Alcohol and HIV-Derived Hepatocyte Apoptotic Bodies Induce Hepatic Stellate Cell Activation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Antibodies
2.2. In Vitro Studies
2.3. Isolation of ABs from RLW Cells
2.4. Generation of TAMRA-Labeled RLW ABs by Prolonged Incubation
2.5. siRNA Transfection of LX2 Cells
2.6. Imaging of ABs by Transmission Electron Microscopy
2.7. In Vivo Studies
- Control-fed mice exposed to (A) uninfected and (B) HIV-infected ABs generated from RLW cells.
- Ethanol-fed mice exposed to (A) uninfected and (B) HIV-infected ABs from RLW cells. Each subgroup contains 3 mice.
2.8. RNA Isolation and RT-PCR
2.9. Immunoblotting
2.10. Immunofluorescence
2.11. Statistical Analyses
3. Results
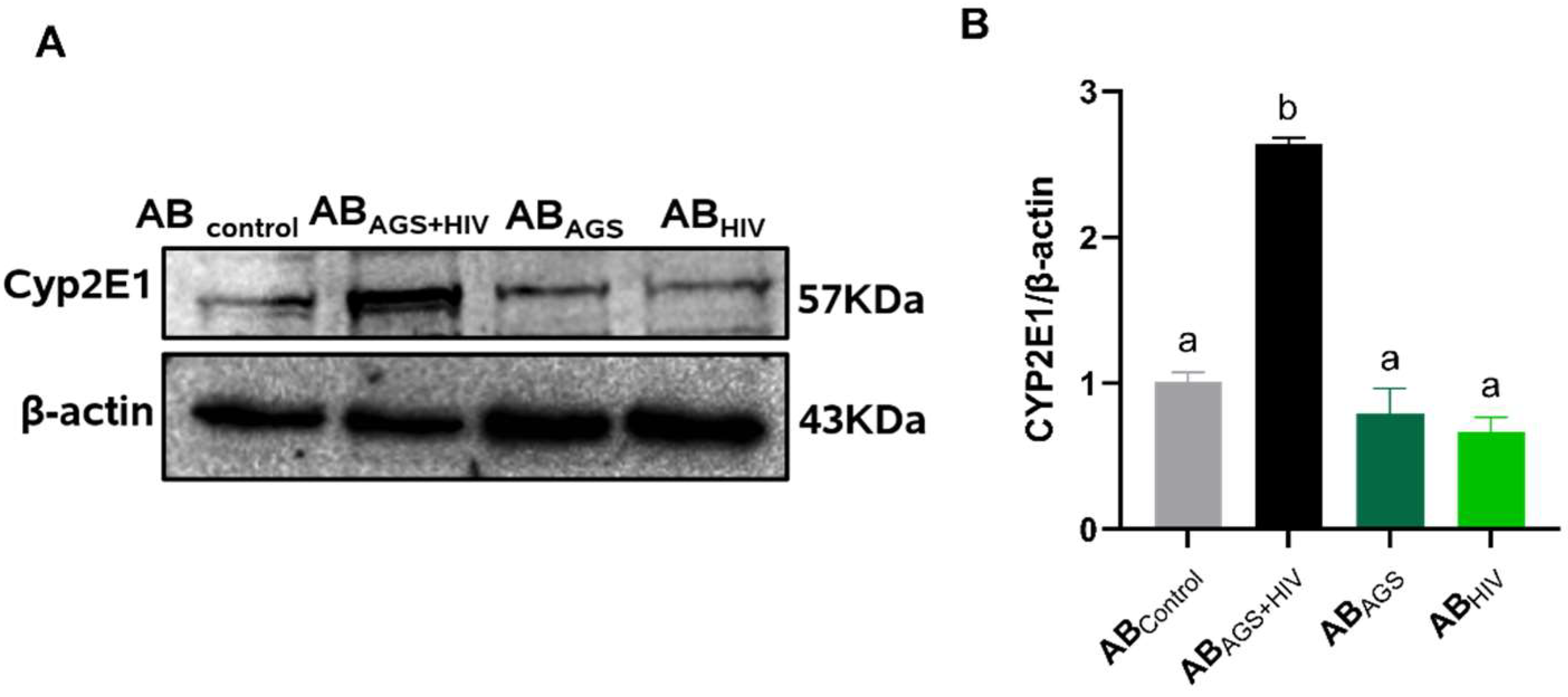
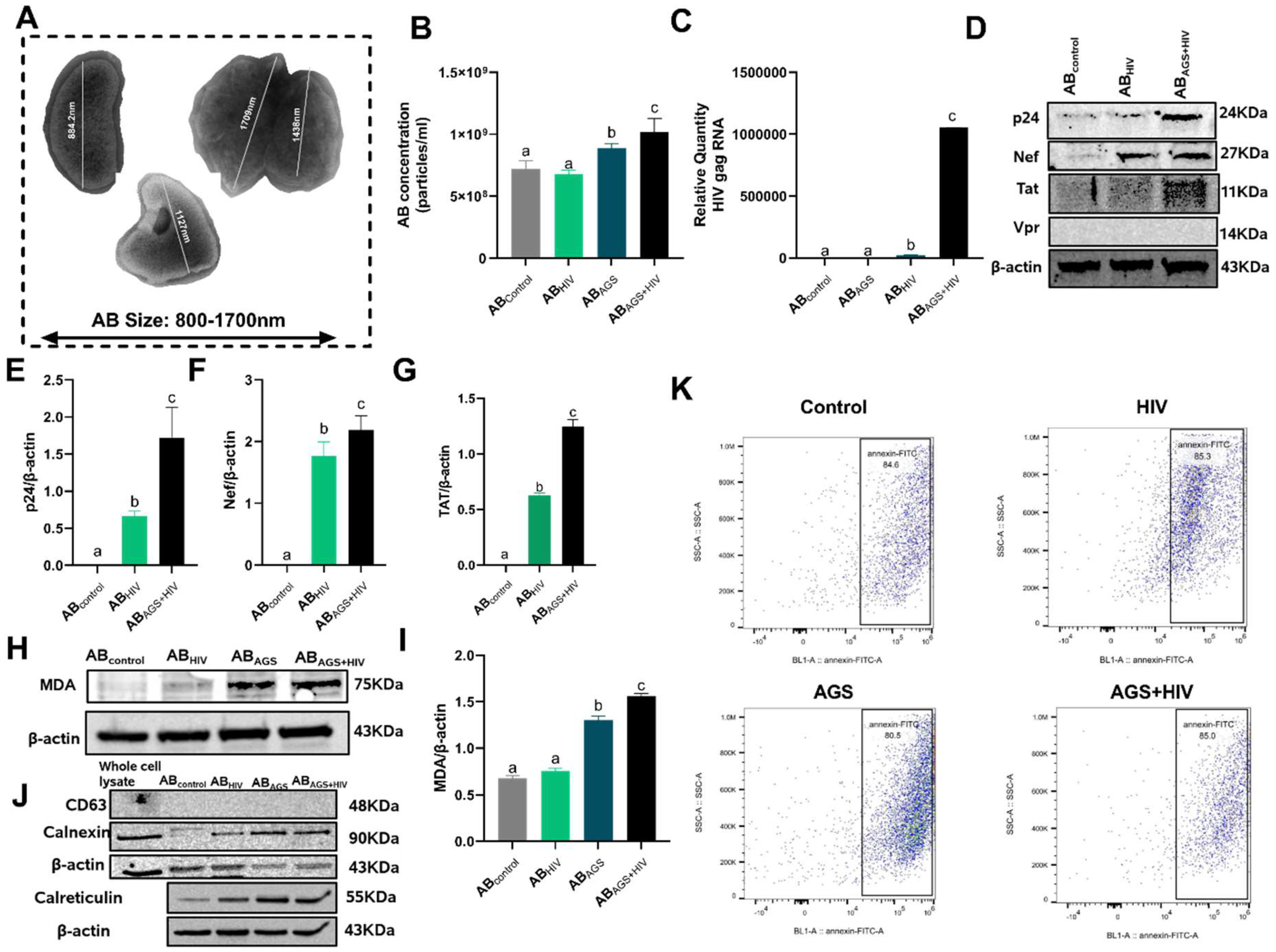
3.1. HIV RNA, HIV Proteins and Malondialdehyde Were Expressed by RLW ABAGS+HIV
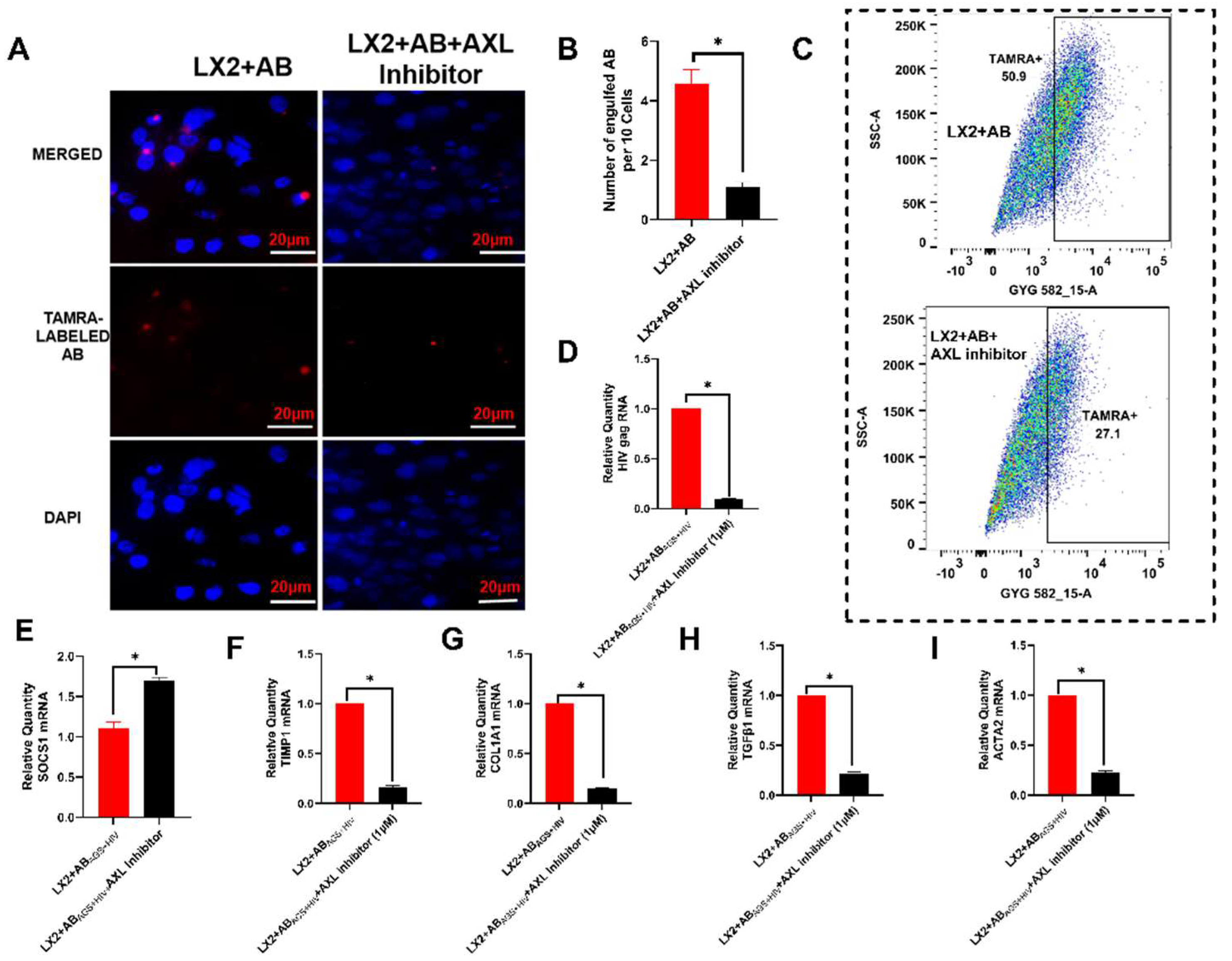
3.2. Engulfment of RLW ABs by LX2 Cells
3.3. Pharmacological Inhibition of Axl Blocks LX2 Engulfment of RLW ABs and Attenuates Atcivation of Profibrotic Genes
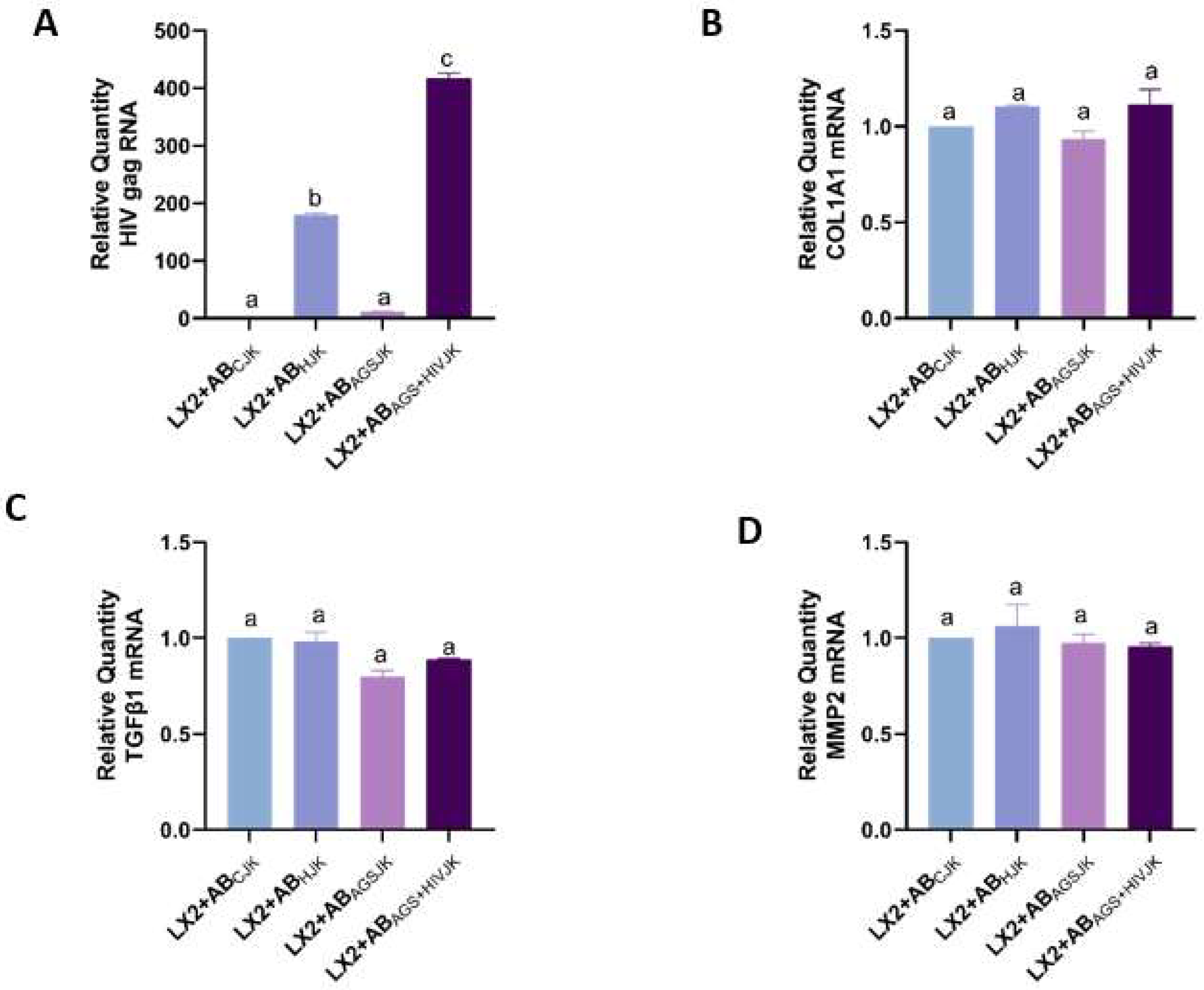
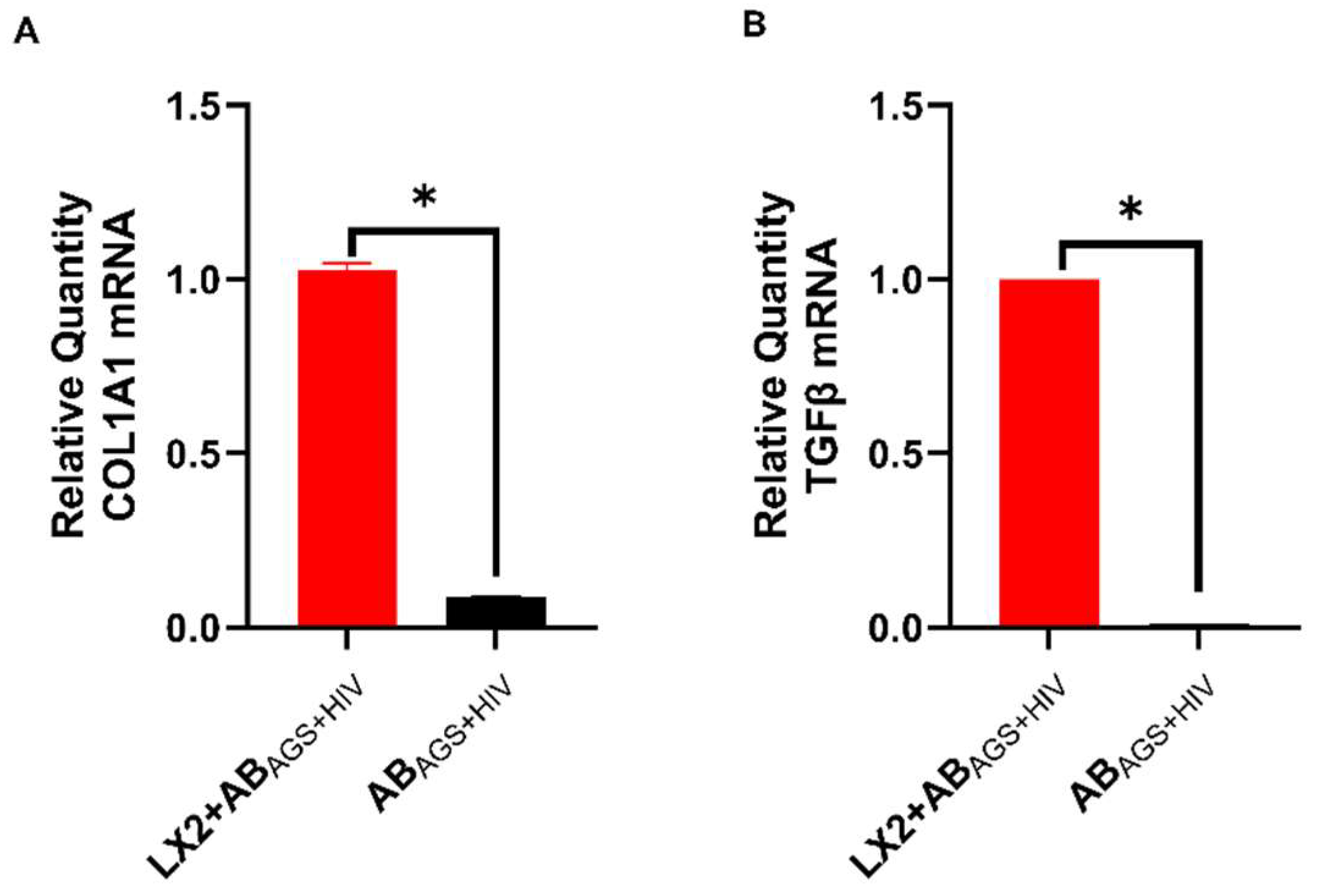
3.4. Engulfment of HIV- and MDA-Containing ABs Induces LX2 Profibrotic Activation
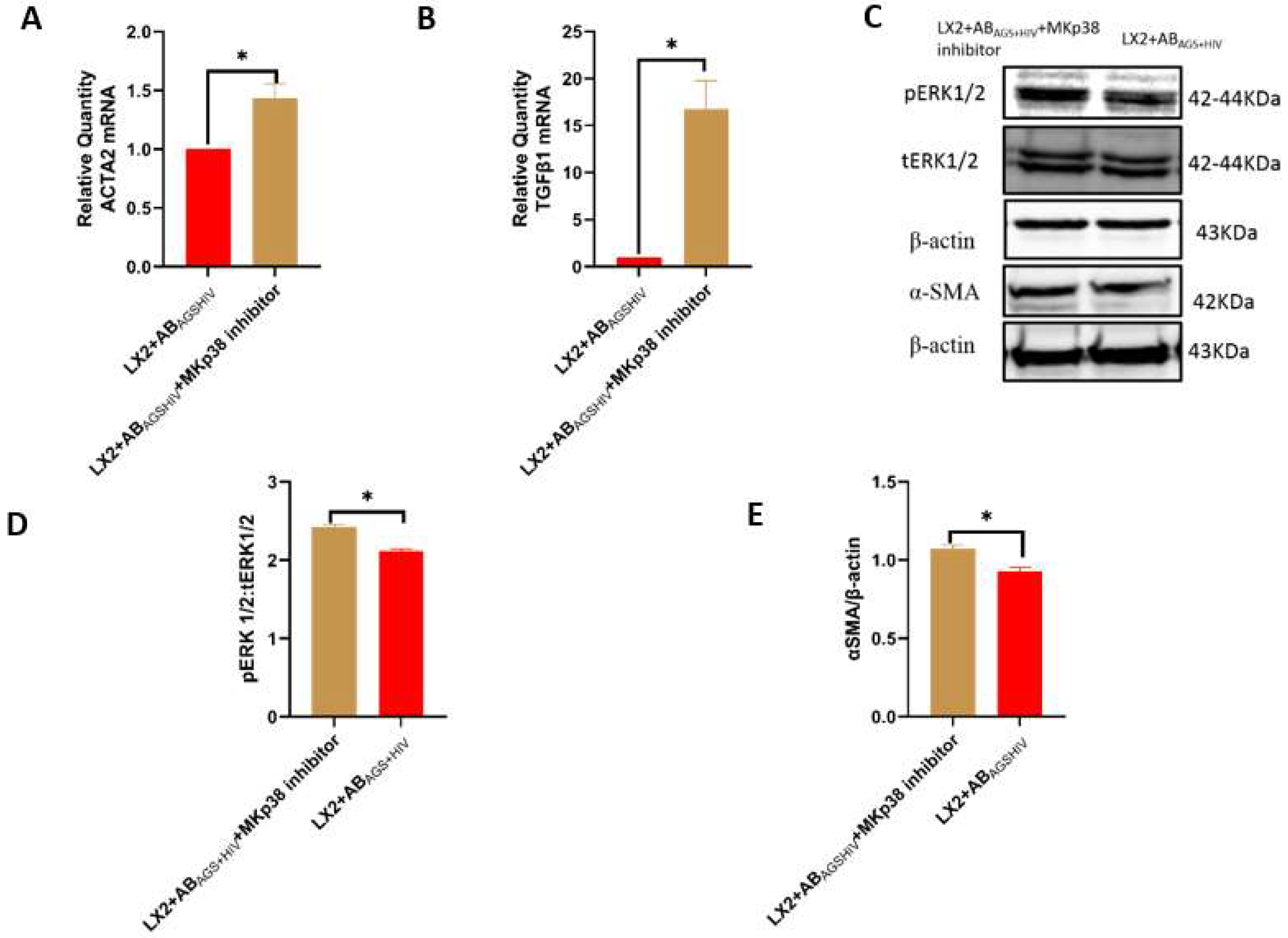
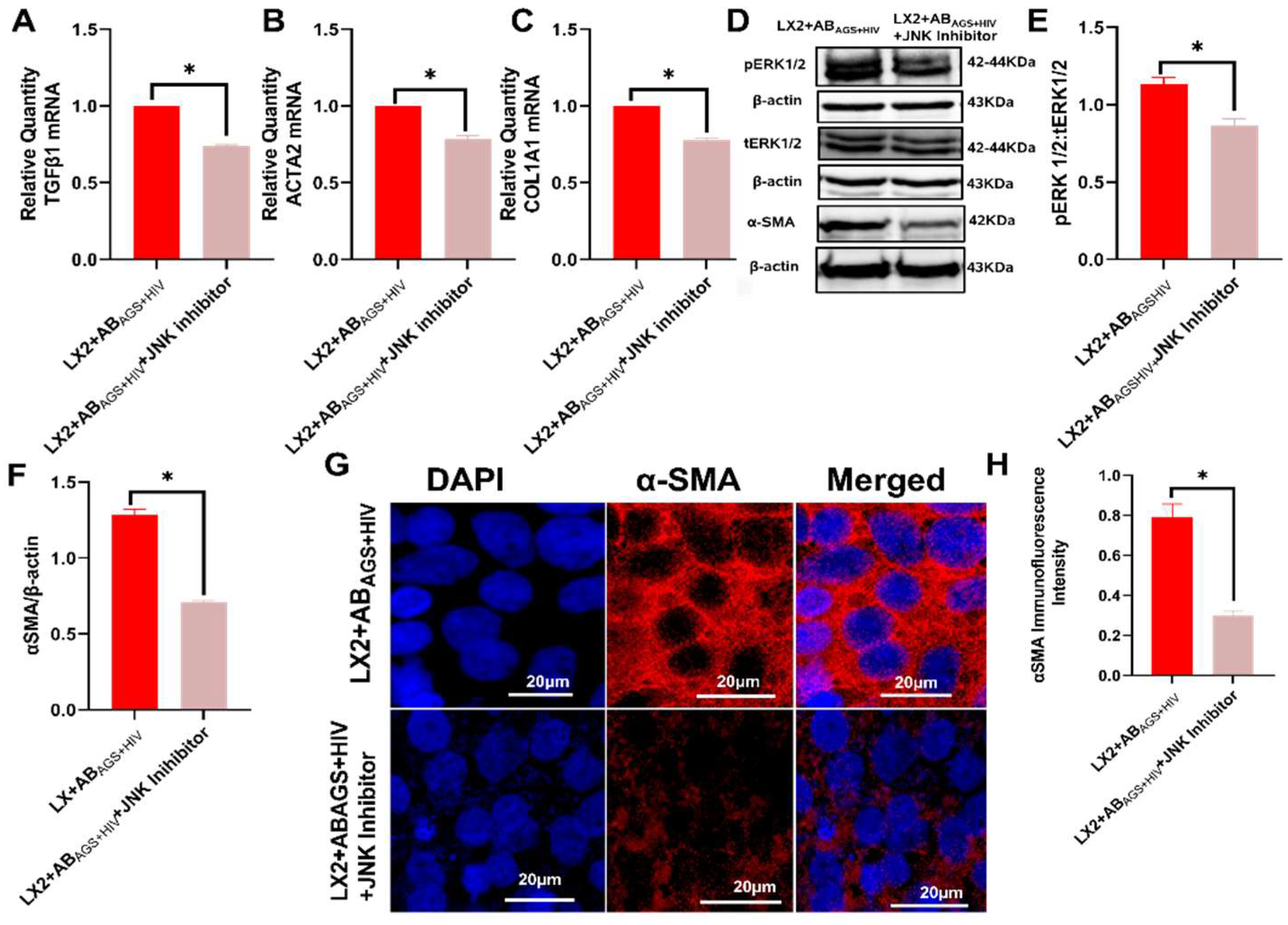
3.5. Pharmacological Inhibition of LX2 Cells Exposed to ABAGS+HIV Attenuates Profibrotic Activation via JNK and ERK1/2 Pathway
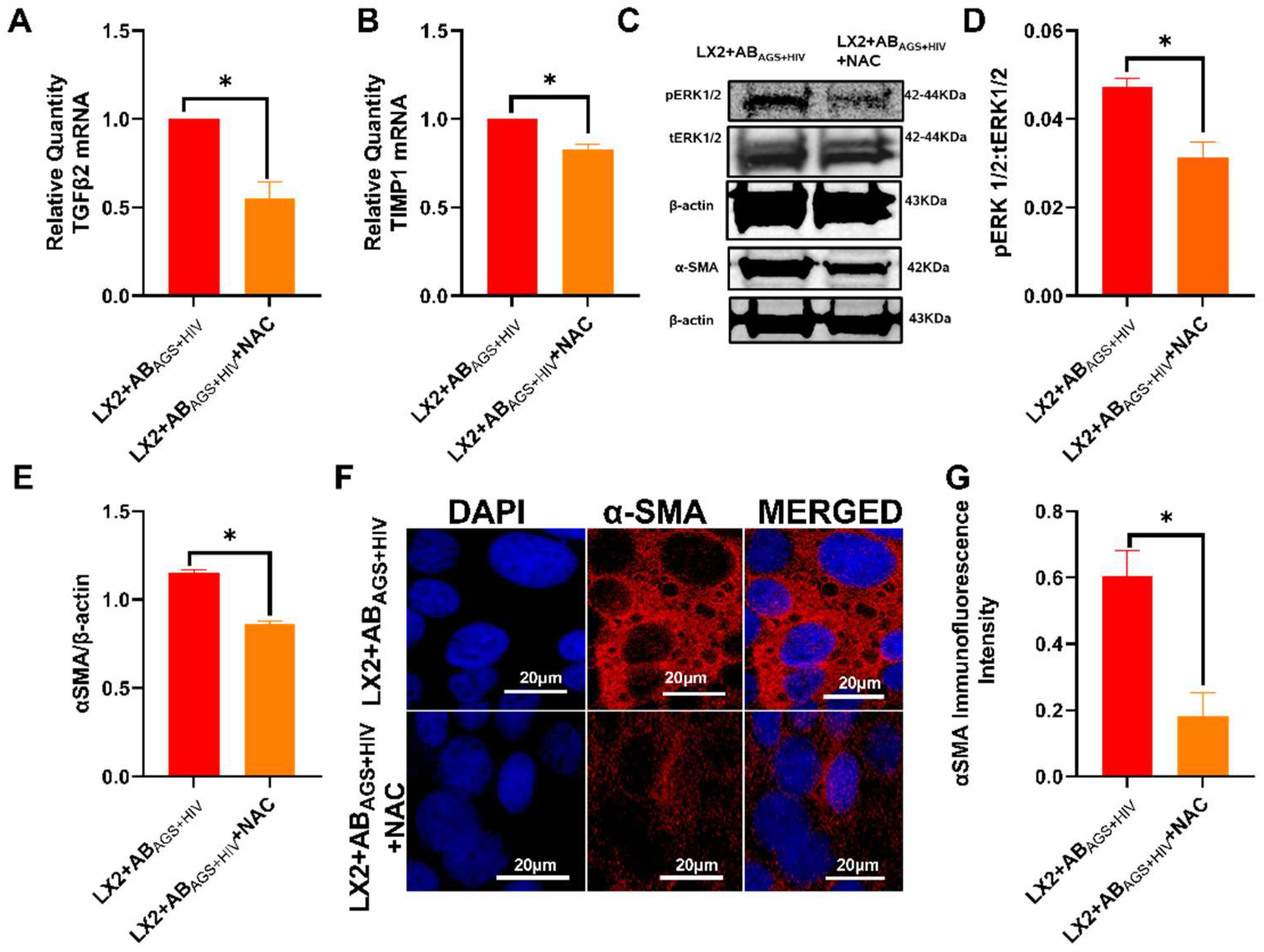
3.6. Oxidative Stress from ABAGS+HIV Activates JNK and ERK1/2 Pathway in LX2 Cells
3.7. ABAGS+HIV Upregulates IL6 mRNA in LX2 Cells
3.8. siRNA STAT3 Transfection Inhibits STAT3 Protein Expressions in LX2 Cells
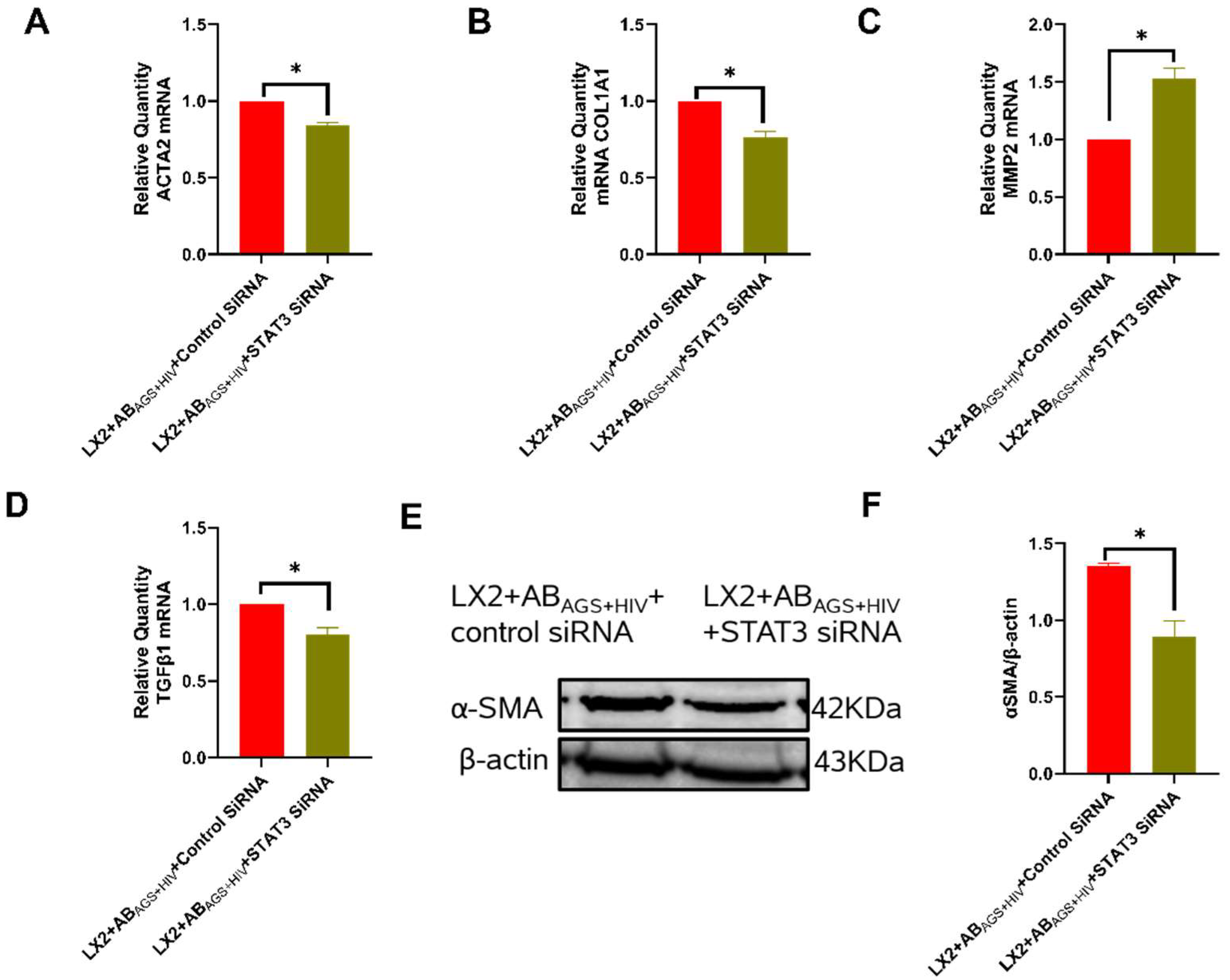
3.9. Silencing STAT3 in LX2 Cells Attenuates ABAGS+HIV Induced Profibrotic Activation
3.10. In Vivo Effects of HIV Containing-Apoptotic Bodies on Ethanol-Fed Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- New-Aaron, M.; Kingi, H.; Meza, J.L.; Goedert, M.H.; Kibusi, S.M.; Mkhoi, M.L.; Mayengo, C.D.; Charles, J.; Shabani, S.; New-Aaron, T.O. Duration on ART, Alcohol Use and HIV Stage May Predict Risky Sexual Behavior in a Resource-limited Environment: A Cross-sectional Study. Curr. HIV Res. 2021, 19, 420–433. [Google Scholar] [CrossRef] [PubMed]
- New-Aaron, M.; Ganesan, M.; Dagur, R.S.; Kharbanda, K.K.; Poluektova, L.Y.; Osna, N.A. Pancreatogenic Diabetes: Triggering Effects of Alcohol and HIV. Biology 2021, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.E.; Nash, S.; Connor, N.; Kirwan, P.D.; Ogaz, D.; Croxford, S.; Angelis, D.; Delpech, V.C. Towards elimination of HIV transmission, AIDS and HIV-related deaths in the UK. HIV Med. 2018, 19, 505–512. [Google Scholar] [CrossRef]
