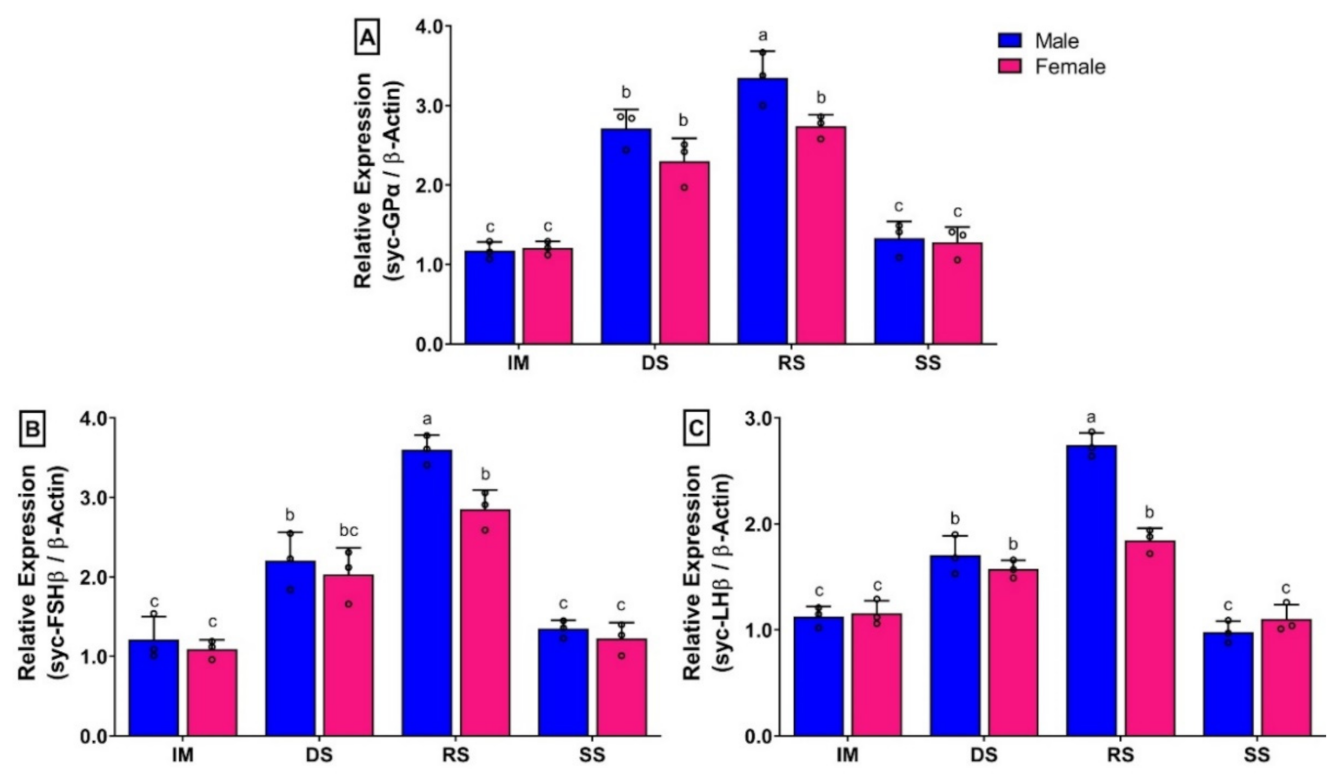
3.9. Changes in mRNA Expression Levels of Three GtH Subunits in Pituitary of Small Yellow Croaker during Gonadal Developmental Stages
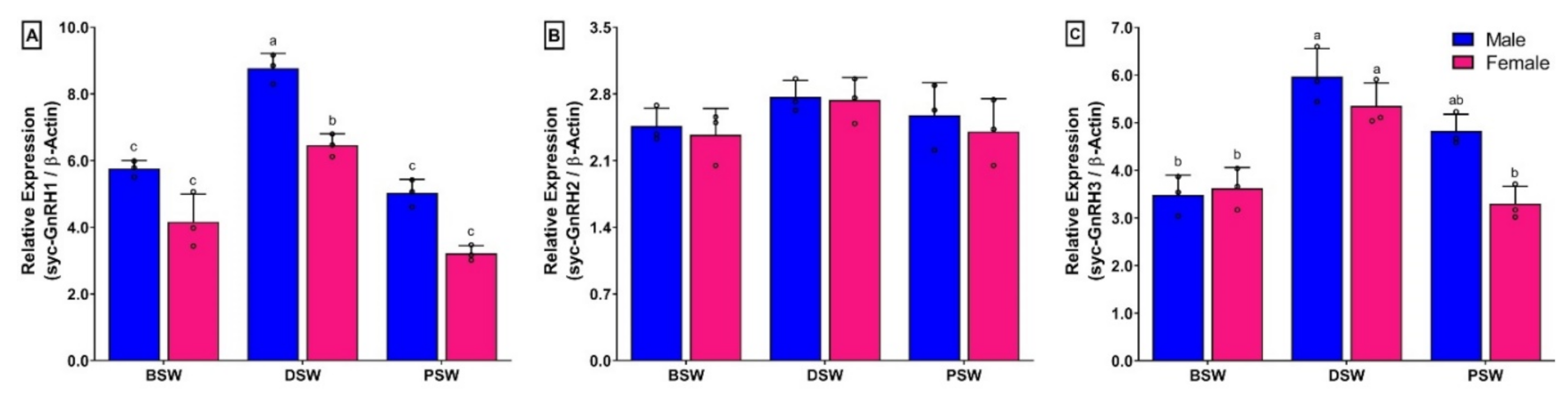
3.10. Changes in mRNA Expression Levels of Three GnRH Isoforms in Brain of Small Yellow Croaker during Induced Spawning Event
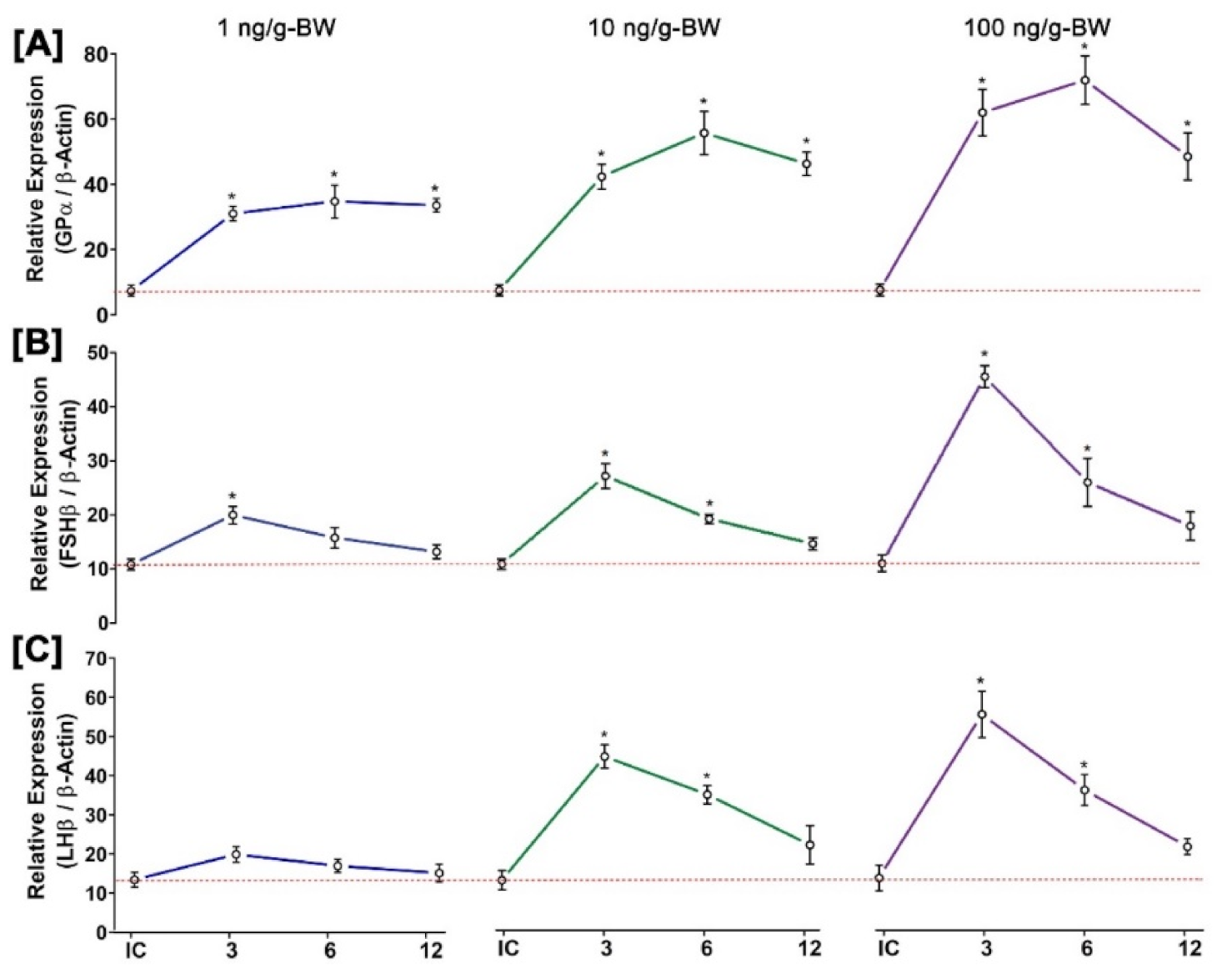
3.11. Time- and Dose-Dependent Effect of GnRH1 on mRNA Expression of Three GtH Subunits in Pituitary after In Vivo Injection of GnRH1 Peptide
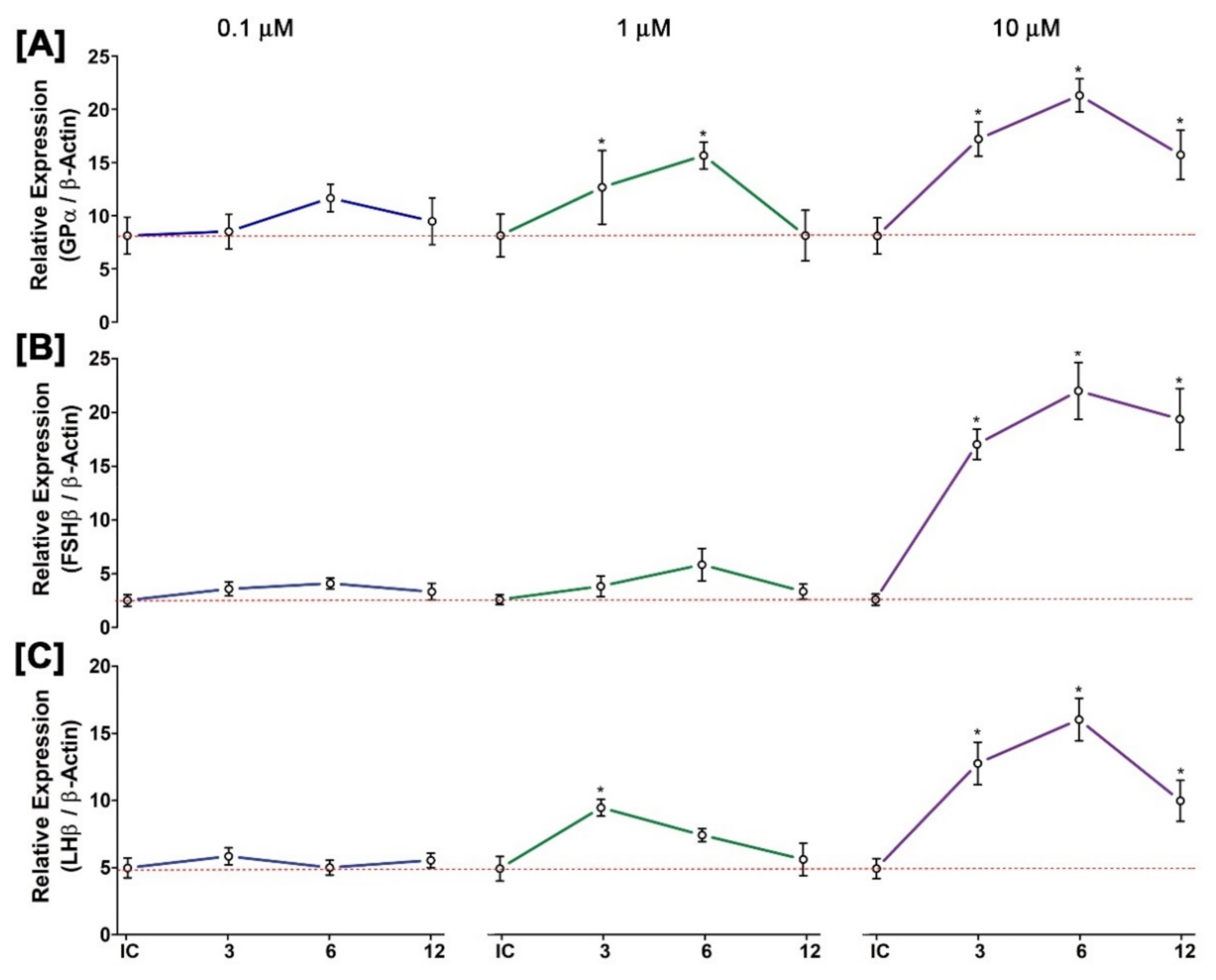
3.12. Time- and Dose-Dependent Effect of GnRH1 on mRNA Expression of Three GtH Subunits in Cultured Pituitary after In Vitro Incubation with GnRH1 Peptide
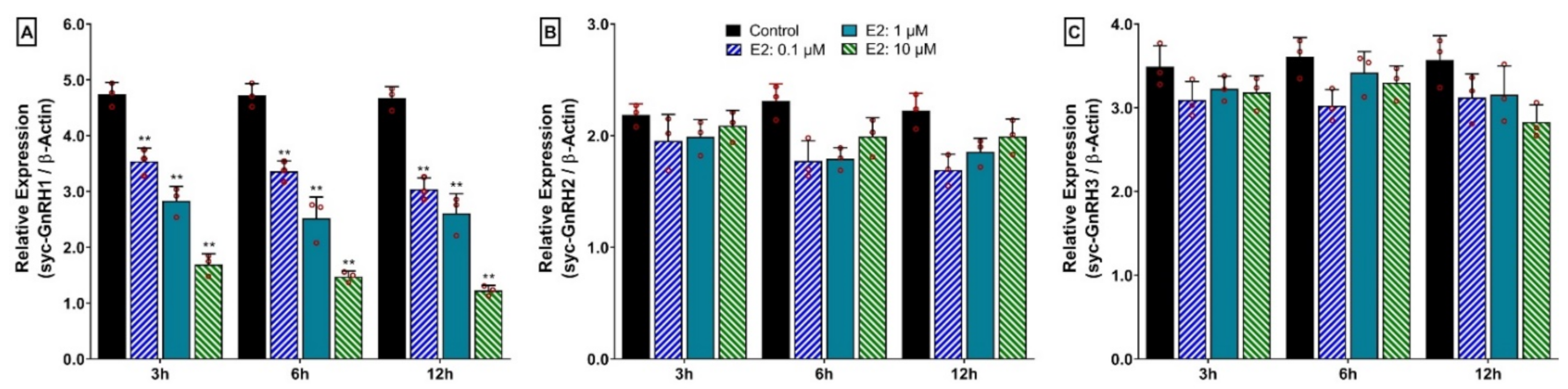
3.13. Time- and Dose-Depended Effect of 17β-Estradiol (E2) on mRNA Expression of Three GnRH Isoforms in Hypothalamus after In Vitro Incubation with E2
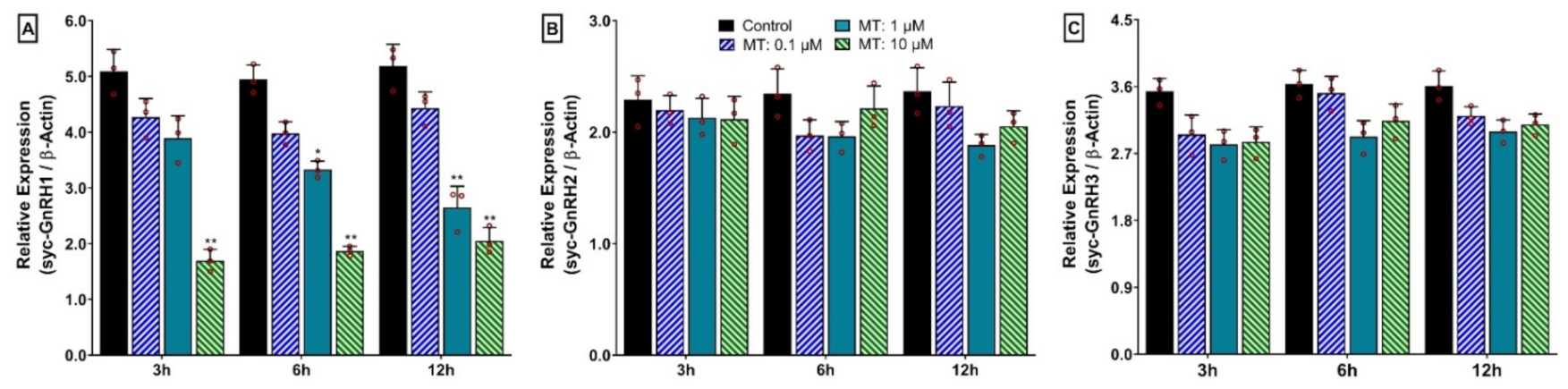
3.14. Time- and Dose-Depended Effect of 17α-Methyltestosterone (MT) on mRNA Expression of Three GnRH Isoforms in Hypothalamus after In Vitro Incubation with MT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reinecke, M. Insulin-like growth factors and fish reproduction. Biol. Reprod. 2010, 82, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Zohar, Y.; Muñoz-Cueto, J.A.; Elizur, A.; Kah, O. Neuroendocrinology of reproduction in teleost fish. Gen. Comp. Endocrinol. 2010, 165, 438–455. [Google Scholar] [CrossRef]
- Dufour, S.; Quérat, B.; Tostivint, H.; Pasqualini, C.; Vaudry, H.; Rousseau, K. Origin and evolution of the neuroendocrine control of reproduction in vertebrates, with special focus on genome and gene duplications. Physiol. Rev. 2020, 100, 869–943. [Google Scholar] [CrossRef]
- Fernald, R.D.; White, R.B. Gonadotropin-releasing hormone genes: Phylogeny, structure, and functions. Front. Neuroendocrinol. 1999, 20, 224–240. [Google Scholar] [CrossRef]
- Millar, R.P. GnRHs and GnRH receptors. Anim. Reprod. Sci. 2005, 88, 5–28. [Google Scholar] [CrossRef]
