Simple Summary
Fluoroquinolones, mainly levofloxacin, are considered an alternative treatment option of Stenotrophomonas maltophilia infections to trimethoprim/sulfamethoxazole. However, an increase in the number of levofloxacin-resistant strains is observed worldwide. The fluoroquinolone resistance in S. maltophilia is usually caused by an overproduction of various multidrug efflux pumps, which are able to extrude antibiotics and chemotherapeutics from the bacterial cells. The purpose of the study was to analyze the contribution of efflux systems to levofloxacin resistance in S. maltophilia clinical strains, isolated in Warsaw, by phenotypic and molecular methods. Previously, the occurrence of genes encoding various ten efflux pumps was shown in 94 studied isolates. Additionally, 44 of 94 isolates demonstrated reduction in susceptibility to levofloxacin. In this study, in the presence of efflux pump inhibitors, an increase in levofloxacin susceptibility was observed in 13 isolates. The overexpression of genes encoding two efflux pump system, such as SmeDEF and Sme VWX (in five and one isolate, respectively), was demonstrated. Sequencing analysis revealed an amino acid change in the local regulators of these efflux pump operons. Our data indicate that the overproduction of the SmeVWX efflux system, unlike SmeDEF, plays a significant role in the levofloxacin resistance of the clinical isolates.
Abstract
Levofloxacin is considered an alternative treatment option of Stenotrophomonas maltophilia infections to trimethoprim/sulfamethoxazole. The fluoroquinolone resistance in S. maltophilia is usually caused by an overproduction of efflux pumps. In this study, the contribution of efflux systems to levofloxacin resistance in S. maltophilia clinical isolates was demonstrated using phenotypic (minimal inhibitory concentrations, MICs, of antibiotics determination ± efflux pump inhibitors, EPIs) and molecular (real-time polymerase-chain-reaction and sequencing) methods. Previously, the occurrence of genes encoding ten efflux pumps was shown in 94 studied isolates. Additionally, 44/94 isolates demonstrated reduction in susceptibility to levofloxacin. Only 5 of 13 isolates (with ≥4-fold reduction in levofloxacin MIC) in the presence of EPIs showed an increased susceptibility to levofloxacin and other antibiotics. The overexpression of smeD and smeV genes (in five and one isolate, respectively) of 5 tested efflux pump operons was demonstrated. Sequencing analysis revealed 20–35 nucleotide mutations in local regulatory genes such as smeT and smeRv. However, mutations leading to an amino acid change were shown only in smeT (Arg123Lys, Asp182Glu, Asp204Glu) for one isolate and in smeRv (Gly266Ser) for the other isolate. Our data indicate that the overproduction of the SmeVWX efflux system, unlike SmeDEF, plays a significant role in the levofloxacin resistance.
1. Introduction
Stenotrophomonas maltophilia is an opportunistic pathogen that is especially dangerous for patients with cystic fibrosis, immunodeficiencies, cancer (particularly lung cancer), and those exposed to immunosuppressive therapy, mechanical ventilation, and catheters. The critically ill patients from intensive care units, as well as patients hospitalized for long periods, especially after broad-spectrum antibiotic therapy, are most exposed to S. maltophilia infections [1,2,3]. It is worth emphasizing that infections caused by S. maltophilia are characterized by a high mortality rate, even up to 69% in patients with bacteremia. Infections occur in both adults and children. Although it is mainly a nosocomial pathogen, community-acquired infections are also observed. An increase in the incidence of S. maltophilia infections in the global population, from 1.3% to 1.7%, was observed between 2007–2012 [4].
The most important problem in the treatment of infections caused by S. maltophilia is an intrinsic resistance of this bacterium to a wide range of antibiotics and chemotherapeutic agents. The drug of choice for the treatment of S. maltophilia infections is trimethoprim/sulfamethoxazole [2,5]. However, a slow but continuous increase in the number of the trimethoprim/sulfamethoxazole-resistant strains is observed worldwide. Resistance rates in the world usually range between 1–20%; however, higher resistance levels have been reported both in Europe (Spain: 27%) and in other continents (Turkey: 10–15%, Taiwan: up to 25%, China: 30–48%) [6,7,8]. Other therapeutic options include the use of fluoroquinolones, minocycline, and ticarcillin/clavulanate. Recently, it has been revealed that fluoroquinolones, an alternative treatment, are equally as effective as trimethoprim/sulfamethoxazole [9,10,11]. Levofloxacin was most frequently used among fluoroquinolones (187 of 331, 56.5%), followed by ciprofloxacin (114 of 331, 34.4%) [10]. Generally, resistance rates to levofloxacin of S. maltophilia strains are relatively similar or higher than that of trimethoprim/sulfamethoxazole and range between 3% and 30% [9,12,13]. In our previous publication, all 94 investigated S. maltophilia clinical isolates were susceptible to trimethoprim/sulfamethoxazole and minocycline, but 7 of them were resistant and 37 were intermediately susceptible to levofloxacin [14].