- Granich, R.; Crowley, S.; Vitoria, M.; Smyth, C.; Kahn, J.G.; Bennett, R.; Lo, Y.R.; Souteyrand, Y.; Williams, B. Highly active antiretroviral treatment as prevention of HIV transmission: Review of scientific evidence and update. Curr. Opin. HIV AIDS 2010, 5, 298–304. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Hernán, M.A.; Ledergerber, B.; Tilling, K.; Weber, R.; Sendi, P.; Rickenbach, M.; Robins, J.M.; Egger, M. Long-term effectiveness of potent antiretroviral therapy in preventing AIDS and death: A prospective cohort study. Lancet 2005, 366, 378–384. [Google Scholar] [CrossRef]
- Debes, J.D.; Bohjanen, P.R.; Boonstra, A. Mechanisms of Accelerated Liver Fibrosis Progression during HIV Infection. J. Clin. Transl. Hepatol. 2016, 4, 328–335. [Google Scholar] [CrossRef]
- Rein, S.M.; Lampe, F.C.; Chaloner, C.; Stafford, A.; Rodger, A.J.; Johnson, M.A.; McDonnell, J.; Burns, F.; Madge, S.; Miners, A.; et al. Causes of hospitalisation among a cohort of people with HIV from a London centre followed from 2011 to 2018. BMC Infect. Dis. 2021, 21, 395. [Google Scholar] [CrossRef]
- Althoff, K.N.; Gebo, K.A.; Moore, R.D.; Boyd, C.M.; Justice, A.C.; Wong, C.; Lucas, G.M.; Klein, M.B.; Kitahata, M.M.; Crane, H.; et al. Contributions of traditional and HIV-related risk factors on non-AIDS-defining cancer, myocardial infarction, and end-stage liver and renal diseases in adults with HIV in the USA and Canada: A collaboration of cohort studies. Lancet HIV 2019, 6, e93–e104. [Google Scholar] [CrossRef]
- Madhombiro, M.; Dube, B.; Dube, M.; Zunza, M.; Chibanda, D.; Rusakaniko, S.; Seedat, S. Intervention for alcohol use disorders at an HIV care clinic in Harare: A pilot and feasibility study. Addict. Sci. Clin. Pract. 2019, 14, 16. [Google Scholar] [CrossRef]
- Cook, J.A.; Burke-Miller, J.K.; Steigman, P.J.; Schwartz, R.M.; Hessol, N.A.; Milam, J.; Merenstein, D.J.; Anastos, K.; Golub, E.T.; Cohen, M.H. Prevalence, Comorbidity, and Correlates of Psychiatric and Substance Use Disorders and Associations with HIV Risk Behaviors in a Multisite Cohort of Women Living with HIV. AIDS Behav. 2018, 22, 3141–3154. [Google Scholar] [CrossRef]
- Petoumenos, K.; Law, M.G. Smoking, alcohol and illicit drug use effects on survival in HIV-positive persons. Curr. Opin. HIV AIDS 2016, 11, 514–520. [Google Scholar] [CrossRef]
- Galvan, F.H.; Bing, E.G.; Fleishman, J.A.; London, A.S.; Caetano, R.; Burnam, M.A.; Longshore, D.; Morton, S.C.; Orlando, M.; Shapiro, M. The prevalence of alcohol consumption and heavy drinking among people with HIV in the United States: Results from the HIV Cost and Services Utilization Study. J. Stud. Alcohol 2002, 63, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, M.; New-Aaron, M.; Dagur, R.S.; Makarov, E.; Wang, W.; Kharbanda, K.K.; Kidambi, S.; Poluektova, L.Y.; Osna, N.A. Alcohol Metabolism Potentiates HIV-Induced Hepatotoxicity: Contribution to End-Stage Liver Disease. Biomolecules 2019, 9, 851. [Google Scholar] [CrossRef] [PubMed]
- New-Aaron, M.; Thomes, P.G.; Ganesan, M.; Dagur, R.S.; Donohue, T.M.; Kusum, K.K.; Poluektova, L.Y.; Osna, N.A. Alcohol-Induced Lysosomal Damage and Suppression of Lysosome Biogenesis Contribute to Hepatotoxicity in HIV-Exposed Liver Cells. Biomolecules 2021, 11, 1497. [Google Scholar] [CrossRef]
