Okra Growth, Yield and Rhizosphere Microbiome Responses to the Encapsulated Bioinoculant Application under Reduced Fertilization Regime
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Bioinoculant Preparation and Soil Application
2.3. Analyses of Soil and Plant Samples
2.4. DNA Extraction, Library Preparation, and 16S rRNA Gene Amplicon Sequencing
2.5. Bioinformatic Analyses of the Rhizosphere Microbiome
2.6. Statistical Analysis
3. Results
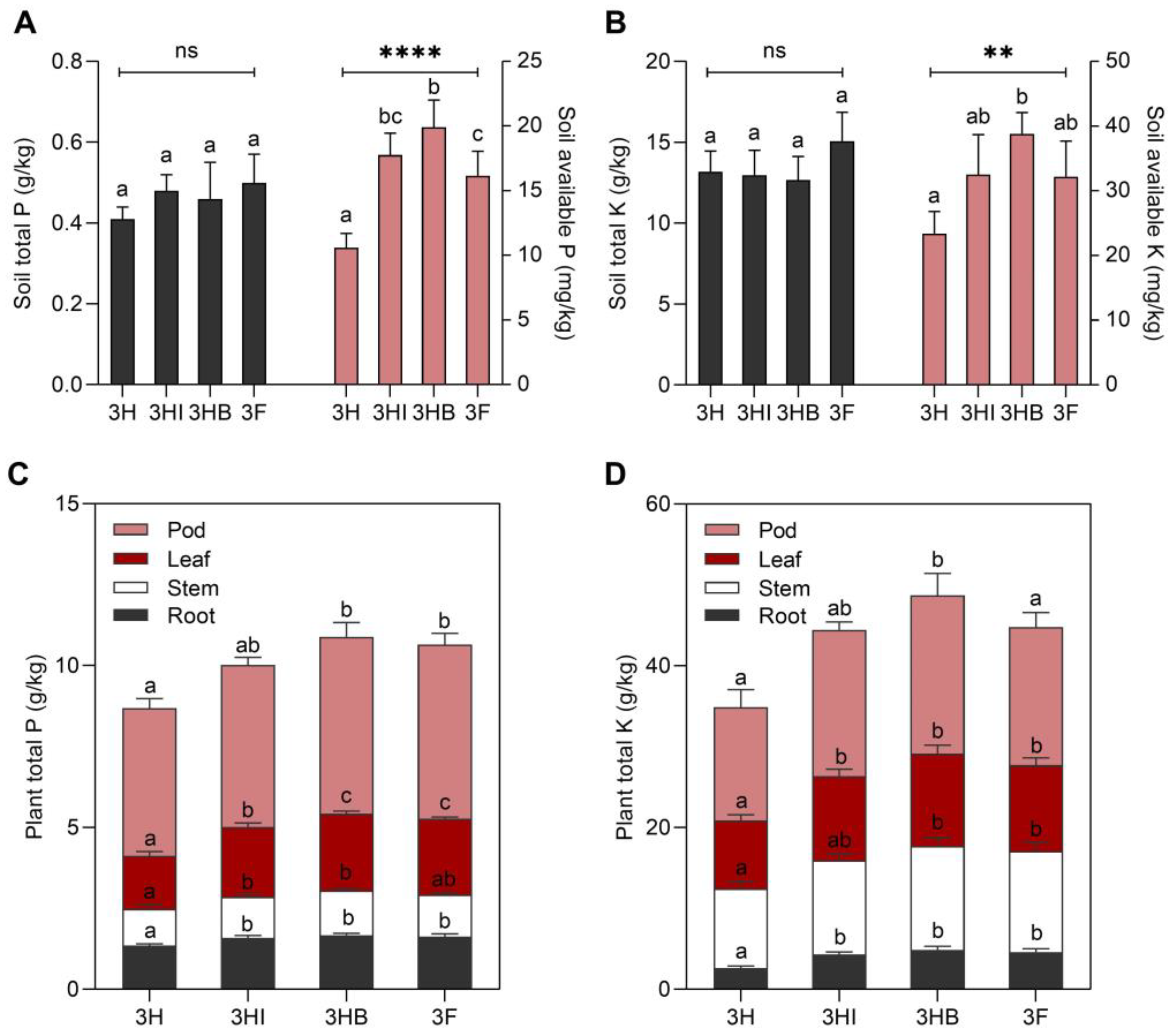