- Kauffman, A.S.; Rissman, E.F. A critical role for the evolutionarily conserved gonadotropin-releasing hormone II: Mediation of energy status and female sexual behavior. Endocrinology 2004, 145, 3639–3646. [Google Scholar] [CrossRef] [PubMed]
- Nishiguchi, R.; Azuma, M.; Yokobori, E.; Uchiyama, M.; Matsuda, K. Gonadotropin-releasing hormone 2 suppresses food intake in the zebrafish, Danio rerio. Front. Endocrinol. 2012, 3, 122. [Google Scholar] [CrossRef] [PubMed]
- Wirsig-Wiechmann, C.R. Function of gonadotropin-releasing hormone in olfaction. Keio J. Med. 2001, 50, 81–85. [Google Scholar] [CrossRef]
- Kim, D.K.; Cho, E.B.; Moon, M.J.; Park, S.; Hwang, J.I.; Kah, O.; Sower, S.A.; Vaudry, H.; Seong, J.Y. Revisiting the evolution of gonadotropin-releasing hormones and their receptors in vertebrates: Secrets hidden in genomes. Gen. Comp. Endocrinol. 2011, 170, 68–78. [Google Scholar] [CrossRef]
- Tostivint, H. Evolution of the gonadotropin-releasing hormone (GnRH) gene family in relation to vertebrate tetraploidizations. Gen. Comp. Endocrinol. 2011, 170, 575–581. [Google Scholar] [CrossRef]
- Yaron, Z.; Gur, G.; Melamed, P.; Rosenfeld, H.; Elizur, A.; Levavi-Sivan, B. Regulation of fish gonadotropins. Int. Rev. Cytol. 2003, 225, 131–185. [Google Scholar] [CrossRef] [PubMed]
- Hassin, S.; Gothilf, Y.; Blaise, O.; Zohar, Y. Gonadotropin-I and -II subunit gene expression of male striped bass (Morone saxatilis) after gonadotropin-releasing hormone analogue injection: Quantitation using an optimized ribonuclease protection assay. Biol. Reprod. 1999, 58, 1233–1240. [Google Scholar] [CrossRef] [PubMed][Green Version]
- An, K.W.; Nelson, E.R.; Habibi, H.R.; Choi, C.Y. Molecular characterization and expression of three GnRH forms mRNA during gonad sex-change process, and effect of GnRHa on GTH subunits mRNA in the protandrous black porgy (Acanthopagrus schlegeli). Gen. Comp. Endocrinol. 2008, 159, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.P.; Cui, X.F.; Wang, Y.R.; Li, Z.Y.; Tian, C.X.; Jiang, D.N.; Zhu, C.H.; Zhang, Y.; Li, S.S.; Li, G.L. Identification, functional characterization, and estrogen regulation on gonadotropin-releasing hormone in the spotted scat, Scatophagus argus. Fish Physiol. Biochem. 2020, 46, 1743–1757. