Differential Radiosensitizing Effect of 50 nm Gold Nanoparticles in Two Cancer Cell Lines
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Receptor Expression
2.3. Cytogenetic Characterization
2.4. Nanoparticles and Internalization
2.5. Irradiation
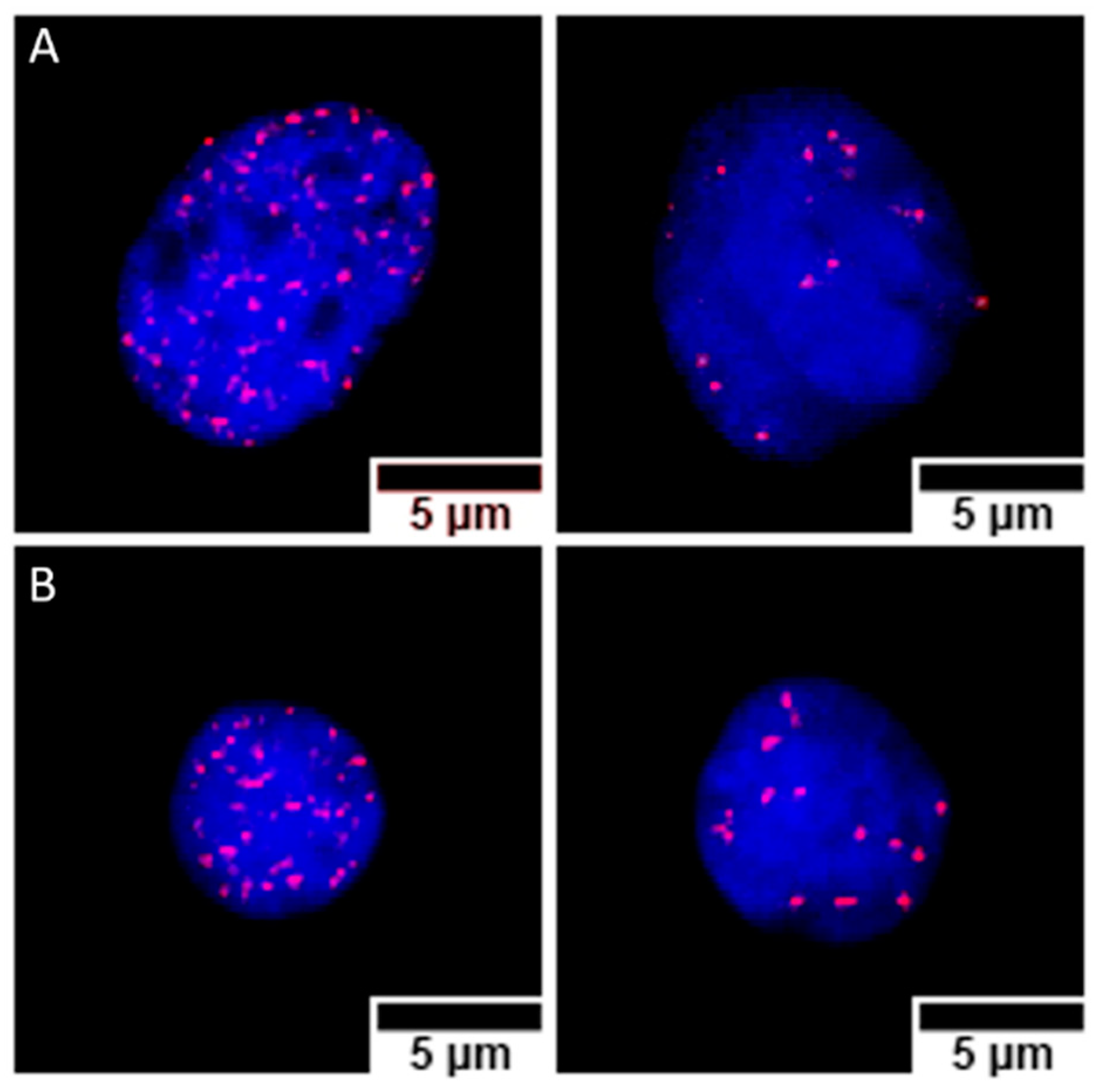
2.6. γ-H2AX Foci Detection
2.7. MTT Cell Viability Assay
2.8. Statistical Analysis
3. Results
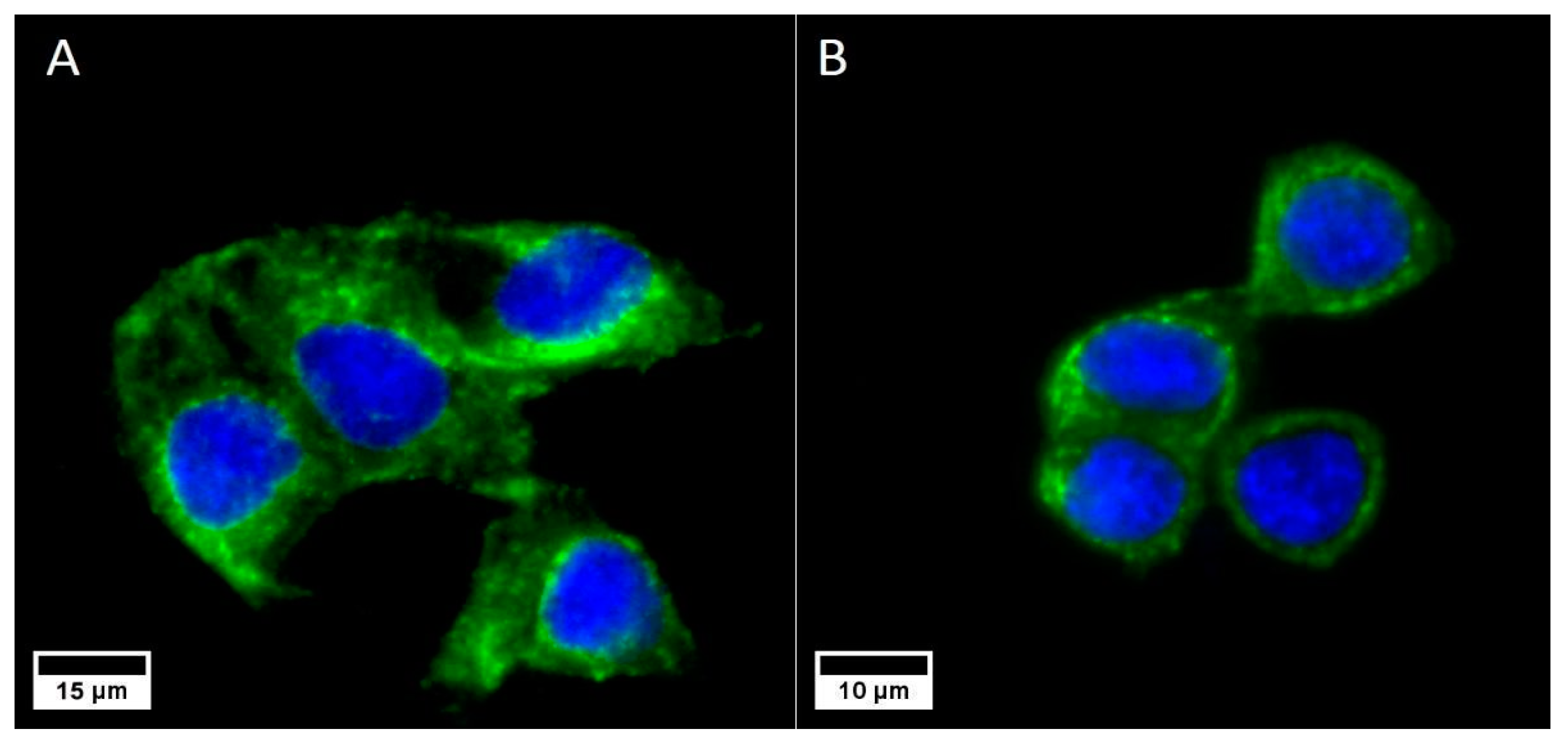
3.1. Receptor Expression
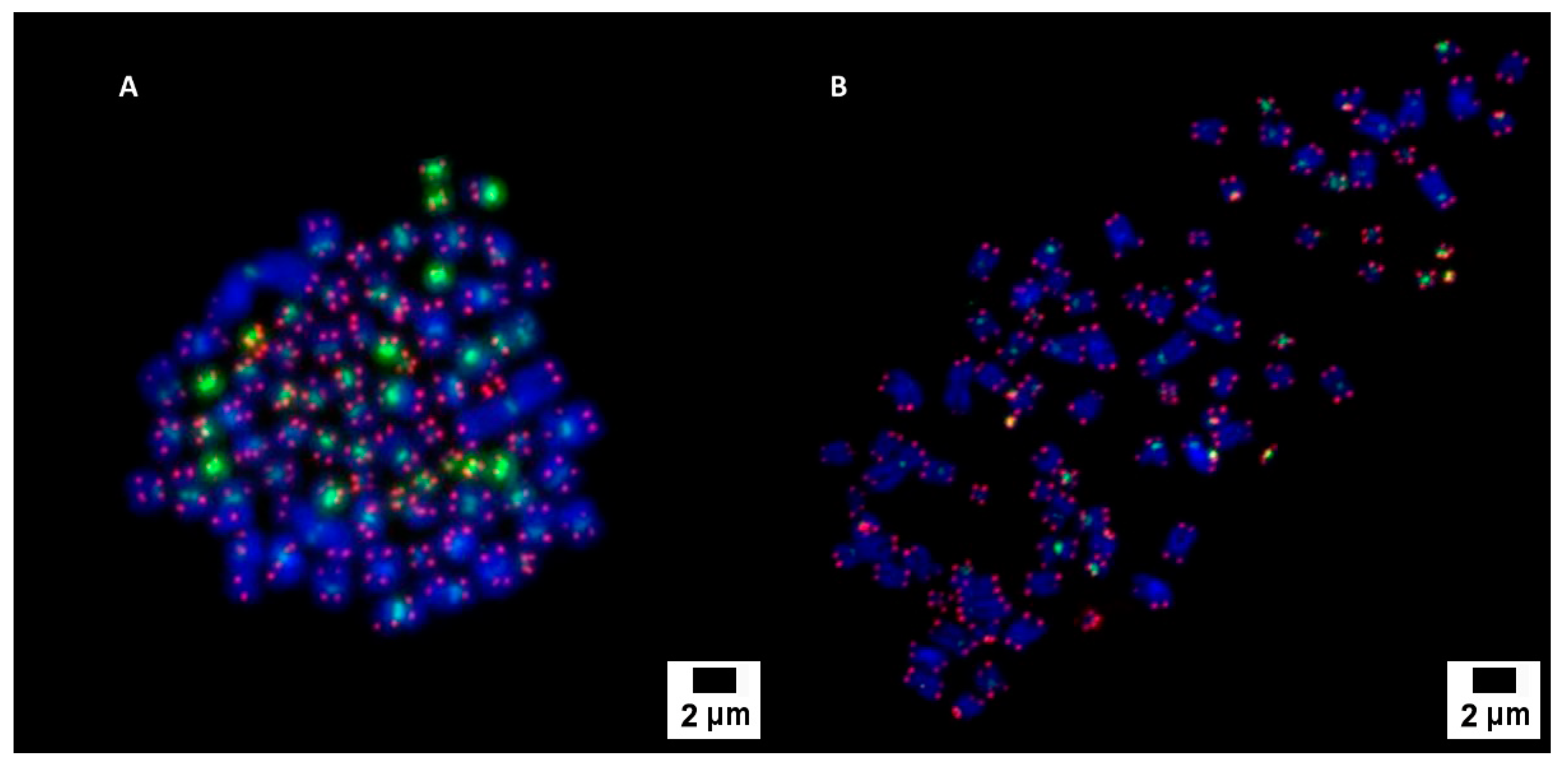
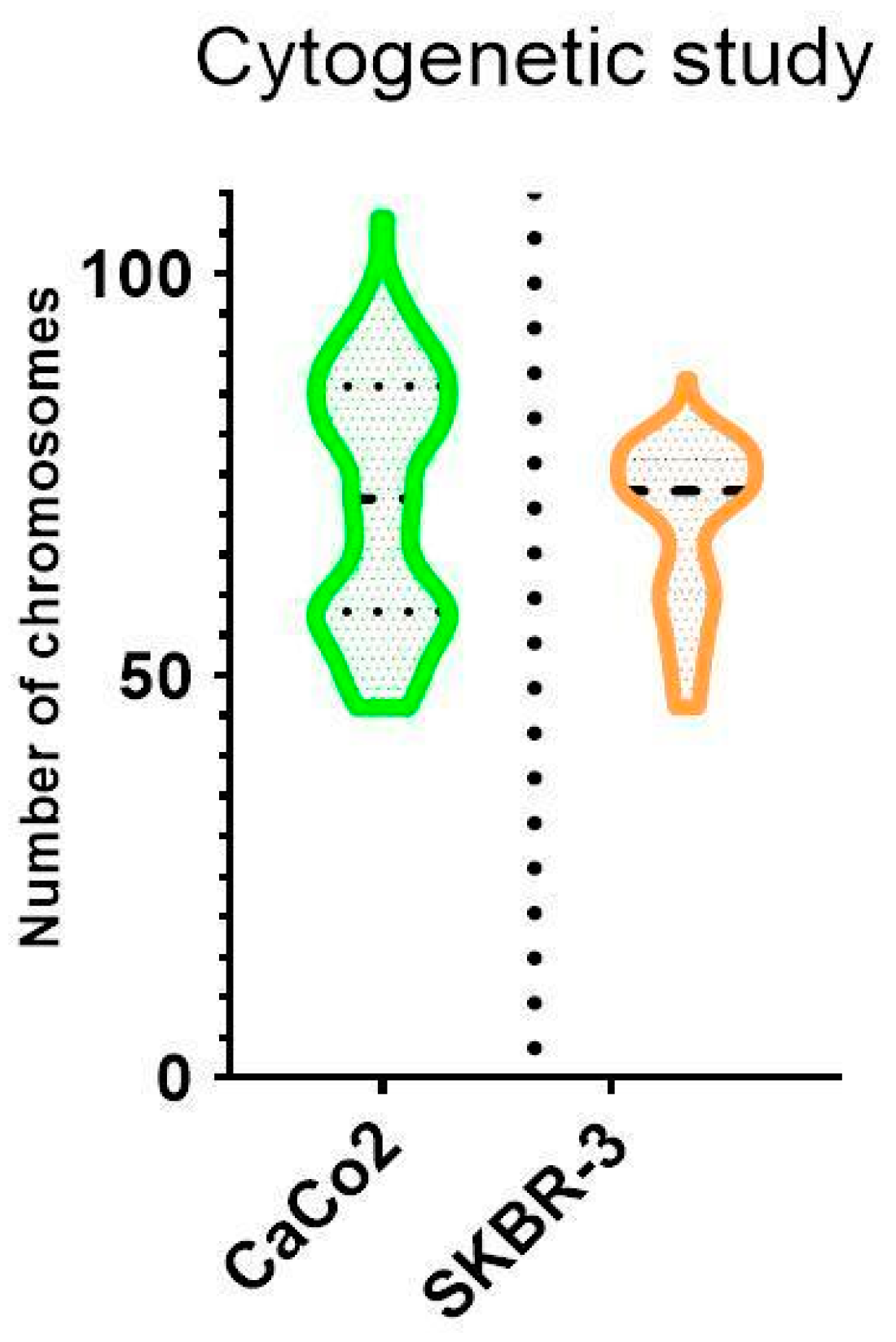
3.2. Cytogenetic Characterization
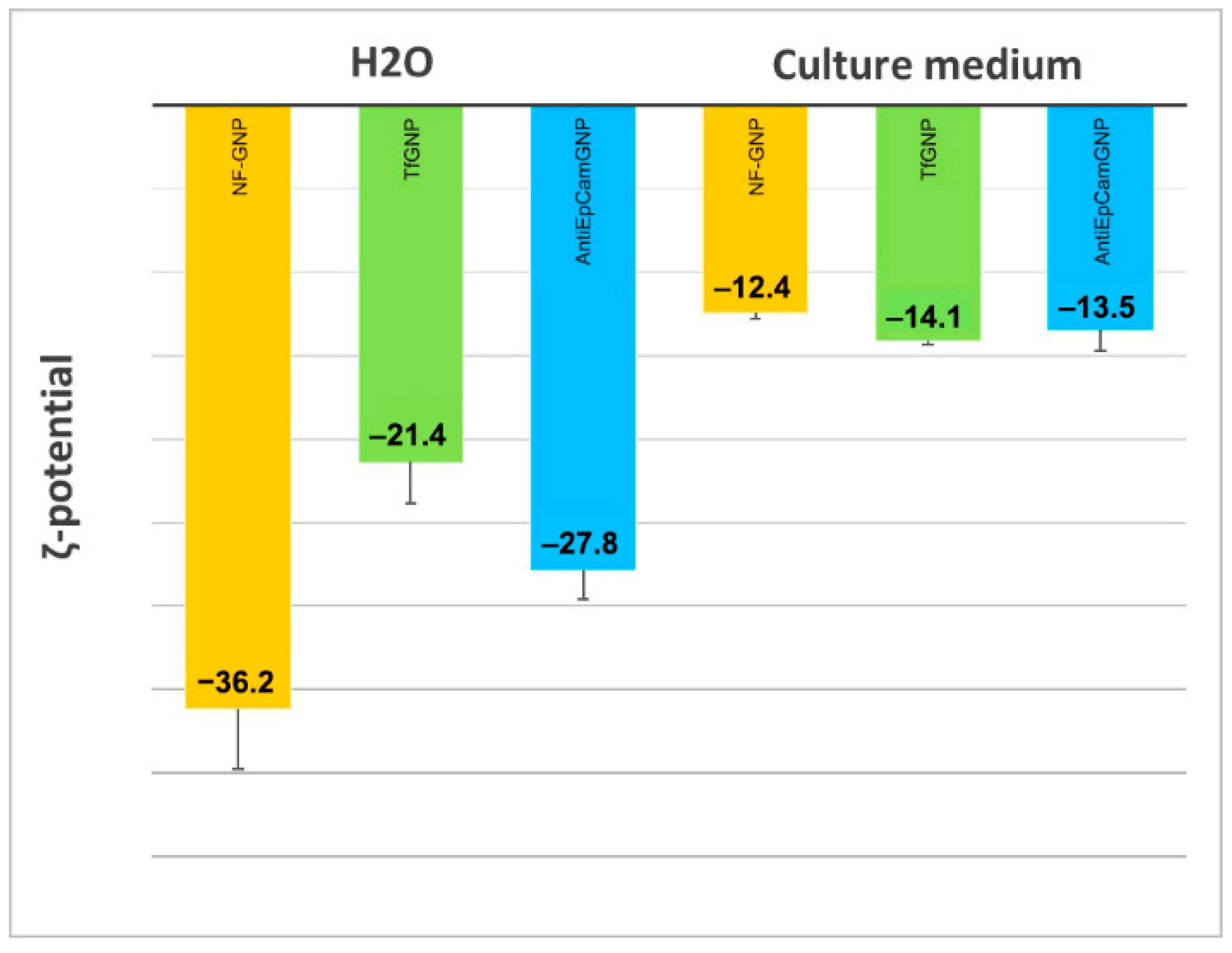
3.3. Nanoparticle Internalization
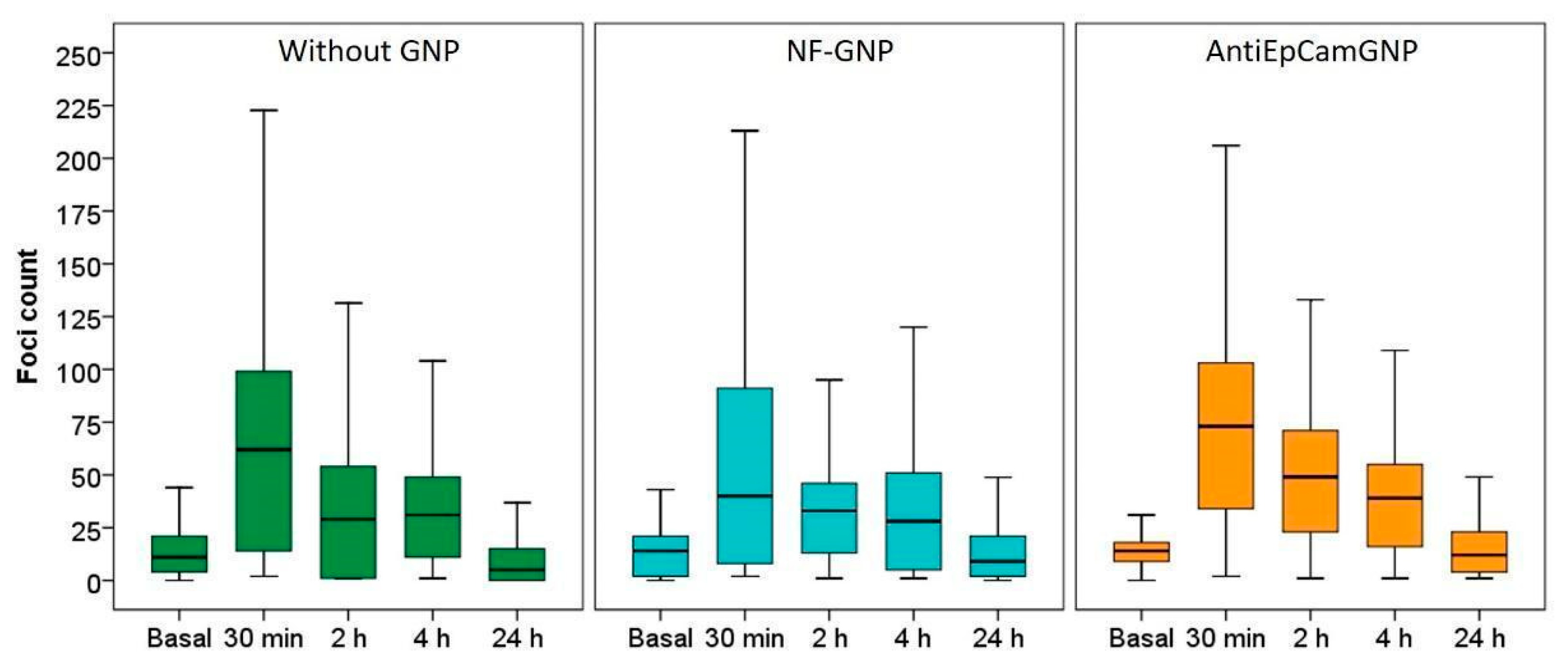
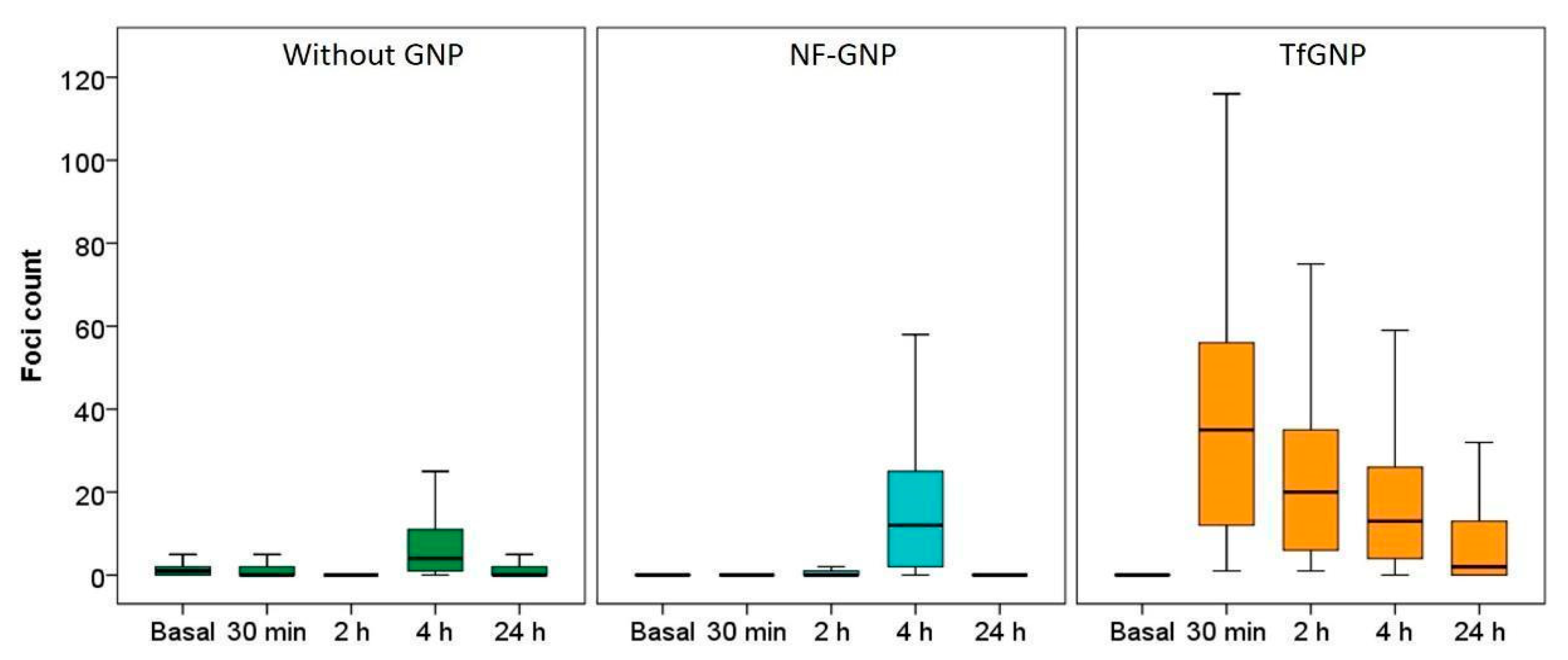
3.4. γ-H2AX Foci Induction and Kinetics
3.5. MTT Cell Viability Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atun, R.; Jaffray, D.A.; Barton, M.B.; Bray, F.; Baumann, M.; Vikram, B.; Hanna, T.P.; Knaul, F.M.; Lievens, Y.; Lui, T.Y.; et al. Expanding global access to radiotherapy. Lancet Oncol. 2015, 16, 1153–1186. [Google Scholar] [CrossRef]
- Berkey, F.J. Managing the Adverse Effects of Radiation Therapy. Am. Fam. Physician 2010, 82, 381–388. [Google Scholar]
- Citrin, D.; Cotrim, A.P.; Hyodo, F.; Baum, B.J.; Krishna, M.C.; Mitchell, J.B. Radioprotectors and Mitigators of Radiation-Induced Normal Tissue Injury. Oncologist 2010, 15, 360–371. [Google Scholar] [CrossRef]
- Roots, R.; Okada, S. Protection of DNA Molecules of Cultured Mammalian Cells from Radiation-Induced Single-Strand Scissions by Various Alcohols and SH Compounds. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1972, 21, 329–342. [Google Scholar] [CrossRef]
- Kamran, M.Z.; Ranjan, A.; Kaur, N.; Sur, S.; Tandon, V. Radioprotective Agents: Strategies and Translational Advances. Med. Res. Rev. 2016, 36, 461–493. [Google Scholar] [CrossRef]
- Smith, T.A.; Kirkpatrick, D.R.; Smith, S.; Smith, T.K.; Pearson, T.; Kailasam, A.; Herrmann, K.Z.; Schubert, J.; Agrawal, D.K. Radioprotective Agents to Prevent Cellular Damage Due to Ionizing Radiation. J. Transl. Med. 2017, 15, 232. [Google Scholar] [CrossRef]
- Bennett, M.; Feldmeier, J.; Smee, R.; Milross, C. Hyperbaric oxygenation for tumour sensitisation to radiotherapy. Cochrane Database Syst. Rev. 2018, 4, CD005007. [Google Scholar] [CrossRef]
- Raviraj, J.; Bokkasam, V.K.; Kumar, V.S.; Reddy, U.S.; Suman, V. Radiosensitizers, Radioprotectors, and Radiation Mitigators. Indian J. Dent. Res. 2014, 25, 83–90. [Google Scholar] [CrossRef]