3.1. Effect of Free-Cell and Encapsulated E. hormaechei 40a on P and K Status of Soil and Plant
3.2. Effect of Free-Cell and Encapsulated E. hormaechei 40a on the Populations of Culturable Bacteria in Soil
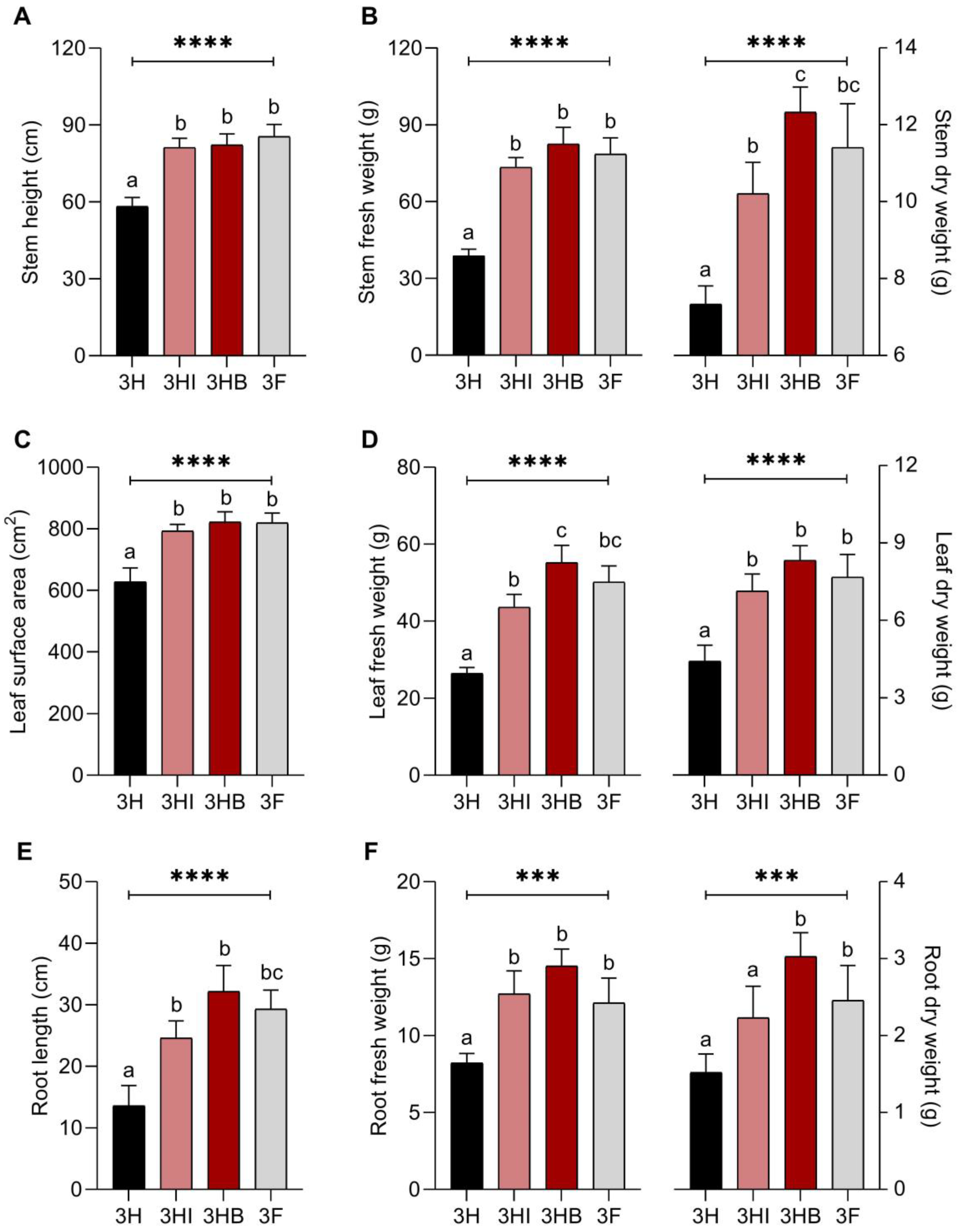
3.3. Effect of Free-Cell and Encapsulated E. hormaechei 40a on Plant Growth
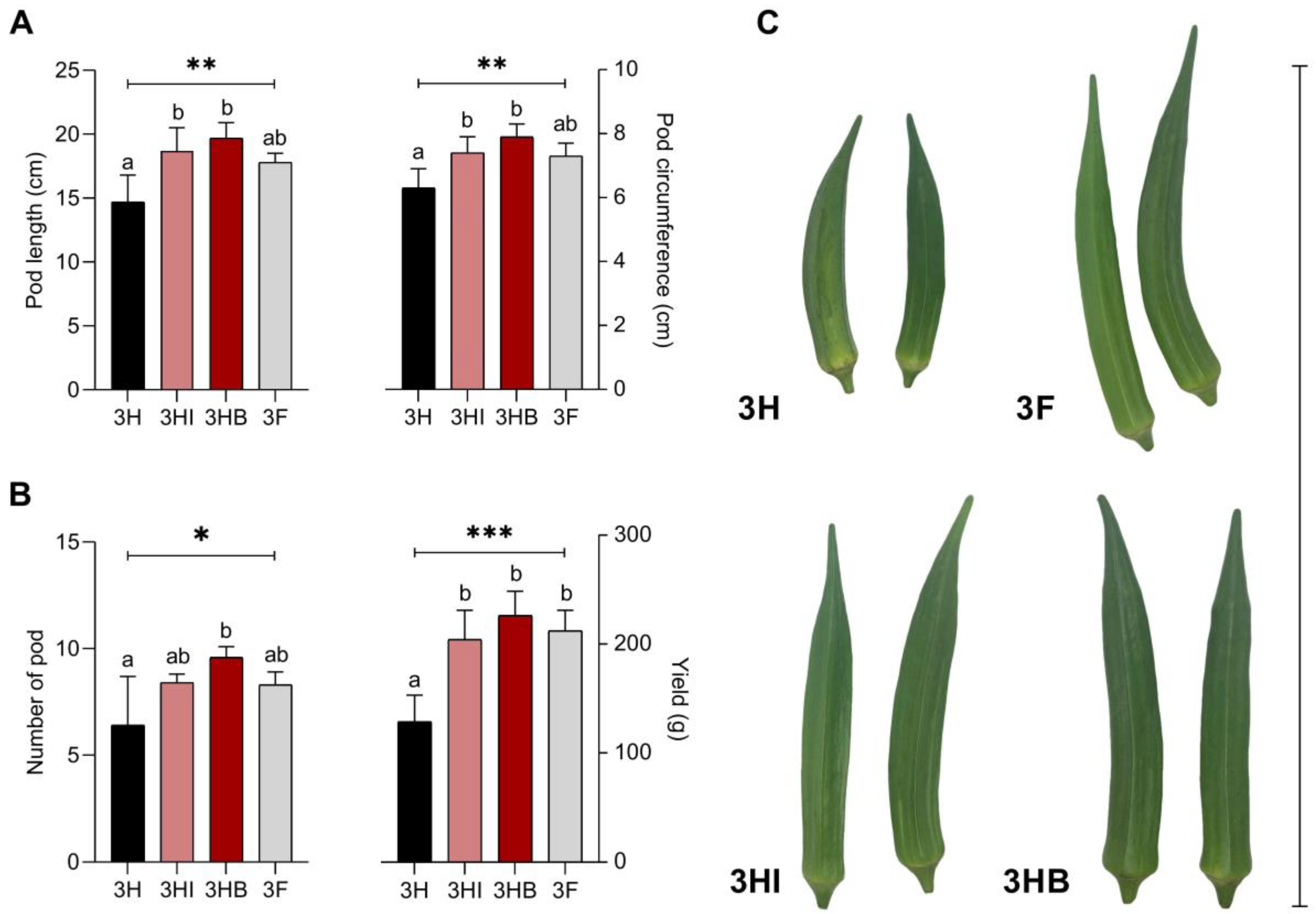
3.4. Effect of Free-Cell and Encapsulated E. hormaechei 40a on Plant Yield
3.5. Correlation between Soil P and K Status with Okra Physiology and Yield