The resistance to fluoroquinolones in S. maltophilia strains is caused by two main mechanisms: an overexpression of multidrug-resistant (MDR) efflux pumps and chromosomally encoded Qnr proteins (SmQnr). Five families (superfamilies) of efflux pumps are described. One of them, ABC (ATP-binding cassette) family, gets its energy from ATP disruption. The other 4 families are proton pumps. Of these, the MATE (multidrug and toxic compound extrusion) family may also take advantage of the differences in sodium ion concentration gradients. It is worth emphasizing that the pumps from the RND (resistance-nodulation-division) family are of the greatest importance for the resistance of Gram-negative bacteria. Unlike the others, pumps from the RND family can extrude antimicrobial agents from various chemical groups from the cells. Twelve MDR efflux systems have been identified in S. maltophilia so far. These systems were classified into three families of MDR efflux pumps: RND, ABC, and MFS (major facilitator superfamily). The following efflux systems, SmeDEF [15], SmeMN [14], SmeABC [16], SmeYZ [17], SmeIJK [17], SmeVWX [18], SmeOP [19], and SmeGH [20], are described from the RND family. The other four efflux pumps, SmrA [21], MacABCsm [22], SmaCDEF, and SmaAB [23], belong to the ABC family. Moreover, in S. maltophilia, the presence of the EmrCABsm system [24] from the MFS family has been described and the FuaABC pump [25] is unclassified as of yet. Fluoroquinolones, including levofloxacin, are substrates for most of these RND efflux systems. The following fluoroquinolone-removing efflux pumps, 5 from the RND family—smeDEF [15], SmeABC [16], SmeVWX [18], SmeGH [20], SmeIJK [17], and one from the ABC family—SmrA [21,26], have been identified so far. The best investigated efflux systems of S. maltophilia are SmeDEF and SmeVWX. Both of these systems are responsible for antibiotics extruding, except fluoroquinolones, as follows: SmeDEF—trimethoprim/sulfamethoxazole, chloramphenicol, macrolides, tetracycline, and tigecycline, SmeVWX—trimethoprim/sulfamethoxazole, and chloramphenicol. The RND efflux systems that occur in S. maltophilia, as in other Gram-negative rods, are encoded by genes organized in operons that are located in bacterial chromosomes. The relationship between operon overexpression of these two systems and the resistance of strains to fluoroquinolones has been demonstrated [15,18,27,28]. Expression of both of the above-mentioned efflux systems operons in S. maltophilia is regulated by local regulatory genes. The smeDEF operon is negatively regulated by the TetR-type transcriptional local repressor SmeT [29], whereas the smeVWX operon is regulated by the LysR-type transcription regulator-SmeRv [18]. Like many LysR-type regulators, SmeRv could act as a negative or positive regulator, depending on the presence of an activator ligand [18]. It has been described that mutations in smeT and smeRv regulatory genes are responsible for the overexpression of smeDEF and smeVWX efflux systems operons, respectively [30]. The third fluoroquinolone-removal efflux system is SmeABC, which also extrudes aminoglycosides and β-lactams [26]. A two-component regulatory system (TCS), SmeRS, is involved in expression of the smeABC operon [31]. The overproduction of the SmeGH efflux system leads to resistance to β-lactams, quinolones, tetracycline, and polymyxin B [20,32], while the SmeIJK efflux system confers resistance to aminoglycosides, tetracycline, and minocycline, but it also has a role in adaptation to envelope stress [33]. The local regulatory systems of SmeGH and SmeIJK have not been characterized yet. The other important fluoroquinolone-resistance mechanism in S. maltophilia is the presence of qnr genes encoding the pentapeptide repeat proteins, which protects topoisomerase IV and gyrase from fluoroquinolones binding [26,34]. The mechanism associated with Smqnr proteins usually contributes to the low level of fluoroquinolone resistance in S. maltophilia [34]. However, Wu et al. demonstrated the high level of qnr genes transcription in 39% of fluoroquinolone-resistant clinical strains of S. maltophilia [35]. It can be underlined that S. maltophilia is the only known bacterium in which resistance to quinolones is not associated with mutations in genes encoding topoisomerases [36].
In the previous article, we performed phenotypic and molecular characterization of the collection of 94 S. maltophilia clinical isolates, including the prevalence of efflux systems. The investigation revealed the presence of smeD and smeH genes in all isolates, smeW in 93 out of 94 isolates, smeI or smeK in 86, and smeA or smeB in 72 isolates [14]. Among them, 7 isolates were resistant, and 37 isolates demonstrated an intermediate level of susceptibility to levofloxacin.
The aim of this study was to investigate the role of RND efflux systems in resistance to levofloxacin in S.maltophilia clinical strains isolated in Warsaw, Poland. The studies were conducted by analyzing the effect of known efflux pump inhibitors (EPIs) on the decrease in minimal inhibitory concentration (MIC) values of levofloxacin and other MDR efflux systems substrates, as well as by analyzing the expression of genes encoding the efflux systems that remove fluoroquinolones. In addition, we studied the local regulatory genes of RND efflux systems by sequencing DNA templates.