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Huang, Y.; Li, X.; Li, Z.; Yang, H.; He, R.; Zhong, H.; Li, G.; Chen, H. Identification and functional characterization of gonadotropin-releasing hormone in pompano (Trachinotus ovatus). Gen. Comp. Endocrinol. 2022, 316, 113958. [Google Scholar] [CrossRef] [PubMed]
- Tilbrook, A.J.; Clarke, I.J. Negative feedback regulation of the secretion and actions of gonadotropin-releasing hormone in males. Biol. Reprod. 2001, 64, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Ng, Y.; Wolfe, A.; Novaira, H.J.; Radovick, S. Estrogen regulation of gene expression in GnRH neurons. Mol. Cell. Endocrinol. 2009, 303, 25–33. [Google Scholar] [CrossRef]
- Petersen, S.L.; Ottem, E.N.; Carpenter, C.D. Direct and indirect regulation of gonadotropin-releasing hormone neurons by estradiol. Biol. Reprod. 2003, 69, 1771–1778. [Google Scholar] [CrossRef]
- Vetillard, A.; Bailhache, T. Effects of 4-n-nonylphenol and tamoxifen on salmon gonadotropin-releasing hormone, estrogen receptor, and vitellogenin gene expression in juvenile rainbow trout. Toxicol. Sci. 2006, 92, 537–544. [Google Scholar] [CrossRef]
- Xiong, Y.; Yang, J.; Jiang, T.; Liu, H.B.; Zhong, X.M.; Tang, J.H. Early life history of the small yellow croaker (Larimichthys polyactis) in sandy ridges of the South Yellow Sea. Mar. Biol. Res. 2017, 13, 993–1002. [Google Scholar] [CrossRef]
- Kim, S.; Lim, J.; Lee, K.; Park, S. Effect of twine thickness on size-selectivity of driftnet for the yellow croaker Larimichthys polyactis in southwestern Sea of Korea. Chin. J. Ocean. Limnol. 2016, 34, 1199–1208. [Google Scholar] [CrossRef]
- Lou, B.; Zhan, W.; Chen, R.Y.; Liu, F.; Wang, L.G.; Xu, D.D.; Mao, G.M. Studies on techniques of the artificial breeding of Larimichthys polyactis. J. Zhejiang Univ. (Nat. Sci.) 2016, 35, 361–365. [Google Scholar]
- Liu, F.; Liu, Y.; Chu, T.; Lou, B.; Zhan, W.; Chen, R. Interspecific hybridization and genetic characterization of Larimichthys polyactis (♀) and L. crocea (♂). Aquacult. Int. 2019, 27, 663–674. [Google Scholar] [CrossRef]