- Fang, J.; Nakamura, H.; Maeda, H. The EPR Effect: Unique Features of Tumor Blood Vessels for Drug Delivery, Factors Involved, and Limitations and Augmentation of the Effect. Adv. Drug Deliv. Rev. 2011, 63, 136–151. [Google Scholar] [CrossRef]
- Li, C. A Targeted Approach to Cancer Imaging and Therapy. Nat. Mater. 2014, 13, 110–115. [Google Scholar] [CrossRef]
- Mora-Espí, I.; Ibáñez, E.; Soriano, J.; Nogués, C.; Gudjonsson, T.; Barrios, L. Cell Internalization in Fluidic Culture Conditions Is Improved When Microparticles Are Specifically Targeted to the Human Epidermal Growth Factor Receptor 2 (HER2). Pharmaceutics 2019, 11, 177. [Google Scholar] [CrossRef]
- Mi, Y.; Shao, Z.; Vang, J.; Kaidar-Person, O.; Wang, A.Z. Application of Nanotechnology to Cancer Radiotherapy. Cancer Nanotechnol. 2016, 7, 11. [Google Scholar] [CrossRef]
- Rosa, S.; Connolly, C.; Schettino, G.; Butterworth, K.T.; Prise, K.M. Biological Mechanisms of Gold Nanoparticle Radiosensitization. Cancer Nanotechnol. 2017, 8, 2. [Google Scholar] [CrossRef]
- Borran, A.A.; Aghanejad, A.; Farajollahi, A.; Barar, J.; Omidi, Y. Gold Nanoparticles for Radiosensitizing and Imaging of Cancer Cells. Radiat. Phys. Chem. 2018, 152, 137–144. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, J.; Fu, S.; Wu, J. Gold Nanoparticles as Radiosensitizers in Cancer Radiotherapy. Int. J. Nanomed. 2020, 15, 9407–9430. [Google Scholar] [CrossRef]
- Penninckx, S.; Heuskin, A.-C.; Michiels, C.; Lucas, S. Gold Nanoparticles as a Potent Radiosensitizer: A Transdisciplinary Approach from Physics to Patient. Cancers 2020, 12, 2021. [Google Scholar] [CrossRef]
- Herold, M.; Herold, I.J.D.; Stobbe, C. Microspheres: A Selective Technique for Producing Biologically Effective Dose Enhancement. Int. J. Radiat. Biol. 2000, 76, 1357–1364. [Google Scholar] [CrossRef]
- Regulla, D.; Schmid, E.; Friedland, W.; Panzer, W.; Heinzmann, U.; Harder, D. Enhanced Values of the RBE and H Ratio for Cytogenetic Effects Induced by Secondary Electrons from an X-Irradiated Gold Gurface. Radiat. Res. 2002, 158, 505–515. [Google Scholar] [CrossRef]
- Butterworth, K.T.; McMahon, S.J.; Currell, F.J.; Prise, K.M. Physical Basis and Biological Mechanisms of Gold Nanoparticle Radiosensitization. Nanoscale 2012, 4, 4830–4838. [Google Scholar] [CrossRef]
- Schuemann, J.; Bagley, A.F.; Berbeco, R.; Bromma, K.; Butterworth, K.T.; Byrne, H.L.; Chithrani, B.D.; Cho, S.H.; Cook, J.R.; Favaudon, V.; et al. Roadmap for Metal Nanoparticles in Radiation Therapy: Current Status, Translational Challenges, and Future Directions. Phys. Med. Biol. 2020, 65, 21RM02. [Google Scholar] [CrossRef]