2. Materials and Methods
2.1. Bacterial Strains
The investigation was performed on the collection of 94 non-duplicate S. maltophilia clinical isolates. Isolates were derived from January 2010 to October 2013 from a variety of clinical materials obtained during routine diagnostic testing of adult patients in microbiology laboratories. All clinical materials from which the tested isolates were derived are listed in our previous publication [14]. Most of the isolates (n = 27) were obtained from blood samples. In our previous article, all of these isolates were characterized by determination of the susceptibility profiles for three anti-S. maltophilia agents (including levofloxacin) and tested for the presence of genes encoding efflux pumps from the RND and ABC families. Moreover, the genetic relationship between clinical isolates and their similarity to the strains isolated in other countries was investigated [14]. All isolates were stored in Luria Bertani broth (BioMaxima SA, Lublin, Poland) with 20% glycerol at −80 °C until analysis. Escherichia coli ATCC 25922, S. maltophilia ATCC 13637, S. maltophilia ATCC 12714, and S. maltophilia 67/2013 clinical isolate, were used as reference strains in this study.
2.2. Determination of the MICs of Antibiotics with and without Efflux Pump Inhibitors
The minimal inhibitory concentrations of levofloxacin and gentamicin (both from Sigma, St. Louis, MO, USA), in the presence or absence of EPIs, were estimated using a 2-fold broth microdilution method in Mueller-Hinton II (MH II) broth medium (Becton, Dickinson and Company, Franklin Lakes, NJ, USA), according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [37]. The MIC values of trimethoprim/sulfamethoxazole, polymyxin B, erythromycin, tigecycline and chloramphenicol, in the presence or absence of EPIs, were estimated using E-tests (Liofilchem srl, Roseto deli Abruzzi, Italy) on Mueller-Hinton II agar medium (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) according to the manufacturer’s instructions [38]. In this study, the following efflux pump inhibitors were used: cyanide 3-chlorophenylhydrazone (CCCP), phenylalanine-arginine β-naphthylamide (PAβN), and reserpine (all from Sigma, St. Louis, MO, USA). CCCP and reserpine were dissolved in dimethyl sulfoxide (DMSO), while PAβN was dissolved in water. The final concentrations of EPIs in both MH II media, broth, and agar were 25 mg/L for PAβN and reserpine and 1 mg/L for CCCP. The used concentrations of EPIs were below a quarter of the MIC values of these inhibitors as determined for the tested isolates.
The results of the susceptibility tests of the studied isolates, in the presence or absence of EPIs, were evaluated after incubation at 35 °C for 18 h. The MICs of antimicrobial agents were interpreted according to the CLSI criteria [39]. Quality control of the MICs determination was performed using the reference strain of Escherichia coli ATCC 25922. At least a 4-fold reduction in the MIC values of antimicrobial agent after addition PAβN, reserpine, or CCCP was considered significant [40]. Such a significant decrease in drug susceptibility in the presence of at least one of the EPIs was interpreted as the likely contribution of efflux pumps to antibiotic resistance of the studied isolate.
2.3. RNA Preparation and Quantitative Real-Time PCR (qPCR)
The expression levels of smeD, smeA, smeI, smeV, and smeG were assessed by real-time PCR using specific primers listed in Table 1. Triplicate cell suspensions were prepared and inoculated in Luria-Bertani (LB) broth (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). After achieving a logarithmic-phase of S. maltophilia growth (optical density, OD600 = 0.6), total RNA was extracted using a RNeasy mini-kit (Qiagen, Germantown, MD, USA). DNA was removed using DNA-free kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). RNA was reverse transcribed to cDNA using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer’s instructions. qPCR was carried out in triplicate on each sample using a CFX96 touch Deep Well real-time PCR system (Bio-Rad, Hercules, CA, USA) and SYBR Green JumpStart Taq Ready Mix kit (Sigma, St. Louis, MO, USA). Amplification conditions were as follows: 94 °C for 10 min, followed by 40 cycles at 94 °C for 15 s and 60 °C for 30 s. The expression levels of target genes were normalized using the 16S rDNA, rpo, and gyr housekeeping genes as endogenous controls and taking primer efficiency into account. Results were obtained as a relative expression in comparison to levofloxacin-susceptible S. maltophilia 67/2013 clinical isolate from Warsaw’s hospital. This isolate was chosen as a reference due to its high susceptibility to levofloxacin (MIClevofloxacin = 1 mg/L), the relatively low MIC value of gentamicin (MICgentamicin = 64 mg/L), and the presence of all eight known efflux systems from the RND family, taking into account pulsed field gel electrophoresis (PFGE) results [14].

Table 1.
Primers used for the analysis of MDR efflux pump gene expression by qPCR and for amplification of the efflux system regulatory genes.