- Zohar, Y.; Mylonas, C.C. Endocrine manipulations of spawning in cultured fish: From hormones to genes. Aquaculture 2001, 197, 99–136. [Google Scholar] [CrossRef]
- Mylonas, C.C.; Papandroulakis, N.; Smboukis, A.; Papadaki, M.; Divanach, P. Induction of spawning of cultured greater amberjack (Seriola dumerili) using GnRHa implants. Aquaculture 2004, 237, 141–154. [Google Scholar] [CrossRef]
- Zhao, C.; Xu, S.; Feng, C.; Liu, Y.; Yang, Y.; Wang, Y.; Xiao, Y.; Song, Z.; Liu, Q.; Li, J. Characterization and differential expression of three GnRH forms during reproductive development in cultured turbot Schophthalmus maximus. J. Oceanol. Limnol. 2018, 36, 1360–1373. [Google Scholar] [CrossRef]
- Imanaga, Y.; Nyuji, M.; Amano, M.; Takahashi, A.; Kitano, H.; Yamaguchi, A.; Matsuyama, M. Characterization of gonadotropin-releasing hormone and gonadotropin in jack mackerel (Trachurus japonicus): Comparative gene expression analysis with respect to reproductive dysfunction in captive and wild fish. Aquaculture 2014, 428, 226–235. [Google Scholar] [CrossRef]
- Honji, R.M.; Caneppele, D.; Pandolfi, M.; Lo Nostro, F.L.; Moreira, R.G. Characterization of the gonadotropin-releasing hormone system in the Neotropical teleost, Steindachneridion parahybae during the annual reproductive cycle in captivity. Gen. Comp. Endocrinol. 2019, 273, 73–85. [Google Scholar] [CrossRef]
- Kim, H.W.; Kim, J.H.; Park, J.W.; Baek, H.J.; Kim, D.J. Effects of photoperiod and water temperature on male sex steroid levels in cultured small yellow croaker (Larimichthys polyactis). J. Life Sci. 2021, 31, 314–320. [Google Scholar] [CrossRef]
- Lim, H.K.; Le, M.H.; An, C.M.; Kim, S.Y.; Park, M.S.; Chang, Y.J. Reproductive cycle of yellow croaker Larimichthys polyactis in southern waters off Korea. Fish. Sci. 2010, 76, 971–980. [Google Scholar] [CrossRef]
- Kang, D.Y.; Jo, K.C.; Lee, J.H.; Kang, H.W.; Kim, H.C.; Kim, G.H. Annual reproductive cycle of wild female yellow croaker, Larimichthys polyactis. J. Aquacul. 2006, 19, 188–196. [Google Scholar]
- Sukhan, Z.P.; Sharker, M.R.; Kho, K.H. Molecular cloning, in silico characterization and expression analysis of axonemal protein 66.0 in Pacific abalone, Haliotis discus hannai. Eur. Zool. J. 2020, 87, 648–658. [Google Scholar] [CrossRef]