3.6. Alpha Diversity Indices of Microbial Community
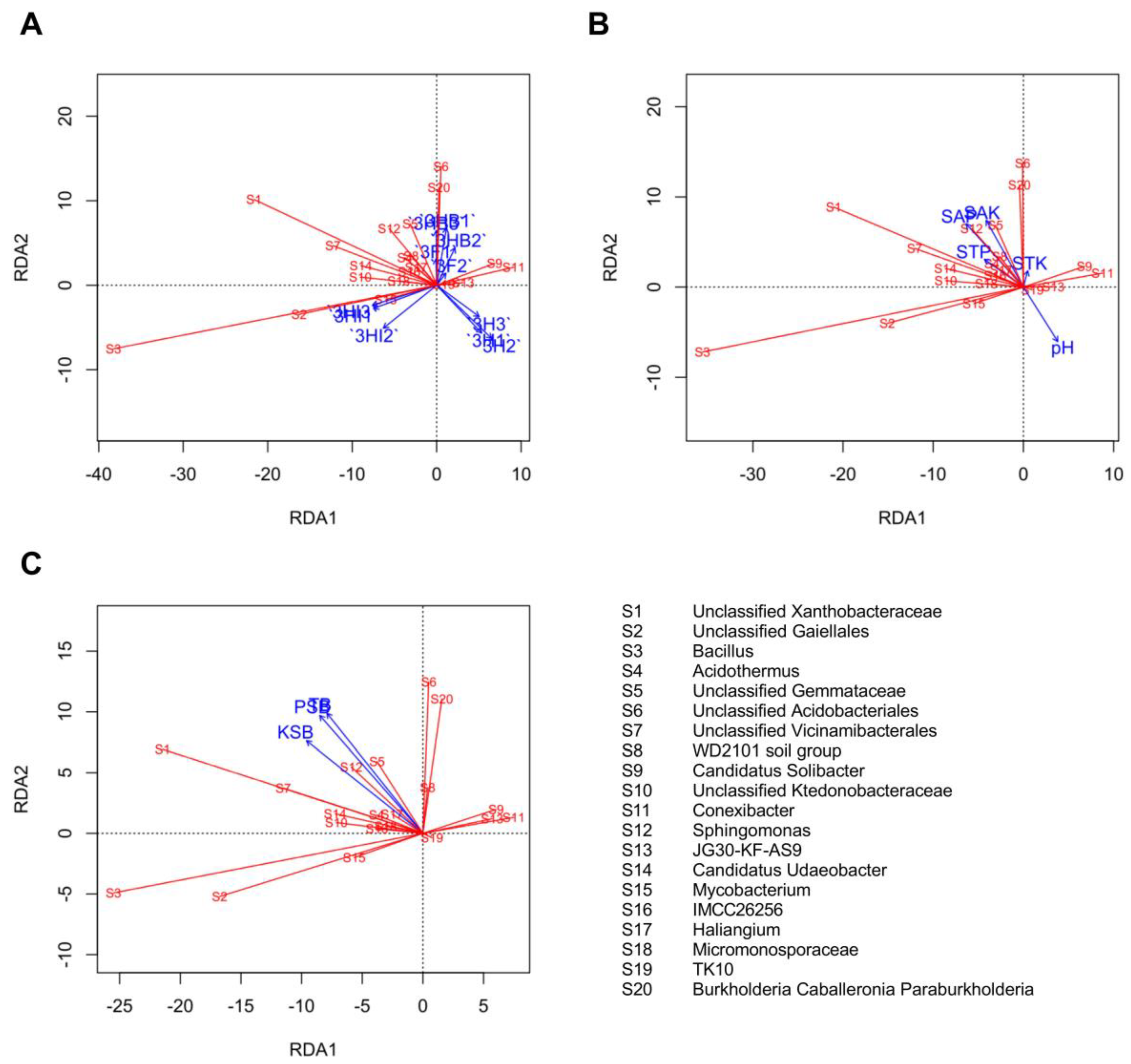
3.7. Beta Diversity Indices and Alteration in Soil Bacterial Composition Structure
3.8. Relationship between Soil Treatments, P and K Status, Culturable Bacteria, and Dominant Genera
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meena, V.K.; Dubey, A.K.; Jain, V.K.; Tiwari, A.; Negi, P. Effect of plant growth promoters on flowering and fruiting attributes of okra [Abelmoschus esculentus (L.) Moench]. Crop Res. 2017, 52, 37–40. [Google Scholar]
- Rahman, M.A.; Akter, F. Effect of NPK fertilizers on growth, yield and yield attributes of okra (Abelmoschus esculentus (L.) Moench). Bangladesh J. Bot. 2012, 41, 131–134. [Google Scholar] [CrossRef]
- FAO. Production-Crops and Livestock Products; Food and Agriculture Organization of the United Nations: 2022. Available online: https://www.fao.org/faostat/en/#compare (accessed on 20 July 2022).