2.4. Amplification and Sequence Analysis of Efflux System Regulatory Genes
The total DNA of selected clinical isolates was extracted using a Genomic Mini Kit (A&A Biotechnology, Gdynia, Poland). The PCR reactions were performed using Maxima Hot Start Taq DNA polymerase (Thermo Scientific, Thermo Fisher Scientific, Waltham, MA, USA) with the following amplification parameters: 95 °C for 4 min, followed by 25 cycles of 30 s at 95 °C, 30 s at 58 °C for smeRv, or 30 s at 59 °C for smeT, 75 s at 72 °C, and a final extension of 5 min at 72 °C. The sequences of the primers were designed for this project based on gene sequences available in the GenBank National Centre for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/, accessed on 1 April 2021) or derived from publications. All primers are listed in Table 1. Sequencing of the obtained DNA templates was carried out in the Laboratory of DNA Sequencing and Oligonucleotide Synthesis Institute of Biochemistry and Biophysics, Polish Academy of Science in Warsaw (IBB PAS). The received DNA sequences were analyzed using Vector NTI Advance 11 software (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) and compared with the efflux regulatory gene sequences of the reference S. maltophilia 67/2013 strain.
2.5. Whole Genome Sequencing (WGS)
Whole genome sequencing was performed in the Laboratory of DNA Sequencing and Oligonucleotide Synthesis IBB PAS in Warsaw using MiSeq system (Illumina Sequencing Technology) (Illumina, San Diego, CA, USA). The detection of sequences encoding smeRv regulatory genes was performed in IBB PAS using PROKKA software v1.14.6. The obtained DNA sequences were compared with the DNA sequences of the reference S. maltophilia 67/2013 strain using Vector NTI Advance 11 software (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA).
3. Results
3.1. Effect of EPIs on the MIC Values of Antibiotics
The results of the impact of three EPIs on the MICs of levofloxacin and gentamicin investigated for 94 clinical isolates are provided in Table S1 in Supplementary Materials. A 4-fold reduction in the MIC values of levofloxacin was observed after the addition of CCCP (for ten isolates), PAβN (for seven isolates), and reserpine (for only one isolate). Moreover, three of the above-mentioned isolates (no. 44/2011, 47/2011, and 52/2012) showed a 4-fold reduction in the MIC values of levofloxacin in the presence of both CCCP and PAβN. For one isolate (no. 20/2011), a 4-fold increase in susceptibility to levofloxacin was observed in the presence of all three EPIs. The addition of CCCP to MH II medium reduced the MIC values of gentamicin (4-fold to at least a 32-fold) in 16 out of 94 isolates. Two of these 16 isolates (no. 15/2010 and 35/2011) also showed a 4-fold reduction in gentamicin MIC values in the presence of a second inhibitor, PAβN. Moreover, only one isolate (no. 42/2011) demonstrated the 4-fold reduction in the MIC values of gentamicin after the addition of reserpine.
The further investigations were performed on the 27 out of 94 isolates, for which at least a 4-fold reduction in the MIC values of levofloxacin or gentamicin was observed in the presence of any inhibitor (Table 2). For them, the effect of CCCP on the MIC values of following agents, trimethoprim/sulfamethoxazole, polymyxin B, chloramphenicol, erythromycin and tigecycline, was determined. The results were presented in Table S2 in Supplementary Materials, and in Table 2. In the presence of CCCP, a significant reduction in the MIC values of polymyxin B, from 5-fold to 16-fold, was observed for 4 out of 27 isolates. Additionally, one of these four isolates (no. 31/2011) also showed a 4-fold reduction in the MIC values of chloramphenicol. Besides, only one out of the group of 27 isolates (no. 41/2011) showed a significant reduction (6-fold) in the MIC values of trimethoprim/sulfamethoxazole after addition of CCCP. No changes in the susceptibility of the tested isolates to erythromycin or to tigecycline were obtained after adding CCCP to the test medium.

Table 2.
Effect of EPIs on the drug susceptibility of S. maltophilia clinical isolates (n = 27).
3.2. Expression of the Efflux Pump Genes
Six S. maltophilia isolates were chosen to analyze the expression level of genes encoding the following efflux systems: SmeDEF, SmeABC, SmeVWX, SmeGH, and SmeIJK. The selected isolates had a different susceptibility level to levofloxacin and also presented a significant reduction in the MIC values of: (I) only levofloxacin in the presence of all three investigated EPIs (no. 20/2011), (II) levofloxacin, and one other antimicrobial in the presence of one or two EPIs (no. 15/2010, 41/2011, and 52/2012), and (III) levofloxacin and two other antimicrobials in the presence of one or two EPIs (no. 31/2011 and 35/2011, respectively). The presence of efflux pump genes in the isolates selected for qPCR were included in Table S3 in Supplementary Materials.
Expression levels of smeD, smeA, smeV, smeG, and smeI genes relative to the susceptibility to levofloxacin in the presence or absence of EPIs are presented in Table 3. At least a 3-fold increase in efflux pump gene expression of tested isolates in comparison to levofloxacin-susceptible S. maltophilia 67/2013 was assessed as significant [35]. Only two out of five tested genes, smeD and smeV, were overexpressed in the selected isolates. The highest expression level of smeV, (60.81) was observed for S. maltophilia 41/2011. This isolate presented the highest MIC values of levofloxacin (MIC 16 mg/L). The highest expression level of smeD gene (21.40) was obtained in the case of S. maltophilia 15/2010, which was susceptible to levofloxacin. The 4-fold reduction in the MIC values of levofloxacin was observed in the presence of one EPI, CCCP for S. maltophilia 41/2011, and PAβN for S. maltophilia 15/2010.