- Sukhan, Z.P.; Hossen, S.; Cho, Y.; Lee, W.K.; Kho, K.H. Hdh-Tektin-4 regulates motility of fresh and cryopreserved sperm in Pacific abalone, Haliotis discus hannai. Front. Cell Dev. Biol. 2022, 10, 870743. [Google Scholar] [CrossRef]
- Nguyen, N.T.T.; Vincens, P.; Dufayard, J.F.; Roest Crollius, H.; Louis, A. Genomicus in 2022: Comparative tools for thousands of genomes and reconstructed ancestors. Nucleic. Acids Res. 2022, 50, D1025–D1031. [Google Scholar] [CrossRef] [PubMed]
- Sukhan, Z.P.; Sharker, M.R.; Cho, Y.; Hossen, S.; Choi, K.S.; Kho, K.H. Thermal stress affects gonadal maturation by regulating GnRH, GnRH Receptor, APGWamide, and Serotonin Receptor gene expression in male Pacific abalone, Haliotis discus hannai during breeding season. Front. Mar. Sci. 2021, 8, 664426. [Google Scholar] [CrossRef]
- Shao, Y.T.; Tseng, Y.C.; Chang, C.H.; Yan, H.Y.; Hwang, P.P.; Borg, B. GnRH mRNA levels in male three-spined sticklebacks, Gasterosteus aculeatus, under different reproductive conditions. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2015, 180, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, J.S.; Thomas, P.; Khan, I.A. Isolation cloning, and expression of three prepro-GnRH mRNAs in Atlantic croaker brain and pituitary. J. Comp. Neurol. 2005, 488, 384–395. [Google Scholar] [CrossRef]
- González-Martínez, D.; Madigou, T.; Zmora, N.; Anglade, I.; Zanuy, S.; Zohar, Y.; Elizur, A.; Muñoz-Cueto, J.A.; Kah, O. Differential expression of three different prepro-GnRH (gonadotrophin-releasing hormone) messengers in the brain of the European sea bass (Dicentrarchus labrax). J. Comp. Neurol. 2001, 429, 144–155. [Google Scholar] [CrossRef]
- Sukhan, Z.P.; Kitano, H.; Selvaraj, S.; Yoneda, M.; Yamaguchi, A.; Matsuyama, M. Identification and distribution of three gonadotropin-releasing hormone (GnRH) isoforms in the brain of a clupeiform fish, Engraulis japonicus. Zool. Sci. 2013, 30, 1081–1091. [Google Scholar] [CrossRef]