- Enferadi, M.; Fu, S.-Y.; Hong, J.-H.; Tung, C.-J.; Chao, T.-C.; Wey, S.-P.; Chiu, C.-H.; Wang, C.-C.; Sadeghi, M. Radiosensitization of Ultrasmall GNP–PEG–CRGDfK in ALTS1C1 Exposed to Therapeutic Protons and Kilovoltage and Megavoltage Photons. Int. J. Radiat. Biol. 2018, 94, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, D.B.; Jelveh, S.; Jalali, F.; Van Prooijen, M.; Allen, C.; Bristow, R.G.; Hill, R.P.; Jaffray, D.A. Gold Nanoparticles as Radiation Sensitizers in Cancer Therapy. Radiat. Res. 2010, 173, 719–728. [Google Scholar] [CrossRef]
- McMahon, S.J.; Hyland, W.B.; Muir, M.F.; Coulter, J.A.; Jain, S.; Butterworth, K.T.; Schettino, G.; Dickson, G.R.; Hounsell, A.R.; O’Sullivan, J.M.; et al. Biological Consequences of Nanoscale Energy Deposition near Irradiated Heavy Atom Nanoparticles. Sci. Rep. 2011, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Penninckx, S.; Heuskin, A.-C.; Michiels, C.; Lucas, S. The Role of Thioredoxin Reductase in Gold Nanoparticle Radiosensitization Effects. Nanomedicine 2018, 13, 2917–2937. [Google Scholar] [CrossRef]
- Taggart, L.E.; McMahon, S.J.; Butterworth, K.T.; Currell, F.J.; Schettino, G.; Prise, K.M. Protein Disulphide Isomerase as a Target for Nanoparticle-Mediated Sensitisation of Cancer Cells to Radiation. Nanotechnology 2016, 27, 215101. [Google Scholar] [CrossRef]
- Penninckx, S.; Heuskin, A.-C.; Michiels, C.; Lucas, S. Thioredoxin Reductase Activity Predicts Gold Nanoparticle Radiosensitization Effect. Nanomaterials 2019, 9, 295. [Google Scholar] [CrossRef]
- Daems, N.; Penninckx, S.; Nelissen, I.; Van Hoecke, K.; Cardinaels, T.; Baatout, S.; Michiels, C.; Lucas, S.; Aerts, A. Gold Nanoparticles Affect the Antioxidant Status in Selected Normal Human Cells. Int. J. Nanomed. 2019, 14, 4991–5015. [Google Scholar] [CrossRef]
- Williams, J.R.; Zhang, Y.; Russell, J.; Koch, C.; Little, J.B. Human Tumor Cells Segregate into Radiosensitivity Groups That Associate with ATM and TP53 Status. Acta Oncol. 2007, 46, 628–638. [Google Scholar] [CrossRef]
- Morini, J.; Babini, G.; Barbieri, S.; Baiocco, G.; Ottolenghi, A. The Interplay between Radioresistant Caco-2 Cells and the Immune System Increases Epithelial Layer Permeability and Alters Signaling Protein Spectrum. Front. Immunol. 2017, 8, 223. [Google Scholar] [CrossRef]
- Xiao, Y.; Zheng, X.; Huang, A.; Liu, T.; Zhang, T.; Ma, H. Deficiency of 53BP1 Inhibits the Radiosensitivity of Colorectal Cancer. Int. J. Oncol. 2016, 49, 1600–1608. [Google Scholar] [CrossRef][Green Version]