- Khandaker, M.M.; Nor, M.F.; Dalorima, T.; Sajili, M.H.; Mat, N. Effect of different rates of inorganic fertilizer on physiology, growth and yield of okra (Abelmoschus esculentus) cultivated on BRIS soil of Terengganu, Malaysia. Aust. J. Crop Sci. 2017, 11, 880–887. [Google Scholar] [CrossRef]
- Ferdous, Z.; Anwar, M.; Uddin, N.; Ullah, H.; Hossain, A. Yield performance of okra (Abelmoschus esculentus) through integrated nutrient management. Int. J. Biosci. 2017, 10, 294–301. [Google Scholar]
- Nagegowda, N.S.; Hebbar, S.S.; Shilpashree, V.M. Performance of mulching and fertigation on nutrient uptake and nutrient use efficiency in okra (Abelmoschus esculentus L. Moench) for seed production. Curr. Agric. Res. J. 2020, 8, 25–29. [Google Scholar] [CrossRef]
- Kaminsky, L.M.; Thompson, G.L.; Trexler, R.V.; Bell, T.H.; Kao-Kniffin, J. Medicago sativa has reduced biomass and nodulation when grown with soil microbiomes conditioned to high phosphorus inputs. Phytobiomes J. 2020, 2, 237–248. [Google Scholar] [CrossRef]
- Pereira, D.G.C.; Santana, I.A.; Megda, M.M.; Megda, M.X.V. Potassium chloride: Impacts on soil microbial activity and nitrogen mineralization. Cienc. Rural 2019, 49, 1–9. [Google Scholar] [CrossRef]
- Elhanafi, L.; Houhou, M.; Rais, C.; Mansouri, I.; Elghadraoui, L.; Greche, H. Impact of excessive nitrogen fertilization on the biochemical quality, phenolic compounds, and antioxidant power of Sesamum indicum L. seeds. J. Food Qual. 2019, 2019, 9428092. [Google Scholar] [CrossRef]
- Alori, E.T.; Babalola, O.O. Microbial inoculants for improving crop quality and human health in Africa. Front. Microbiol. 2018, 9, 2213. [Google Scholar] [CrossRef]
- Etesami, H.; Emami, S.; Alikhani, H.A. Potassium solubilizing bacteria (KSB): Mechanisms, promotion of plant growth, and future prospects—A review. J. Soil Sci. Plant Nutr. 2017, 17, 897–911. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; McInroy, J.; Kloepper, J. The interactions of rhizodeposits with plant growth-promoting rhizobacteria in the rhizosphere: A review. Agriculture 2019, 9, 142. [Google Scholar] [CrossRef]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef]
- Xiong, Q.; Hu, J.; Wei, H.; Zhang, H.; Zhu, J. Relationship between plant roots, rhizosphere microorganisms, and nitrogen and its special focus on rice. Agriculture 2021, 11, 234. [Google Scholar] [CrossRef]
- Khan, N.; Ali, S.; Shahid, M.A.; Mustafa, A.; Sayyed, R.Z.; Curá, J.A. Insights into the interactions among roots, rhizosphere, and rhizobacteria for improving plant growth and tolerance to abiotic stresses: A review. Cells 2021, 10, 1551. [Google Scholar] [CrossRef] [PubMed]
- Lazcano, C.; Boyd, E.; Holmes, G.; Hewavitharana, S.; Pasulka, A.; Ivors, K. The rhizosphere microbiome plays a role in the resistance to soil-borne pathogens and nutrient uptake of strawberry cultivars under field conditions. Sci. Rep. 2021, 11, 3188. [Google Scholar] [CrossRef]