- Chen, M.; Liu, J.; Yang, W.; Ling, W. Lipopolysaccharide mediates hepatic stellate cell activation by regulating autophagy and retinoic acid signaling. Autophagy 2017, 13, 1813–1827. [Google Scholar] [CrossRef]
- Ramos-Tovar, E.; Muriel, P. Molecular Mechanisms That Link Oxidative Stress, Inflammation, and Fibrosis in the Liver. Antioxidants 2020, 9, 1279. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.-C.; Tseng, C.-P.; Liao, M.-H.; Peng, C.-Y.; Yu, J.-S.; Chuang, P.-H.; Huang, J.-T.; Chen, J.J.W. Activation of hepatic stellate cells by the ubiquitin C-terminal hydrolase 1 protein secreted from hepatitis C virus-infected hepatocytes. Sci. Rep. 2017, 7, 4448. [Google Scholar] [CrossRef]
- Gäbele, E.; Mühlbauer, M.; Dorn, C.; Weiss, T.S.; Froh, M.; Schnabl, B.; Wiest, R.; Schölmerich, J.; Obermeier, F.; Hellerbrand, C. Role of TLR9 in hepatic stellate cells and experimental liver fibrosis. Biochem. Biophys. Res. Commun. 2008, 376, 271–276. [Google Scholar] [CrossRef]
- Kiziltas, S. Toll-like receptors in pathophysiology of liver diseases. World J. Hepatol. 2016, 8, 1354. [Google Scholar] [CrossRef]
- Nakamoto, N.; Kanai, T. Role of Toll-Like Receptors in Immune Activation and Tolerance in the Liver. Front. Immunol. 2014, 5, 221. [Google Scholar] [CrossRef]
- Watanabe, A.; Hashmi, A.; Gomes, D.A.; Town, T.; Badou, A.; Flavell, R.A.; Mehal, W.Z. Apoptotic hepatocyte DNA inhibits hepatic stellate cell chemotaxis via toll-like receptor 9. Hepatology 2007, 46, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, X.; Vikash, V.; Ye, Q.; Wu, D.; Liu, Y.; Dong, W. ROS and ROS-Mediated Cellular Signaling. Oxid. Med. Cell. Longev. 2016, 2016, 4350965. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; Li, S.P.; Westermarck, J. Phosphatase-mediated crosstalk between MAPK signaling pathways in the regulation of cell survival. FASEB J. 2008, 22, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.; Cheong, Y.K.; Kim, N.H.; Chung, H.T.; Kang, D.G.; Pae, H.O. Mitogen-Activated Protein Kinases and Reactive Oxygen Species: How Can ROS Activate MAPK Pathways? J. Signal. Transduct. 2011, 2011, 792639. [Google Scholar] [CrossRef]
- Ichijo, H. From receptors to stress-activated MAP kinases. Oncogene 1999, 18, 6087–6093. [Google Scholar] [CrossRef]
- Simon, A.R.; Rai, U.; Fanburg, B.L.; Cochran, B.H. Activation of the JAK-STAT pathway by reactive oxygen species. Am. J. Physiol.-Cell Physiol. 1998, 275, C1640–C1652. [Google Scholar] [CrossRef]
- Butturini, E.; Carcereri de Prati, A.; Mariotto, S. Redox Regulation of STAT1 and STAT3 Signaling. Int. J. Mol. Sci. 2020, 21, 7034. [Google Scholar] [CrossRef]
- Charras, A.; Arvaniti, P.; Le Dantec, C.; Dalekos, G.N.; Zachou, K.; Bordron, A.; Renaudineau, Y. JAK Inhibitors and Oxidative Stress Control. Front. Immunol. 2019, 10, 2814. [Google Scholar] [CrossRef]