- Ruka, W.; Taghian, A.; Gioioso, D.; Fletcher, J.A.; Preffer, F.; Suit, H.D. Comparison Between the In Vitro Intrinsic Radiation Sensitivity of Human Soft Tissue Sarcoma and Breast Cancer Cell Lines. J. Surg. Oncol. 1996, 61, 290–292. [Google Scholar] [CrossRef]
- Hou, J.; Zhou, Z.; Chen, X.; Zhao, R.; Yang, Z.; Wei, N.; Ni, Q.; Feng, Y.; Yu, X.; Ma, J.; et al. HER2 Reduces Breast Cancer Radiosensitivity by Activating Focal Adhesion Kinase in Vitro and in Vivo. Oncotarget 2016, 7, 45186–45198. [Google Scholar] [CrossRef]
- He, H.-T.; Fokas, E.; You, A.; Engenhart-Cabillic, R.; An, H.-X. Siah1 Proteins Enhance Radiosensitivity of Human Breast Cancer Cells. BMC Cancer 2010, 10, 403. [Google Scholar] [CrossRef] [PubMed]
- Hainfeld, J.F.; Slatkin, D.N.; Smilowitz, H.M. The Use of Gold Nanoparticles to Enhance Radiotherapy in Mice. Phys. Med. Biol. 2004, 49, N309–N315. [Google Scholar] [CrossRef]
- Joiner, M.C.; van der Koegel, A.J. Basic Clinical Radiobiology, 5th ed.; CRC Press: London, UK, 2018; p. 235. [Google Scholar] [CrossRef]
- Mestres, M.; Caballín, M.R.; Barrios, L.; Ribas, M.; Barquinero, J.F. RBE of X-rays of Different Energies: A Cytogenetic Evaluation by FISH. Radiat. Res. 2008, 170, 93–100. [Google Scholar] [CrossRef]
- Schmid, E.; Regulla, D.; Kramer, H.-M.; Harder, D. The Effect of 29 KV X-rays on the Dose Response of Chromosome Aberrations in Human Lymphocytes. Radiat. Res. 2002, 158, 771–777. [Google Scholar] [CrossRef]
- Gomolka, M.; Rössler, U.; Hornhardt, S.; Walsh, L.; Panzer, W.; Schmid, E. Measurement of the Initial Levels of DNA Damage in Human Lymphocytes Induced by 29 KV X-rays (Mammography X-rays) Relative to 220 KV X Rays and γ Rays. Radiat. Res. 2005, 163, 510–519. [Google Scholar] [CrossRef]
- Frankenberg, D.; Kelnhofer, K.; Bär, K.; Frankenberg-Schwager, M. Enhanced Neoplastic Transformation by Mammography X Rays Relative to 200 KVp X-rays: Indication for a Strong Dependence on Photon Energy of the RBE M for Various End Points. Radiat. Res. 2002, 157, 99–105. [Google Scholar] [CrossRef]
- Göggelmann, W.; Jacobsen, C.; Panzer, W.; Walsh, L.; Roos, H.; Schmid, E. Re-Evaluation of the RBE of 29 kV X-rays (Mammography X-rays) Relative to 220 kV X-rays Using Neoplastic Transformation of Human CGL1-Hybrid Cells. Radiat. Environ. Biophys. 2003, 42, 175–182. [Google Scholar] [CrossRef]
- Heyes, G.J.; Mill, A.J. The Neoplastic Transformation Potential of Mammography X-rays and Atomic Bomb Spectrum Radiation. Radiat. Res. 2004, 162, 120–127. [Google Scholar] [CrossRef][Green Version]