- Selvaraj, S.; Kitano, H.; Amano, M.; Nyuji, M.; Kaneko, K.; Yamaguchi, A.; Matsuyama, M. Molecular characterization and expression profiles of three GnRH forms in the brain and pituitary of adult chub mackerel (Scomber japonicus) maintained in captivity. Aquaculture 2012, 356, 200–210. [Google Scholar] [CrossRef]
- Shahjahan, M.; Hamabata, T.; Motohashi, E.; Doi, H.; Ando, H. Differential expression of three types of gonadotropin-releasing hormone genes during the spawning season in grass puffer, Takifugu niphobles. Gen. Comp. Endocrinol. 2010, 167, 153–163. [Google Scholar] [CrossRef]
- Servili, A.; Lethimonier, C.; Lareyre, J.J.; López-Olmeda, J.F.; Sánchez-Vázquez, F.J.; Kah, O.; Muñoz-Cueto, J.A. The Highly conserved gonadotropin-releasing hormone-2 form acts as a melatonin-releasing factor in the pineal of a teleost fish, the European sea bass Dicentrarchus labrax. Endocrinology 2010, 151, 2265–2275. [Google Scholar] [CrossRef]
- Maruska, K.P.; Tricas, T.C. Gonadotropin-releasing hormone (GnRH) modulates auditory processing in the fish brain. Horm. Behav. 2011, 59, 451–464. [Google Scholar] [CrossRef]
- Okubo, K.; Nagahama, Y. Structural and functional evolution of gonadotropin-releasing hormone in vertebrates. Acta. Physiol. 2008, 193, 3–15. [Google Scholar] [CrossRef]
- Ogawa, S.; Akiyama, G.; Kato, S.; Soga, T.; Sakuma, Y.; Parhar, I.S. Immunoneutralization of gonadotropin-releasing hormone type-III suppresses male reproductive behavior of cichlids. Neurosci. Lett. 2006, 403, 201–205. [Google Scholar] [CrossRef]
- Yamamoto, N.; Oka, Y.; Kawashima, S. Lesions of gonadotropin-releasing hormone-immunoreactive terminal nerve cells: Effects on the reproductive behavior of male dwarf gouramis. Neuroendocrinology 1997, 65, 403–412. [Google Scholar] [CrossRef]
- Shahjahan, M.; Kitahashi, T.; Parhar, I.S. Central pathways integrating metabolism and reproduction in teleosts. Front. Endocrinol. 2014, 5, 36. [Google Scholar] [CrossRef]
- Bauer, C.M.; Fudickar, A.M.; Anderson-Buckingham, S.; Abolins-Abols, M.; Atwell, J.W.; Ketterson, E.D.; Greives, T.J. Seasonally sympatric but allochronic: Differential expression of hypothalamic genes in a songbird during gonadal development. Proc. Biol. Sci. B 2018, 285, 20181735. [Google Scholar] [CrossRef]