- New-Aaron, M.; Ganesan, M.; Dagur, R.S.; Kharbanda, K.K.; Poluektova, L.Y.; Osna, N.A. Obeticholic acid attenuates human immunodeficiency virus/alcohol metabolism-induced pro-fibrotic activation in liver cells. World J. Hepatol. 2020, 12, 965–975. [Google Scholar] [CrossRef]
- Phan, T.K.; Poon, I.K.; Atkin-Smith, G.K. Detection and Isolation of Apoptotic Bodies to High Purity. J. Vis. Exp. 2018, 138, e58317. [Google Scholar] [CrossRef]
- Powers, J.; Zhang, H.; Battrell, L.; Meadows, G.G.; Trobridge, G.D. Establishment of an immunodeficient alcohol mouse model to study the effects of alcohol on human cells in vivo. J. Stud. Alcohol Drugs 2012, 73, 933–937. [Google Scholar] [CrossRef][Green Version]
- Melhem, A.; Muhanna, N.; Bishara, A.; Alvarez, C.E.; Ilan, Y.; Bishara, T.; Horani, A.; Nassar, M.; Friedman, S.L.; Safadi, R. Anti-fibrotic activity of NK cells in experimental liver injury through killing of activated HSC. J. Hepatol. 2006, 45, 60–71. [Google Scholar] [CrossRef]
- Muthiah, M.D.; Huang, D.Q.; Zhou, L.; Jumat, N.H.; Choolani, M.; Chan, J.K.Y.; Wee, A.; Lim, S.G.; Dan, Y.Y. A murine model demonstrating reversal of structural and functional correlates of cirrhosis with progenitor cell transplantation. Sci. Rep. 2019, 9, 15446. [Google Scholar] [CrossRef] [PubMed]
- Maacha, S.; Bhat, A.A.; Jimenez, L.; Raza, A.; Haris, M.; Uddin, S.; Grivel, J.-C. Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Mol. Cancer 2019, 18, 55. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.J.; Kim, O.Y.; Gho, Y.S. Extracellular vesicles as emerging intercellular communicasomes. BMB Rep. 2014, 47, 531–539. [Google Scholar] [CrossRef]
- Battistelli, M.; Falcieri, E. Apoptotic bodies: Particular extracellular vesicles involved in intercellular communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef]
- Naeini, M.B.; Bianconi, V.; Pirro, M.; Sahebkar, A. The role of phosphatidylserine recognition receptors in multiple biological functions. Cell. Mol. Biol. Lett. 2020, 25, 23. [Google Scholar] [CrossRef]
- Miyanishi, M.; Tada, K.; Koike, M.; Uchiyama, Y.; Kitamura, T.; Nagata, S. Identification of Tim4 as a phosphatidylserine receptor. Nature 2007, 450, 435–439. [Google Scholar] [CrossRef]
- Lemke, G. Phosphatidylserine Is the Signal for TAM Receptors and Their Ligands. Trends Biochem. Sci. 2017, 42, 738–748. [Google Scholar] [CrossRef]
- Shao, W.H.; Zhen, Y.; Eisenberg, R.A.; Cohen, P.L. The Mer receptor tyrosine kinase is expressed on discrete macrophage subpopulations and mainly uses Gas6 as its ligand for uptake of apoptotic cells. Clin. Immunol. 2009, 133, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Raggi, F.; Pelassa, S.; Pierobon, D.; Penco, F.; Gattorno, M.; Novelli, F.; Eva, A.; Varesio, L.; Giovarelli, M.; Bosco, M.C. Regulation of Human Macrophage M1–M2 Polarization Balance by Hypoxia and the Triggering Receptor Expressed on Myeloid Cells-1. Front. Immunol. 2017, 8, 1097. [Google Scholar] [CrossRef] [PubMed]