- Panteleeva, A.; Slonina, D.; Brankovic, K.; Spekl, K.; Pawelke, J.; Hoinkis, C.; Dorr, W. Clonogenic Survival of Human Keratinocytes and Rodent Fibroblasts after Irradiation with 25 kV X-rays. Radiat. Environ. Biophys. 2003, 42, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Frankenberg-Schwager, M.; Garg, I.; Frankenberg, D.; Greve, B.; Severin, E.; Uthe, D.; Göhde, W. Mutagenicity of Low-Filtered 30 KVp X-rays, Mammography X-rays and Conventional X-rays in Cultured Mammalian Cells. Int. J. Radiat. Biol. 2002, 78, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Verhaegen, F.; Vral, A. Sensitivity of Micronucleus Induction in Human Lymphocytes to Low-LET Radiation Qualities: RBE and Correlation of RBE and LET. Radiat. Res. 1994, 139, 208. [Google Scholar] [CrossRef]
- Slonina, D.; Spekl, K.; Panteleeva, A.; Brankovic, K.; Hoinkis, C.; Dörr, W. Induction of Micronuclei in Human Fibroblasts and Keratinocytes by 25 KV X-Rays. Radiat. Environ. Biophys. 2003, 42, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Lehnert, A.; Lessmann, E.; Pawelke, J.; Dörr, W. RBE of 25 KV X-rays for the Survival and Induction of Micronuclei in the Human Mammary Epithelial Cell Line MCF-12A. Radiat. Environ. Biophys. 2006, 45, 253–260. [Google Scholar] [CrossRef]
- Virsik, R.P.; Harder, D.; Hansmann, I. The RBE of 30 KV X-rays for the Induction of Dicentric Chromosomes in Human Lymphocytes. Radiat. Environ. Biophys. 1977, 14, 109–121. [Google Scholar] [CrossRef]
- Stewart, F.A.; Dörr, W. Milestones in Normal Tissue Radiation Biology over the Past 50 Years: From Clonogenic Cell Survival to Cytokine Networks and Back to Stem Cell Recovery. Int. J. Radiat. Biol. 2009, 85, 574–586. [Google Scholar] [CrossRef]
- Huber, R.; Braselmann, H.; Geinitz, H.; Jaehnert, I.; Baumgartner, A.; Thamm, R.; Figel, M.; Molls, M.; Zitzelsberger, H. Chromosomal Radiosensitivity and Acute Radiation Side Effects after Radiotherapy in Tumour Patients—A Follow-up Study. Radiat. Oncol. 2011, 6, 32. [Google Scholar] [CrossRef]
- Bailey, S.M.; Bedford, J.S. Studies on Chromosome Aberration Induction: What Can They Tell Us about DNA Repair? DNA Repair 2006, 5, 1171–1181. [Google Scholar] [CrossRef]
- Beaton, L.A.; Marro, L.; Samiee, S.; Malone, S.; Grimes, S.; Malone, K.; Wilkins, R.C. Investigating Chromosome Damage Using Fluorescent in Situ Hybridization to Identify Biomarkers of Radiosensitivity in Prostate Cancer Patients. Int. J. Radiat. Biol. 2013, 89, 1087–1093. [Google Scholar] [CrossRef]