Table 3.
The expression level of efflux pump genes with regard to the susceptibility to levofloxacin in the presence or absence of EPIs.
3.3. Analysis of Efflux Systems Regulatory Genes
Analysis was performed for selected isolates, which presented the overexpression of smeD and smeV genes in comparison to levofloxacin-susceptible S. maltophilia 67/2013. Molecular detection by PCR revealed the presence of the smeT regulatory gene of the SmeDEF system in three studied isolates. The nucleotide sequences of smeT genes of isolates no. 31/2011, 35/2011, and 52/2012, in comparison to DNA of the reference isolate S. maltophilia 67/2013, are shown in Figure 1. Sequencing analysis revealed in all isolates from 27 to 35 nucleotide mutations in smeT in comparison to the reference isolate. However, the nucleotide mutations only led to the amino acid changes in one isolate, no. 35/2011 (AGG → AAG [Arg123Lys], GAC → GAA [Asp182Glu], and GAC → GAA [Asp204Glu]; Figure 1). The identical nucleotide sequences of smeT gene, as presented in isolates no. 35/2011 and 67/2013, were previously deposited in the GenBank NCBI database (accession no. CP067993.1, CP040440.1, and CP060026.1, respectively).

Figure 1.
The point mutations of smeT gene in studied isolates in comparison to reference S. maltophilia 67/2013. Nucleotide mutations that led to amino acids changes are shown in the red boxes.
The amplification of the smeRv regulatory gene of the SmeVWX system failed. Thus, the whole genome sequencing of S. maltophilia 41/2011 presented overexpression of smeV gene and the reference strain of S. maltophilia 67/2013 was performed. Sequencing analysis of the smeRv gene revealed 21 nucleotide mutations in the DNA of isolate no. 41/2011 in comparison to the reference strain, but only one of these mutations resulted in an amino acid change GGC → AGC (Gly266Ser). The nucleotide sequences of the smeRv gene of isolate no. 41/2011, in comparison to DNA of S. maltophilia 67/2013, are shown in Figure 2. The identical nucleotide sequence of smeRv as in reference strain 67/2013, is presented in the GenBank database (accession No. CP049956.1, CP040440.1, CP022053.2, CP060024.1, CP060026.1, AP021908.1, AM743169.1). The smeRv nucleotide sequence from 41/2011 strain has not been previously described.
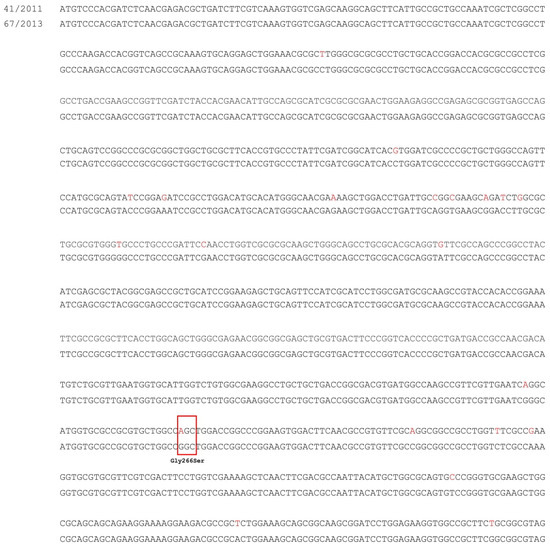
Figure 2.
The point mutations of smeRv gene in studied S. maltophilia 41/2011 in comparison to reference S. maltophilia 67/2013. The nucleotide mutations that led to amino acids changes are shown in the red boxes.