- de la Fuente Cantó, C.; Simonin, M.; King, E.; Moulin, L.; Bennett, M.J.; Castrillo, G.; Laplaze, L. An extended root phenotype: The rhizosphere, its formation and impacts on plant fitness. Plant J. 2020, 103, 951–964. [Google Scholar] [CrossRef]
- Niu, H.; Pang, Z.; Fallah, N.; Zhou, Y.; Zhang, C.; Hu, C.; Lin, W.; Yuan, Z. Diversity of microbial communities and soil nutrients in sugarcane rhizosphere soil under water soluble fertilizer. PLoS ONE 2021, 16, e0245626. [Google Scholar] [CrossRef]
- Shi, X.; Zhao, X.; Ren, J.; Dong, J.; Zhang, H.; Dong, Q.; Jiang, C.; Zhong, C.; Zhou, Y.; Yu, H. Influence of peanut, sorghum, and soil salinity on microbial community composition in interspecific interaction zone. Front. Microbiol. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Wang, J.; Li, R.; Zhang, H.; Wei, G.; Li, Z. Beneficial bacteria activate nutrients and promote wheat growth under conditions of reduced fertilizer application. BMC Microbiol. 2020, 20, 38. [Google Scholar] [CrossRef]
- Suleman, M.; Yasmin, S.; Rasul, M.; Yahya, M.; Atta, B.M.; Mirza, M.S. Phosphate solubilizing bacteria with glucose dehydrogenase gene for phosphorus uptake and beneficial effects on wheat. PLoS ONE 2018, 13, e0204408. [Google Scholar] [CrossRef] [PubMed]
- Roslan MA, M.; Zulkifli, N.N.; Sobri, Z.M.; Zuan AT, K.; Cheak, S.C.; Abdul Rahman, N.A. Seed biopriming with P- and K-solubilizing Enterobacter hormaechei sp. improves the early vegetative growth and the P and K uptake of okra (Abelmoschus esculentus) seedling. PLoS ONE 2020, 15, e0232860. [Google Scholar] [CrossRef] [PubMed]
- Roslan, M.A.M.; Sobri, Z.M.; Zuan, A.T.K.; Cheak, S.C.; Rahman, N.A.A. Bioprospecting microwave-alkaline hydrolysate cocktail of defatted soybean meal and jackfruit peel biomass as carrier additive of molasses-alginate-bead biofertilizer. Sci. Rep. 2022, 12, 254. [Google Scholar] [CrossRef]
- Schoebitz, M.; López, M.D.; Roldán, A. Bioencapsulation of microbial inoculants for better soil-plant fertilization. A review. Agron. Sustain. Dev. 2013, 33, 751–765. [Google Scholar] [CrossRef]
- Barrera, M.C.; Jakobs-Schoenwandt, D.; Gómez, M.I.; Serrato, J.; Ruppel, S.; Patel, A.V. Formulating bacterial endophyte: Pre-conditioning of cells and the encapsulation in amidated pectin beads. Biotechnol. Rep. 2020, 26, e00463. [Google Scholar] [CrossRef]
- John, R.P.; Tyagi, R.D.; Brar, S.K.; Surampalli, R.Y.; Prévost, D. Bio-encapsulation of microbial cells for targeted agricultural delivery. Crit. Rev. Biotechnol. 2011, 31, 211–226. [Google Scholar] [CrossRef]
- Panichikkal, J.; Prathap, G.; Nair, R.A.; Krishnankutty, R.E. Evaluation of plant probiotic performance of Pseudomonas sp. encapsulated in alginate supplemented with salicylic acid and zinc oxide nanoparticles. Int. J. Biol. Macromol. 2020, 166, 138–143. [Google Scholar] [CrossRef]
- Bremner, J.M. Methods of Soil Analysis; Soil Science Society of America, American Society of Agronomy: Madison, WI, USA, 1996. [Google Scholar] [CrossRef]
- Olsen, S.R.; Cole, V.C.; Watanabe, F.S.; Dean, A.L. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate; Department of Agriculture: Washington, DC, USA, 1954.