- Rothkamm, K.; Löbrich, M. Evidence for a Lack of DNA Double-Strand Break Repair in Human Cells Exposed to Very Low X-ray Doses. Proc. Natl. Acad. Sci. USA 2003, 100, 5057–5062. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Boon, C.; Redon, C.; Bonner, W.M. Megabase Chromatin Domains Involved in DNA Double-Strand Breaks in Vivo. J. Cell Biol. 1999, 146, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Ivashkevich, A.; Redon, C.E.; Nakamura, A.J.; Martin, R.F.; Martin, O.A. Use of the γ-H2AX Assay to Monitor DNA Damage and Repair in Translational Cancer Research. Cancer Lett. 2012, 327, 123–133. [Google Scholar] [CrossRef]
- Willers, H.; Gheorghiu, L.; Liu, Q.; Efstathiou, J.A.; Wirth, L.J.; Krause, M.; von Neubeck, C. DNA Damage Response Assessments in Human Tumor Samples Provide Functional Biomarkers of Radiosensitivity. Semin. Radiat. Oncol. 2015, 25, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Riss, T.; Moravec, R.A.; Niles, A.; Duellman, S.; Benink, H.; Worxella, T. Cell Viability Assays. In Assay Guidance; Markossian, S., Grossman, A., Brimacombe, K., Eds.; NIH: Bethesda, MD, USA, 2004. Available online: https://www.ncbi.nlm.nih.gov/books/NBK144065/ (accessed on 16 March 2022).
- Coulter, J. Cell Type-Dependent Uptake, Localization, and Cytotoxicity of 1.9 Nm Gold Nanoparticles. Int. J. Nanomed. 2012, 2673–2685. [Google Scholar] [CrossRef]
- Pan, Y.; Leifert, A.; Ruau, D.; Neuss, S.; Bornemann, J.; Schmid, G.; Brandau, W.; Simon, U.; Jahnen-Dechent, W. Gold Nanoparticles of Diameter 1.4 Nm Trigger Necrosis by Oxidative Stress and Mitochondrial Damage. Small 2009, 5, 2067–2076. [Google Scholar] [CrossRef]
- Patiño, T.; Nogués, C.; Ibáñez, E.; Barrios, L. Enhancing Microparticle Internalization by Nonphagocytic Cells through the Use of Noncovalently Conjugated Polyethyleneimine. Int. J. Nanomed. 2012, 5671–5682. [Google Scholar] [CrossRef]
- Wang, F.; Yu, L.; Monopoli, M.P.; Sandin, P.; Mahon, E.; Salvati, A.; Dawson, K.A. The Biomolecular Corona Is Retained during Nanoparticle Uptake and Protects the Cells from the Damage Induced by Cationic Nanoparticles until Degraded in the Lysosomes. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 1159–1168. [Google Scholar] [CrossRef]
- Patiño, T.; Soriano, J.; Barrios, L.; Ibáñez, E.; Nogués, C. Surface Modification of Microparticles Causes Differential Uptake Responses in Normal and Tumoral Human Breast Epithelial Cells. Sci. Rep. 2015, 5, 11371. [Google Scholar] [CrossRef]
- Yan, Y.; Gause, K.T.; Kamphuis, M.M.J.; Ang, C.-S.; O’Brien-Simpson, N.M.; Lenzo, J.C.; Reynolds, E.C.; Nice, E.C.; Caruso, F. Differential Roles of the Protein Corona in the Cellular Uptake of Nanoporous Polymer Particles by Monocyte and Macrophage Cell Lines. ACS Nano 2013, 7, 10960–10970. [Google Scholar] [CrossRef]