4. Discussion
Levofloxacin, along with trimethoprim/sulfamethoxazole, is the main drug used in the treatment of S. maltophilia infection [9,10,11]. However, an increase in the number of resistant strains or strains with reduced susceptibility to this fluoroquinolone has been observed worldwide in the last two decades [9,12,13]. When considering the molecular basis of S. maltophilia clinical strains resistant to levofloxacin, the overproduction of RND efflux systems should be taken into account [15,18,26,27,35]. In this study, the contribution of efflux systems to levofloxacin resistance in 94 S. maltophilia clinical strains isolated in Warsaw was determined using phenotypic and molecular methods. Previously, the presence of genes encoding various ten efflux pumps from both RND and ABC families was shown in the majority of the 94 studied isolates [14]. To assess the role of efflux systems in antimicrobial resistance of S. maltophilia, three efflux pump inhibitors were used: CCCP, reserpine, and PAβN [42,43,44,45,46]. Among the 13 isolates for which a significant reduction in the MIC value of levofloxacin was demonstrated in the presence of EPIs, the majority of the isolates gave a positive result after the addition of CCCP. In the case of three isolates, the increase in susceptibility to this fluoroquinolone was obtained only after the addition of PAβN. It is known that CCCP inhibits the activity of all proton pumps in Gram-negative rods, including S. maltophilia. CCCP, as one of the protonophores, reduces ATP production and increases membrane permeability in bacteria by interfering with the transmembrane electrochemical gradient and proton motive force [47]. Unlike CCCP, other inhibitors used, such as PAβN and reserpine, have a narrower spectrum of activity. PAβN is an inhibitor of RND efflux pumps [43,45]. Reserpine is an indole alkaloid due to low specificity and affinity of reserpine-membrane glycoprotein binding. A high dose is required to inhibit some efflux pumps [44]. It should be emphasized that the activity of MDR efflux systems could be decreased by EPIs, but the relationship between the antibiotic MIC reduction in the presence of EPIs and efflux system overexpression is not always provided [42,43,44,45]. In addition, it has recently been shown that the simultaneous overexpression of the two efflux systems does not necessarily increase the resistance level to levofloxacin but results in the resistance to the extended spectrum of substrates of these systems [35]. Taking into account the above statements, the widespread occurrence of MDR efflux pumps in the tested isolates, up to 100%, and the possibility of overproduction of more than one efflux system in one isolate, we investigated the effect of CCCP, as a universal EPI, on the susceptibility of S. maltophilia to a wide spectrum of antibiotics. Only 5 out of 13 isolates (with the 4-fold reduction in levofloxacin MIC) showed an increased susceptibility to both levofloxacin and other antibiotics such as gentamicin, polymyxin B, or chloramphenicol in the presence of EPIs. On the other hand, for the majority of the isolates showing a significant reduction in the gentamicin MIC values in the presence of CCCP, such decreases were not obtained by testing other antibiotics, including levofloxacin.
The investigation of the role of MDR efflux pumps in resistance to antimicrobial agents is usually conducted by analyzing the expression levels of the efflux pump genes and studying regulatory genes [18,30,36]. In our study, five out of six investigated isolates overexpressed the smeDEF efflux system operon, but there was no correlation between the overexpression level, the resistance to levofloxacin, and the reduction of the MIC value of levofloxacin in the presence of EPIs. CCCP was the most effective EPI, which significantly reduced the levofloxacin MIC in each isolate with the overexpression of the smeD gene. It has been described that overexpression of the smeDEF operon is associated with mutations in its local regulatory gene, smeT [30,48]. The nucleotide mutation, which led to the amino acid change Arg123Lys, present in isolate no. 35/2011 in comparison to 67/2013, was previously described by Sanchez in urinary and sputum S. maltophilia isolates (C357 and E923, respectively) with an overproducing SmeDEF efflux system [48]. The second change in the amino acid sequence of SmeT found in isolate 35/2011 compared to isolate 67/2013 was Asp204Glu, while a similar change, Ala204Glu, was previously described in the smeDEF-overexpressed clinical isolate no. C357 [48]. Thus, it appears that the presence of Lys at position 123 and Glu at position 204 of the regulator protein contributes to the overexpression of the smeDEF operon. The third revealed amino acid change in SmeT of isolate no. 35/2011, in comparison to isolate no. 67/2013, was Asp182Glu. However, this change is not significant for the susceptibility profile of the strain. The presence of both Asp and Glu at position 182, previously observed in various isolates, was not associated with overexpression of the smeDEF operon [48]. An identical smeT nucleotide sequence, as present in 35/2011, was deposited in the GenBank NCBI database (accession no. CP067993.1). On the other hand, the same smeT gene sequence was demonstrated for the reference levofloxacin-sensitive isolate no. 67/2013 as well as for the levofloxacin-resistant smeD-overexpression clinical isolates. Therefore, the expression of the pump efflux operons in S. maltophilia is controlled not only by local regulators but also by global regulators. In other non-fermentative Gram-negative bacterium such as Psudomonas aeruginosa, the global regulator, SoxR, which affects the expression of genes encoding Mex efflux systems, has been described [49]. Moreover, in S. maltophilia strains, the presence of two-component regulatory systems and their influence on the expression of efflux pump operons was demonstrated. SmeSR involved in smeABC expression and the second TCS, SmeSyRy, regulated the smeYZ operon [26]. Recently, Wu et al. proved that inactivation of smeRySy not only decreased the expression of smeYZ but also increased the expression of the smeDEF operon [50]. Additionally, the obtained results of the expression levels of the smeDEF operons did not correlate with the susceptibility of the tested S. maltophilia isolates to levofloxacin. The levofloxacin-susceptible isolate no. 15/2010 showed the highest level of the smeD gene expression. On the other hand, the levofloxacin-resistant isolate no. 31/2011 exhibited the lowest level of this gene expression. Our results seem to confirm the observations of Wu et al. [35]. They indicated that simply assessing the overexpression of smeDEF operon by RT-qPCR did not significantly contribute to fluoroquinolone’s resistance. Moreover, for several investigated strains, deletion of smeDEF operons had no impact on their susceptibility.