- Chua, B.A.; Ngo, J.A.; Situ, K.; Ramirez, C.M.; Nakano, H.; Morizono, K. Protein S and Gas6 induce efferocytosis of HIV-1-infected cells. Virology 2018, 515, 176–190. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Lahair, M.M.; Franklin, R.A. Reactive oxygen species-induced activation of the MAP kinase signaling pathways. Antioxid. Redox Signal. 2006, 8, 1775–1789. [Google Scholar] [CrossRef]
- Xiang, D.M.; Sun, W.; Ning, B.F.; Zhou, T.F.; Li, X.F.; Zhong, W.; Cheng, Z.; Xia, M.Y.; Wang, X.; Deng, X.; et al. The HLF/IL-6/STAT3 feedforward circuit drives hepatic stellate cell activation to promote liver fibrosis. Gut 2018, 67, 1704–1715. [Google Scholar] [CrossRef]
- Bica, I.; McGovern, B.; Dhar, R.; Stone, D.; McGowan, K.; Scheib, R.; Snydman, D.R. Increasing Mortality Due to End-Stage Liver Disease in Patients with Human Immunodeficiency Virus Infection. Clin. Infect. Dis. 2001, 32, 492–497. [Google Scholar] [CrossRef]
- Ganesan, M.; Poluektova, L.Y.; Kharbanda, K.K.; Osna, N.A. Liver as a target of human immunodeficiency virus infection. World J. Gastroenterol. 2018, 24, 4728–4737. [Google Scholar] [CrossRef]
- Sastry, J.; Mohammed, H.; Campos, M.M.; Uetrecht, J.; Abu-Asab, M. Nevirapine-induced liver lipid-SER inclusions and other ultrastructural aberrations. Ultrastruct. Pathol. 2018, 42, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Gwag, T.; Meng, Z.; Sui, Y.; Helsley, R.N.; Park, S.H.; Wang, S.; Greenberg, R.N.; Zhou, C. Non-nucleoside reverse transcriptase inhibitor efavirenz activates PXR to induce hypercholesterolemia and hepatic steatosis. J. Hepatol. 2019, 70, 930–940. [Google Scholar] [CrossRef]
- Ganesan, M.; Poluektova, L.Y.; Enweluzo, C.; Kharbanda, K.K.; Osna, N.A. Hepatitis C Virus-Infected Apoptotic Hepatocytes Program Macrophages and Hepatic Stellate Cells for Liver Inflammation and Fibrosis Development: Role of Ethanol as a Second Hit. Biomolecules 2018, 8, 113. [Google Scholar] [CrossRef]
- Nevola, R.; Rinaldi, L.; Giordano, M.; Marrone, A.; Adinolfi, L.E. Mechanisms and clinical behavior of hepatocellular carcinoma in HBV and HCV infection and alcoholic and non-alcoholic fatty liver disease. Hepatoma Res. 2018, 4, 55. [Google Scholar] [CrossRef]
- Kaspar, M.B.; Sterling, R.K. Mechanisms of liver disease in patients infected with HIV. BMJ Open Gastroenterol. 2017, 4, e000166. [Google Scholar] [CrossRef]
- Chakraborty, J.B.; Oakley, F.; Walsh, M.J. Mechanisms and biomarkers of apoptosis in liver disease and fibrosis. Int. J. Hepatol. 2012, 2012, 648915. [Google Scholar] [CrossRef] [PubMed]
- Witek, R.P.; Stone, W.C.; Karaca, F.G.; Syn, W.K.; Pereira, T.A.; Agboola, K.M.; Omenetti, A.; Jung, Y.; Teaberry, V.; Choi, S.S.; et al. Pan-caspase inhibitor VX-166 reduces fibrosis in an animal model of nonalcoholic steatohepatitis. Hepatology 2009, 50, 1421–1430. [Google Scholar] [CrossRef] [PubMed]