- Chithrani, B.D.; Ghazani, A.A.; Chan, W.C.W. Determining the Size and Shape Dependence of Gold Nanoparticle Uptake into Mammalian Cells. Nano Lett. 2006, 6, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Patiño, T.; Soriano, J.; Amirthalingam, E.; Durán, S.; González-Campo, A.; Duch, M.; Ibáñez, E.; Barrios, L.; Plaza, J.A.; Pérez-García, L.; et al. Polysilicon-Chromium-Gold Intracellular Chips for Multi-Functional Biomedical Applications. Nanoscale 2016, 8, 8773–8783. [Google Scholar] [CrossRef] [PubMed]
- Mora-Espí, I.; Barrios, L.; Ibáñez, E.; Soriano, J.; Nogués, C. Membrane Reorganization after Photochemical Internalization to Release Transferrin-Biofunctionalized Polystyrene Microparticles. Sci. Rep. 2018, 8, 17617. [Google Scholar] [CrossRef]
- Leonard, F.; Collnot, E.-M.; Lehr, C.-M. A Three-Dimensional Coculture of Enterocytes, Monocytes and Dendritic Cells to Model Inflamed Intestinal Mucosa in Vitro. Mol. Pharm. 2010, 7, 2103–2119. [Google Scholar] [CrossRef] [PubMed]
- Janer, G.; Mas del Molino, E.; Fernández-Rosas, E.; Fernández, A.; Vázquez-Campos, S. Cell Uptake and Oral Absorption of Titanium Dioxide Nanoparticles. Toxicol. Lett. 2014, 228, 103–110. [Google Scholar] [CrossRef]
- Maltez-da Costa, M.; de la Escosura-Muñiz, A.; Nogués, C.; Barrios, L.; Ibáñez, E.; Merkoçi, A. Simple Monitoring of Cancer Cells Using Nanoparticles. Nano Lett. 2012, 12, 4164–4171. [Google Scholar] [CrossRef]
- Maltez-da Costa, M.; de la Escosura-Muñiz, A.; Nogués, C.; Barrios, L.; Ibáñez, E.; Merkoçi, A. Detection of Circulating Cancer Cells Using Electrocatalytic Gold Nanoparticles. Small 2012, 8, 3605–3612. [Google Scholar] [CrossRef] [PubMed]
- Kucki, M.; Diener, L.; Bohmer, N.; Hirsch, C.; Krug, H.F.; Palermo, V.; Wick, P. Uptake of Label-Free Graphene Oxide by Caco-2 Cells Is Dependent on the Cell Differentiation Status. J. Nanobiotechnol. 2017, 15, 46. [Google Scholar] [CrossRef]
- Song, Z.-M.; Chen, N.; Liu, J.-H.; Tang, H.; Deng, X.; Xi, W.-S.; Han, K.; Cao, A.; Liu, Y.; Wang, H. Biological Effect of Food Additive Titanium Dioxide Nanoparticles on Intestine: An in Vitro Study: Bioeffects of Food Additive TiO2 Nanoparticles to Intestinal Cells. J. Appl. Toxicol. 2015, 35, 1169–1178. [Google Scholar] [CrossRef]
- Rothkamm, K.; Horn, S. Gamma-H2AX as Protein Biomarker for Radiation Exposure. Ann. Ist. Super. Sanita 2009, 45, 265–271. [Google Scholar]