- Thomas, G.W. Exchangeable Cations. In Handbook of Soil Analysis; American Society of Agronomy, Inc.; Soil Science Society of America, Inc.: Madison, WI, USA, 2015; Volume 9, pp. 159–165. [Google Scholar]
- Uka, U.; Chukwuka, K.; Iwuagwu, M. Relative effect of organic and inorganic fertilizers on the growth of okra [Abelmoschus esculentus (L.) Moench]. J. Agric. Sci. Belgrade 2013, 58, 159–166. [Google Scholar] [CrossRef][Green Version]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef]
- Roslan, M.A.M.; Amirudin, N.A.; Abidin, Z.A.Z.; Omar, S.M. Isolation of bacteria from the acidic peat swamp forest soil and their lignin degradation potential. J. Teknol. 2015, 77, 77–81. [Google Scholar]
- Roslan, M.A.M.; Zainal Abidin, Z.A.; Omar, S.M. Screening of ligninase-producing bacteria from south east Pahang peat swamp forest soil. Malays. J. Microbiol. 2016, 12, 433–437. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Roslan, M.A.M.; Naim Mohamad, M.A.; Mohd Omar, S. High-quality DNA from peat soil for metagenomic studies: A minireview on DNA extraction methods. Sci. Herit. J. 2017, 1, 1–6. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Roslan, M.A.M.; Sohedein, I.; Ling, P.S.; Sobri, Z.M.; Zuan, A.T.K.; Cheak, S.C.; Rahman, N.A.A. Sustainable agronomic valorization of unsulfured molasses and defatted soybean meal as an optimized formulation of bio-organic fertilizer enriched with high cell density P-solubilizing bacteria. Agronomy 2021, 11, 996. [Google Scholar] [CrossRef]
- Pérez-Rodriguez, M.M.; Piccoli, P.; Anzuay, M.S.; Baraldi, R.; Neri, L.; Taurian, T.; Ureche, M.A.L.; Segura, D.M.; Cohen, A.C. Native bacteria isolated from roots and rhizosphere of Solanum lycopersicum L. increase tomato seedling growth under a reduced fertilization regime. Sci. Rep. 2020, 10, 15642. [Google Scholar] [CrossRef]
- FAMA. Standard and Grade Specifications of Okra Pods; Ministry of Agriculture and Food Industries: Kuala Lumpur, Malaysia, 2001; pp. 1–4. Available online: https://www.fama.gov.my/documents/20143/548645/FS024-2001+KACANG+BENDI.pdf/6a7cd7f7-7b73-d780-3c11-0846ef123ef8?version=1.0 (accessed on 20 July 2022).
- Lu, K.; Jin, Q.; Lin, Y.; Lu, W.; Li, S.; Zhou, C.; Jin, J.; Jiang, Q.; Ling, L.; Xiao, M. Cell-free fermentation broth of Bacillus velezensis strain S3-1 improves Pak choi nutritional quality and changes the bacterial community structure of the rhizosphere soil. Front. Microbiol. 2020, 11, 2043. [Google Scholar] [CrossRef] [PubMed]
- Msimbira, L.A.; Smith, D.L. The Roles of Plant Growth Promoting Microbes in Enhancing Plant Tolerance to Acidity and Alkalinity Stresses. Front. Sustain. Food Syst. 2020, 4, 106. [Google Scholar] [CrossRef]
- Shen, H.; He, X.; Liu, Y.; Chen, Y.; Tang, J.; Guo, T. A complex inoculant of N2-fixing, P- and K-solubilizing bacteria from a purple soil improves the growth of kiwifruit (Actinidia chinensis) plantlets. Front. Microbiol. 2016, 7, 841. [Google Scholar] [CrossRef]
- Garcha, S.; Kansal, R.; Gosal, S.K. Molasses growth medium for production of Rhizobium sp. Based biofertilizer. Indian J. Biochem. Biophys. 2019, 56, 378–383. [Google Scholar]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S. Encapsulation of Bacillus salmalaya 139SI using double coating biopolymer technique. Lett. Appl. Microbiol. 2019, 68, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, N.; Vassileva, M.; Martos, V.; Garcia del Moral, L.F.; Kowalska, J.; Tylkowski, B.; Malusá, E. Formulation of microbial inoculants by encapsulation in natural polysaccharides: Focus on beneficial properties of carrier additives and derivatives. Front. Plant Sci. 2020, 11, 270. [Google Scholar] [CrossRef]
- He, Y.; Wu, Z.; Ye, B.C.; Wang, J.; Guan, X.; Zhang, J. Viability evaluation of alginate-encapsulated Pseudomonas putida Rs-198 under simulated salt-stress conditions and its effect on cotton growth. Eur. J. Soil Biol. 2016, 75, 135–141. [Google Scholar] [CrossRef]