- Barreyro, F.J.; Holod, S.; Finocchietto, P.V.; Camino, A.M.; Aquino, J.B.; Avagnina, A.; Carreras, M.C.; Poderoso, J.J.; Gores, G.J. The pan-caspase inhibitor Emricasan (IDN-6556) decreases liver injury and fibrosis in a murine model of non-alcoholic steatohepatitis. Liver Int. 2015, 35, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Shiffman, M.L.; Pockros, P.; McHutchison, J.G.; Schiff, E.R.; Morris, M.; Burgess, G. Clinical trial: The efficacy and safety of oral PF-03491390, a pancaspase inhibitor—A randomized placebo-controlled study in patients with chronic hepatitis C. Aliment. Pharmacol. Ther. 2010, 31, 969–978. [Google Scholar] [CrossRef]
- Canbay, A.; Feldstein, A.; Baskin-Bey, E.; Bronk, S.F.; Gores, G.J. The caspase inhibitor IDN-6556 attenuates hepatic injury and fibrosis in the bile duct ligated mouse. J. Pharmacol. Exp. Ther. 2004, 308, 1191–1196. [Google Scholar] [CrossRef]
- Guo, R.; Ren, J. Alcohol and acetaldehyde in public health: From marvel to menace. Int. J. Environ. Res. Public Health 2010, 7, 1285–1301. [Google Scholar] [CrossRef]
- Teschke, R.; Neuman, M.G.; Liangpunsakul, S.; Seitz, H.-K. Alcoholic Liver Disease and the co-triggering Role of MEOS with Its CYP 2E1 Catalytic Cycle and ROS. Arch. Gastroenterol. Res. 2021, 2, 9–25. [Google Scholar]
- Doody, E.E.; Groebner, J.L.; Walker, J.R.; Frizol, B.M.; Tuma, D.J.; Fernandez, D.J.; Tuma, P.L. Ethanol metabolism by alcohol dehydrogenase or cytochrome P(450) 2E1 differentially impairs hepatic protein trafficking and growth hormone signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 313, G558–G569. [Google Scholar] [CrossRef]
- Koop, D.R. Alcohol metabolism’s damaging effects on the cell: A focus on reactive oxygen generation by the enzyme cytochrome P450 2E1. Alcohol Res. Health 2006, 29, 274–280. [Google Scholar] [PubMed]
- Urtasun, R.; de la Rosa, L.C.; Nieto, N. Oxidative and nitrosative stress and fibrogenic response. Clin. Liver Dis. 2008, 12, 769–790. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, M.; Natarajan, S.K.; Zhang, J.; Mott, J.L.; Poluektova, L.I.; McVicker, B.L.; Kharbanda, K.K.; Tuma, D.J.; Osna, N.A. Role of apoptotic hepatocytes in HCV dissemination: Regulation by acetaldehyde. Am. J. Physiol.-Gastrointest. Liver Physiol. 2016, 310, G930–G940. [Google Scholar] [CrossRef] [PubMed]
- Poon, I.K.; Parkes, M.A.; Jiang, L.; Atkin-Smith, G.K.; Tixeira, R.; Gregory, C.D.; Ozkocak, D.C.; Rutter, S.F.; Caruso, S.; Santavanond, J.P. Moving beyond size and phosphatidylserine exposure: Evidence for a diversity of apoptotic cell-derived extracellular vesicles in vitro. J. Extracell. Vesicles 2019, 8, 1608786. [Google Scholar] [CrossRef]
- Johnston, D.E.; Kroening, C. Mechanism of early carbon tetrachloride toxicity in cultured rat hepatocytes. Pharm. Toxicol. 1998, 83, 231–239. [Google Scholar] [CrossRef]