The second best-known efflux pump that contributes to the acquired fluoroquinolones-resistance of S. maltophilia strains is SmeVWX [18,36]. The overexpression of the smeV gene has been demonstrated by us in one isolate, no. 41/2011. Furthermore, this isolate was the most resistant to levofloxacin from the studied collection, and the 4-fold reduction in levofloxacin MIC in the presence of PAβN was shown. Recently, it was described that overexpression of the smeVWX operon is associated with mutations in the smeRv gene encoding the local regulator [27,30,36]. The change of SmeRv amino acid sequence in position 266, Gly266Ser, present in isolate no. 41/2011, has been previously described in several clinical strains isolated in Spain and in mutants obtained in vitro with the overproduction of the SmeVWX efflux system [27,30,36]. Moreover, the following changes in the amino acid sequence of the SmeRv regulator in smeVWX-overexpressing strains have been shown so far: Gly46Asp, Thr222Pro, Glu256Asp, Ala265Thr, Gly266Asp, Gly266Cys, and Cys310Phe. [27,30,36]. It should be emphasized that mutations leading to an amino acid change at position 266 of the regulatory protein were the most often detected mutation. They are extremely important for smeVWX overexpression, which contributes to drug resistance of such a strain. The strain PUC101, isolated in Spain (GenBank accession No. WP_180835982.1) with the same SmeRv amino acid sequence as Polish isolate no. 41/2011, was characterized by resistance to levofloxacin, colistin, ceftazidime, and ticarcillin-clavulanate, but there was no information concerning expression of the efflux system operon in PUC101 [51]. Besides that, only isolate no. 41/2011 showed a significant reduction in trimethoprim/sulfamethoxazole MIC in the presence of CCCP. This indicated that SmeVWX may also play an important role in resistance to trimethoprim/sulfamethoxazole, which is a known substrate of this efflux system.
As previously demonstrated, MDR efflux pumps are commonly found in clinical isolates [14]. Importantly, levofloxacin is a known substrate for 5 out of the 8 RND efflux systems. It is possible that the use of fluoroquinolones in monotherapy may lead to mutations in local and global regulatory genes. As a consequence, there are amino acid changes in the regulatory proteins, which causes the overexpression of the efflux system operons and the resistance of the strains.
5. Conclusions
Our data indicate that the SmeVWX efflux system plays a significant role in the resistance of clinical isolates to levofloxacin. Until now, mutations resulting in amino acid changes in the SmeRv regulatory protein have been reported in strains isolated in Spain. Analysis of the smeRv gene of the Polish isolate showed many point mutations and only one of them caused a significant change in the amino acid sequence, i.e., Gly266Ser. It should be emphasized that mutations leading to an amino acid change at position 266 of the regulatory protein were extremely important for the high level of smeVWX overexpression, which contributes to drug resistance of such a strain, especially in resistance to levofloxacin. However, our studies did not show a direct correlation between the level of susceptibility/resistance to levofloxacin of clinical isolates, the relative level of the smeD gene expression, and the sequence of the SmeT protein that regulates the activity of the smeDEF efflux system operon. Our data indicate that overexpression of SmeDEF did not play a significant role in levofloxacin resistance. It is likely that this system could be responsible for resistance to other antimicrobials.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biology11071044/s1, Table S1: Effect of EPIs on the susceptibility of S. maltophilia clinical isolates (n = 94) to levofloxacin and gentamycin, Table S2: Effect of CCCP on the drug susceptibility of S. maltophilia clinical isolates (n = 27), Table S3: Presence of efflux pump genes in S. maltophilia isolates selected for qPCR.
Author Contributions
Conceptualization, O.M.Z. and A.E.L.; methodology, O.M.Z. and A.E.L.; software, O.M.Z.; validation, O.M.Z., S.T. and A.E.L.; formal analysis, O.M.Z., S.T. and A.E.L.; investigation, O.M.Z. and A.E.L.; resources, O.M.Z.; data curation, O.M.Z. and A.E.L.; writing—original draft preparation, O.M.Z. and A.E.L.; writing—review and editing, O.M.Z., S.T. and A.E.L.; visualization, O.M.Z.; supervision, S.T. and A.E.L.; project administration, O.M.Z. and A.E.L.; funding acquisition, O.M.Z., S.T. and A.E.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded the grant from the National Science Center, Poland, no. 2016/21/N/NZ7/03336.
Institutional Review Board Statement
The clinical samples were part of the routine diagnostic procedure in microbiology laboratories and therefore the Institutional Review Board Statement was not applicable.
Informed Consent Statement
The clinical samples were part of the routine diagnostic procedure in microbiology laboratories and therefore the informed consent statement was not applicable.