- Andersson, E.; Borg, B.; Goos, H.J. Temperature; but not photoperiod, influences gonadotropin-releasing hormone binding in the pituitary of the three-spined stickleback, Gasterosteus aculeatus. Gen. Comp. Endocrinol. 1992, 88, 111–116. [Google Scholar] [CrossRef]
- Nyuji, M.; Selvaraj, S.; Kitano, H.; Ohga, H.; Yoneda, M.; Shimizu, A.; Kaneko, K.; Yamaguchi, A.; Matsuyama, M. Changes in the expression of pituitary gonadotropin subunits during reproductive cycle of multiple spawning female chub mackerel Scomber japonicus. Fish Physiol. Biochem. 2012, 38, 883–897. [Google Scholar] [CrossRef]
- Gomez, J.M.; Weil, C.; Ollitrault, M.; Le Bail, P.Y.; Breton, B.; Le Gac, F. Growth hormone (GH) and gonadotropin subunit gene expression and pituitary and plasma changes during spermatogenesis and oogenesis in rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 1999, 113, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Gen, K.; Okuzawa, K.; Senthilkumaran, B.; Tanaka, H.; Moriyama, S.; Kagawa, H. Unique expression of gonadotropin-I and-II subunit genes in male and female red seabream (Pagrus major) during sexual maturation. Biol. Reprod. 2000, 63, 308–319. [Google Scholar] [CrossRef]
- Khakoo, Z.; Bhatia, A.; Gedamu, L.; Habibi, H.R. Functional specificity for salmon gonadotropin-releasing hormone (GnRH) and chicken GnRH-II coupled to the gonadotropin release and subunit messenger ribonucleic acid level in the goldfish pituitary. Endocrinology 1994, 134, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Mateos, J.; Mañanos, E.; Carrillo, M.; Zanuy, S. Regulation of follicle-stimulating hormone (FSH) and luteinizing hormone (LH) gene expression by gonadotropin-releasing hormone (GnRH) and sexual steroids in the Mediterranean Sea bass. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2002, 132, 75–86. [Google Scholar] [CrossRef]
- Dickey, J.T.; Swanson, P. Effects of salmon gonadotropin-releasing hormone on follicle stimulating hormone secretion and subunit gene expression in coho salmon (Oncorhynchus kisutch). Gen. Comp. Endocrinol. 2000, 118, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Schulz, R.W.; Miura, T. Spermatogenesis and its endocrine regulation. Fish Physiol. Biochem. 2002, 26, 43–56. [Google Scholar] [CrossRef]