- Perquin, M.; Oster, T.; Maul, A.; Froment, N.; Untereiner, M.; Bagrel, D. The Glutathione-Related Detoxification Pathway in the Human Breast: A Highly Coordinated System Disrupted in the Tumour Tissues. Cancer Lett. 2000, 158, 7–16. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Hayama, K.; Sasagawa, I.; Okada, Y.; Kawase, T.; Tsubokawa, N.; Tsuchimochi, M. HER2-Targeted Multifunctional Silica Nanoparticles Specifically Enhance the Radiosensitivity of HER2-Overexpressing Breast Cancer Cells. Int. J. Mol. Sci. 2018, 19, 8. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Yook, S.; Lu, Y.; Bergstrom, D.; Winnik, M.A.; Pignol, J.-P.; Reilly, R.M. Local Radiation Treatment of HER2-Positive Breast Cancer Using Trastuzumab-Modified Gold Nanoparticles Labeled with 177Lu. Pharm. Res. 2017, 34, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Ionita, P.; Conte, M.; Gilbert, B.C.; Chechik, V. Gold Nanoparticle-Initiated Free Radical Oxidations and Halogen Abstractions. Org. Biomol. Chem. 2007, 5, 3504–3509. [Google Scholar] [CrossRef] [PubMed]



| AntiEpCamGNP vs. | TfGNP vs. | |||
|---|---|---|---|---|
| Postirradiation Time | w/o GNP | NF-GNP | w/o GNP | NF-GNP |
| Foci SER | Foci SER | |||
| 30 min | 1.10 | 1.30 | - | - |
| 2 h | 1.46 | 1.55 | - | - |
| 4 h | 1.20 | 1.18 | 2.05 | 1.07 |
| 24 h | 1.87 | 1.18 | 4.98 | 22.57 |
| Viability SER | Viability SER | |||
| 1.11 | 1.07 | 1.12 | 1.02 | |
| Irradiation Conditions | K (h−1) | Foci Half-Life (h) | |
|---|---|---|---|
| CaCo2 | w/o GNPs | 0.35 | 2.00 |
| NFGNP | 0.32 | 2.17 | |
| AntiEpCamGNP | 0.28 | 2.44 | |
| SKBR3 | w/o GNPs | - | - |
| NFGNP | - | - | |
| TfGNP | 0.30 | 2.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Amor, M.Á.; Barrios, L.; Armengol, G.; Barquinero, J.F. Differential Radiosensitizing Effect of 50 nm Gold Nanoparticles in Two Cancer Cell Lines. Biology 2022, 11, 1193. https://doi.org/10.3390/biology11081193
Pérez-Amor MÁ, Barrios L, Armengol G, Barquinero JF. Differential Radiosensitizing Effect of 50 nm Gold Nanoparticles in Two Cancer Cell Lines. Biology. 2022; 11(8):1193. https://doi.org/10.3390/biology11081193
Chicago/Turabian StylePérez-Amor, Miguel Ángel, Leonardo Barrios, Gemma Armengol, and Joan Francesc Barquinero. 2022. "Differential Radiosensitizing Effect of 50 nm Gold Nanoparticles in Two Cancer Cell Lines" Biology 11, no. 8: 1193. https://doi.org/10.3390/biology11081193
APA StylePérez-Amor, M. Á., Barrios, L., Armengol, G., & Barquinero, J. F. (2022). Differential Radiosensitizing Effect of 50 nm Gold Nanoparticles in Two Cancer Cell Lines. Biology, 11(8), 1193. https://doi.org/10.3390/biology11081193