- Liffourrena, A.S.; Lucchesi, G.I. Alginate-perlite encapsulated Pseudomonas putida A (ATCC 12633) cells: Preparation, characterization and potential use as plant inoculants. J. Biotechnol. 2018, 278, 28–33. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Hacquard, S. Disentangling the factors shaping microbiota composition across the plant holobiont. New Phytol. 2016, 209, 454–457. [Google Scholar] [CrossRef]
- Wang, J.; Liu, L.; Gao, X.; Hao, J.; Wang, M. Elucidating the effect of biofertilizers on bacterial diversity in maize rhizosphere soil. PLoS ONE 2021, 16, e0249834. [Google Scholar] [CrossRef]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.S.; Kumar, A.; Rai, A.N.; Singh, D.P. Cyanobacteria: A precious bio-resource in agriculture, ecosystem, and environmental sustainability. Front. Microbiol. 2016, 7, 529. [Google Scholar] [CrossRef] [PubMed]
- Köberl, M.; Dita, M.; Martinuz, A.; Staver, C.; Berg, G. Members of Gammaproteobacteria as indicator species of healthy banana plants on Fusarium wilt-infested fields in Central America. Sci. Rep. 2017, 7, 45318. [Google Scholar] [CrossRef] [PubMed]
- Effendi, Y.; Aini, N.; Pambudi, A.; Sasaerila, H.Y. Metagenomics analysis of soil microbial communities in plant agroforestry system rubber tree (Hevea brasiliensis)—Ganyong (Canna sp.). IOP Conf. Ser. Earth Environ. Sci. 2020, 468, 012045. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Kalam, S.; Das, S.N.; Basu, A.; Podile, A.R. Population densities of indigenous Acidobacteria change in the presence of plant growth promoting rhizobacteria (PGPR) in rhizosphere. J. Basic Microbiol. 2017, 57, 376–385. [Google Scholar] [CrossRef]
- Karmakar, J.; Goswami, S.; Pramanik, K.; Maiti, T.K.; Kar, R.K.; Dey, N. Growth promoting properties of Mycobacterium and Bacillus on rice plants under induced drought. Plant Sci. Today 2021, 8, 49–57. [Google Scholar] [CrossRef]
- Saxena, A.K.; Kumar, M.; Chakdar, H.; Anuroopa, N.; Bagyaraj, D.J. Bacillus species in soil as a natural resource for plant health and nutrition. J. Appl. Microbiol. 2020, 128, 1583–1594. [Google Scholar] [CrossRef]
- Okutani, F.; Hamamoto, S.; Aoki, Y.; Nakayasu, M.; Nihei, N.; Nishimura, T.; Yazaki, K.; Sugiyama, A. Rhizosphere modelling reveals spatiotemporal distribution of daidzein shaping soybean rhizosphere bacterial community. Plant Cell Environ. 2020, 43, 1036–1046. [Google Scholar] [CrossRef]




| Treatment | Total Bacteria, TB (Log CFU/g Soil) | P-Solubilizing Bacteria, PSB (Log CFU/g Soil) | K-Solubilizing Bacteria, KSB (Log CFU/g Soil) |
|---|---|---|---|
| 3H | 7.02 ± 0.52 a | 3.23 ± 0.35 a | 2.83 ± 0.66 a |
| 3HI | 8.27 ± 0.53 b | 4.23 ± 0.42 b | 4.15 ± 0.37 b |
| 3HB | 8.75 ± 0.43 b | 4.58 ± 0.33 b | 4.29 ± 0.32 b |
| 3F | 8.21 ± 0.46 b | 4.11 ± 0.21 b | 3.75 ± 0.51 ab |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roslan, M.A.M.; Sobri, Z.M.; Zuan, A.T.K.; Abdul Rahman, N.A. Okra Growth, Yield and Rhizosphere Microbiome Responses to the Encapsulated Bioinoculant Application under Reduced Fertilization Regime. Biology 2022, 11, 1107. https://doi.org/10.3390/biology11081107
Roslan MAM, Sobri ZM, Zuan ATK, Abdul Rahman NA. Okra Growth, Yield and Rhizosphere Microbiome Responses to the Encapsulated Bioinoculant Application under Reduced Fertilization Regime. Biology. 2022; 11(8):1107. https://doi.org/10.3390/biology11081107
Chicago/Turabian StyleRoslan, Muhamad Aidilfitri Mohamad, Zulfazli M. Sobri, Ali Tan Kee Zuan, and Nor Aini Abdul Rahman. 2022. "Okra Growth, Yield and Rhizosphere Microbiome Responses to the Encapsulated Bioinoculant Application under Reduced Fertilization Regime" Biology 11, no. 8: 1107. https://doi.org/10.3390/biology11081107
APA StyleRoslan, M. A. M., Sobri, Z. M., Zuan, A. T. K., & Abdul Rahman, N. A. (2022). Okra Growth, Yield and Rhizosphere Microbiome Responses to the Encapsulated Bioinoculant Application under Reduced Fertilization Regime. Biology, 11(8), 1107. https://doi.org/10.3390/biology11081107