- Imamura, S.; Hur, S.P.; Takeuchi, Y.; Badruzzaman, M.; Mahardini, A.; Rizky, D.; Takemura, A. The mRNA expression patterns of kisspeptins, GnRHs, and gonadotropins in the brain and pituitary gland of a tropical damselfish, Chrysiptera cyanea, during the reproductive cycle. Fish Physiol. Biochem. 2020, 46, 277–291. [Google Scholar] [CrossRef]
- Lee, Y.H.; Du, J.L.; Shih, Y.S.; Jeng, S.R.; Sun, L.T.; Chang, C.F. In vivo and in vitro sex steroids stimulate seabream gonadotropin-releasing hormone content and release in the protandrous black porgy, Acanthopagrus schlegeli. Gen. Comp. Endocrinol. 2004, 139, 12–19. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sukhan, Z.P.; Cho, Y.; Hossen, S.; Yang, S.-W.; Hwang, N.-Y.; Lee, W.K.; Kho, K.H. Functional Characterization of Three GnRH Isoforms in Small Yellow Croaker Larimichthys polyactis Maintained in Captivity: Special Emphasis on Reproductive Dysfunction. Biology 2022, 11, 1200. https://doi.org/10.3390/biology11081200
Sukhan ZP, Cho Y, Hossen S, Yang S-W, Hwang N-Y, Lee WK, Kho KH. Functional Characterization of Three GnRH Isoforms in Small Yellow Croaker Larimichthys polyactis Maintained in Captivity: Special Emphasis on Reproductive Dysfunction. Biology. 2022; 11(8):1200. https://doi.org/10.3390/biology11081200
Chicago/Turabian StyleSukhan, Zahid Parvez, Yusin Cho, Shaharior Hossen, Seok-Woo Yang, Nam-Yong Hwang, Won Kyo Lee, and Kang Hee Kho. 2022. "Functional Characterization of Three GnRH Isoforms in Small Yellow Croaker Larimichthys polyactis Maintained in Captivity: Special Emphasis on Reproductive Dysfunction" Biology 11, no. 8: 1200. https://doi.org/10.3390/biology11081200
APA StyleSukhan, Z. P., Cho, Y., Hossen, S., Yang, S.-W., Hwang, N.-Y., Lee, W. K., & Kho, K. H. (2022). Functional Characterization of Three GnRH Isoforms in Small Yellow Croaker Larimichthys polyactis Maintained in Captivity: Special Emphasis on Reproductive Dysfunction. Biology, 11(8), 1200. https://doi.org/10.3390/biology11081200