- Higuchi, H.; Kurose, I.; Kato, S.; Miura, S.; Ishii, H. Ethanol-induced apoptosis and oxidative stress in hepatocytes. Alcohol Clin. Exp. Res. 1996, 20, 340a–346a. [Google Scholar] [CrossRef]
- Jiang, J.X.; Mikami, K.; Venugopal, S.; Li, Y.; Török, N.J. Apoptotic body engulfment by hepatic stellate cells promotes their survival by the JAK/STAT and Akt/NF-kappaB-dependent pathways. J. Hepatol. 2009, 51, 139–148. [Google Scholar] [CrossRef]
- Xu, X.; Lai, Y.; Hua, Z.-C. Apoptosis and apoptotic body: Disease message and therapeutic target potentials. Biosci. Rep. 2019, 39, BSR20180992. [Google Scholar] [CrossRef]
- Depraetere, V. “Eat me” signals of apoptotic bodies. Nat. Cell Biol. 2000, 2, E104. [Google Scholar] [CrossRef]
- Fadok, V.A.; De Cathelineau, A.; Daleke, D.L.; Henson, P.M.; Bratton, D.L. Loss of phospholipid asymmetry and surface exposure of phosphatidylserine is required for phagocytosis of apoptotic cells by macrophages and fibroblasts. J. Biol. Chem. 2001, 276, 1071–1077. [Google Scholar] [CrossRef]
- Canbay, A.; Taimr, P.; Torok, N.; Higuchi, H.; Friedman, S.; Gores, G.J. Apoptotic body engulfment by a human stellate cell line is profibrogenic. Lab. Investig. 2003, 83, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.K.; Wilhelm, A.; Antoniades, C.G. TAM receptor tyrosine kinase function and the immunopathology of liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G899–G905. [Google Scholar] [CrossRef]
- Myers, K.V.; Amend, S.R.; Pienta, K.J. Targeting Tyro3, Axl and MerTK (TAM receptors): Implications for macrophages in the tumor microenvironment. Mol. Cancer 2019, 18, 94. [Google Scholar] [CrossRef]
- Ortmayr, G.; Brunnthaler, L.; Pereyra, D.; Huber, H.; Santol, J.; Rumpf, B.; Najarnia, S.; Smoot, R.; Ammon, D.; Sorz, T.; et al. Immunological Aspects of AXL/GAS-6 in the Context of Human Liver Regeneration. Hepatol. Commun. 2022, 6, 576–592. [Google Scholar] [CrossRef] [PubMed]
- Kluwe, J.; Pradere, J.P.; Gwak, G.Y.; Mencin, A.; De Minicis, S.; Osterreicher, C.H.; Colmenero, J.; Bataller, R.; Schwabe, R.F. Modulation of hepatic fibrosis by c-Jun-N-terminal kinase inhibition. Gastroenterology 2010, 138, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Hong, I.H.; Park, S.J.; Goo, M.J.; Lee, H.R.; Park, J.K.; Ki, M.R.; Kim, S.H.; Lee, E.M.; Kim, A.Y.; Jeong, K.S. JNK1 and JNK2 regulate α-SMA in hepatic stellate cells during CCl4-induced fibrosis in the rat liver. Pathol. Int. 2013, 63, 483–491. [Google Scholar] [CrossRef]
- Parola, M.; Robino, G.; Marra, F.; Pinzani, M.; Bellomo, G.; Leonarduzzi, G.; Chiarugi, P.; Camandola, S.; Poli, G.; Waeg, G. HNE interacts directly with JNK isoforms in human hepatic stellate cells. J. Clin. Investig. 1998, 102, 1942–1950. [Google Scholar] [CrossRef]
- Novo, E.; Marra, F.; Zamara, E.; Valfrè di Bonzo, L.; Monitillo, L.; Cannito, S.; Petrai, I.; Mazzocca, A.; Bonacchi, A.; De Franco, R.S.; et al. Overexpression of Bcl-2 by activated human hepatic stellate cells: Resistance to apoptosis as a mechanism of progressive hepatic fibrogenesis in humans. Gut 2006, 55, 1174–1182. [Google Scholar] [CrossRef]
- Novo, E.; Marra, F.; Zamara, E.; Valfrè di Bonzo, L.; Caligiuri, A.; Cannito, S.; Antonaci, C.; Colombatto, S.; Pinzani, M.; Parola, M. Dose dependent and divergent effects of superoxide anion on cell death, proliferation, and migration of activated human hepatic stellate cells. Gut 2006, 55, 90–97. [Google Scholar] [CrossRef]
- Pessayre, D.; Fromenty, B.; Mansouri, A. Mitochondrial injury in steatohepatitis. Eur. J. Gastroenterol. Hepatol. 2004, 16, 1095–1105. [Google Scholar] [CrossRef]
- Gandhi, C.R. Oxidative Stress and Hepatic Stellate Cells: A Paradoxical Relationship. Trends Cell Mol. Biol. 2012, 7, 1–10. [Google Scholar] [PubMed]
- Jiang, J.X.; Venugopal, S.; Serizawa, N.; Chen, X.; Scott, F.; Li, Y.; Adamson, R.; Devaraj, S.; Shah, V.; Gershwin, M.E.; et al. Reduced nicotinamide adenine dinucleotide phosphate oxidase 2 plays a key role in stellate cell activation and liver fibrogenesis in vivo. Gastroenterology 2010, 139, 1375–1384. [Google Scholar] [CrossRef]
- Schmidt-Arras, D.; Rose-John, S. IL-6 pathway in the liver: From physiopathology to therapy. J. Hepatol. 2016, 64, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; van Boxel-Dezaire, A.H.; Cheon, H.; Yang, J.; Stark, G.R. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc. Natl. Acad. Sci. USA 2013, 110, 16975–16980. [Google Scholar] [CrossRef] [PubMed]
- Flowers, L.O.; Subramaniam, P.S.; Johnson, H.M. A SOCS-1 peptide mimetic inhibits both constitutive and IL-6 induced activation of STAT3 in prostate cancer cells. Oncogene 2005, 24, 2114–2120. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
New-Aaron, M.; Dagur, R.S.; Koganti, S.S.; Ganesan, M.; Wang, W.; Makarov, E.; Ogunnaike, M.; Kharbanda, K.K.; Poluektova, L.Y.; Osna, N.A. Alcohol and HIV-Derived Hepatocyte Apoptotic Bodies Induce Hepatic Stellate Cell Activation. Biology 2022, 11, 1059. https://doi.org/10.3390/biology11071059
New-Aaron M, Dagur RS, Koganti SS, Ganesan M, Wang W, Makarov E, Ogunnaike M, Kharbanda KK, Poluektova LY, Osna NA. Alcohol and HIV-Derived Hepatocyte Apoptotic Bodies Induce Hepatic Stellate Cell Activation. Biology. 2022; 11(7):1059. https://doi.org/10.3390/biology11071059
Chicago/Turabian StyleNew-Aaron, Moses, Raghubendra Singh Dagur, Siva Sankar Koganti, Murali Ganesan, Weimin Wang, Edward Makarov, Mojisola Ogunnaike, Kusum K. Kharbanda, Larisa Y. Poluektova, and Natalia A. Osna. 2022. "Alcohol and HIV-Derived Hepatocyte Apoptotic Bodies Induce Hepatic Stellate Cell Activation" Biology 11, no. 7: 1059. https://doi.org/10.3390/biology11071059
APA StyleNew-Aaron, M., Dagur, R. S., Koganti, S. S., Ganesan, M., Wang, W., Makarov, E., Ogunnaike, M., Kharbanda, K. K., Poluektova, L. Y., & Osna, N. A. (2022). Alcohol and HIV-Derived Hepatocyte Apoptotic Bodies Induce Hepatic Stellate Cell Activation. Biology, 11(7), 1059. https://doi.org/10.3390/biology11071059