Acknowledgments
Research was carried out with the use of the CePT infrastructure financed by the European Union through the European Regional Development Fund as part of the Operational Program “Innovative Economy” for 2007–2013.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Al-Anazi, K.A.; Al-Jasser, A.M. Infections caused by Stenotrophomonas maltophilia in recipients of hematopoietic stem cell transplantation. Front. Oncol. 2014, 4, 232. [Google Scholar] [CrossRef] [PubMed]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.M.; Wagner, J.L.; Chastain, D.B.; Bookstaver, P.B.; Stover, K.; Lin, J.; Matson, H.; White, N.; Motesh, M.; Bland, C.M. Real-world, multicentre evaluation of the incidence and risk factors for non-susceptible Stenotrophomonas maltophilia isolates. J. Glob. Antimicrob. Resist. 2022, 28, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.T.; Lin, C.Y.; Chen, Y.H.; Hsueh, P.R. Update of infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front. Microbiol. 2015, 6, 893. [Google Scholar] [CrossRef]
- Andelković, M.V.; Janković, S.M.; Kostić, M.J.; Živković Zarić, R.S.; Opančina, V.D.; Živić, M.Ž.; Milosavljević, M.J.; Pejčić, A.V. Antimicrobial treatment of Stenotrophomonas maltophilia invasive infections: Systematic review. J. Chemother. 2019, 31, 297–306. [Google Scholar]
- Brooke, J.S. New strategies against Stenotrophomonas maltophilia: A serious worldwide intrinsically drug-resistant opportunistic pathogen. Expert Rev. Anti. Infect. Ther. 2014, 12, 1–4. [Google Scholar] [CrossRef]
- Juhász, E.; Pongrácz, J.; Iván, M.; Kristóf, K. Antibiotic susceptibility of sulfamethoxazole-trimethoprim resistant Stenotrophomonas maltophilia strains isolated at a tertiary care centre in Hungary. Acta Microbiol. Immunol. Hung. 2015, 62, 295–305. [Google Scholar] [CrossRef][Green Version]
- Al-Jasser, A.M. Stenotrophomonas maltophilia resistant to trimethoprim–sulfamethoxazole: An increasing problem. Ann. Clin. Microbiol. Antimicrob. 2006, 5, 23. [Google Scholar] [CrossRef]
- Gajdacs, M.; Urban, E. Epidemiological trends and resistance associated with Stenotrophomonas maltophilia bacteraemia: A 10-year retrospective cohort study in a tertiary-care hospital in Hungary. Diseases 2019, 7, 41. [Google Scholar] [CrossRef]
- Ko, J.H.; Kang, C.I.; Cornejo-Juárez, P.; Yeh, K.M.; Wang, C.H.; Cho, S.Y.; Gözel, M.G.; Kim, S.H.; Hsueh, P.R.; Sekiya, N.; et al. Fluoroquinolones versus trimethoprim-sulfamethoxazole for the treatment of Stenotrophomonas maltophilia infections: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2019, 25, 546–554. [Google Scholar] [CrossRef]
- Sarzynski, S.H.; Warner, S.; Sun, J.; Matsouaka, R.; Dekker, J.P.; Babiker, A.; Li, W.; Lai, Y.L.; Danner, R.L.; Fowler, V.G., Jr.; et al. Trimethoprim-sulfamethoxazole versus levofloxacin for Stenotrophomonas maltophilia infections: A retrospective comparative effectiveness study of electronic health records from 154 US hospitals. Open Forum Infect. Dis. 2022, 9, ofab644. [Google Scholar] [CrossRef]
- Duan, Z.; Qin, J.; Liu, Y.; Li, C.; Ying, C. Molecular epidemiology and risk factors of Stenotrophomonas maltophilia infections in a Chinese teaching hospital. BMC Microbiol. 2020, 20, 294. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, Y.; Liu, Y.; Wang, D.; Ni, W.; Wang, R.; Liu, Y.; Zhang, B. Frequency and genetic determinants of tigecycline resistance in clinically isolated Stenotrophomonas maltophilia in Beijing, China. Front. Microbiol. 2018, 9, 549. [Google Scholar] [CrossRef]
- Zając, O.M.; Tyski, S.; Laudy, A.E. Phenotypic and molecular characteristics of the MDR efflux pump gene-carrying Stenotrophomonas maltophilia strains isolated in Warsaw, Poland. Biology 2022, 11, 105. [Google Scholar] [CrossRef]
- Alonso, A.; Martinez, J.L. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2000, 44, 3079–3086. [Google Scholar] [CrossRef]
- Chang, L.L.; Chen, H.F.; Chang, C.Y.; Lee, T.M.; Wu, W.J. Contribution of integrons, and SmeABC and SmeDEF efflux pumps to multidrug resistance in clinical isolates of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2004, 53, 518–521. [Google Scholar] [CrossRef]
- Gould, V.C.; Okazaki, A.; Avison, M.B. Coordinate hyperproduction of SmeZ and SmeJK efflux pumps extends drug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2013, 57, 655–657. [Google Scholar] [CrossRef]
- Chen, C.H.; Huang, C.C.; Chung, T.C.; Hu, R.M.; Huang, Y.W.; Yang, T.C. Contribution of resistance-nodulation-division efflux pump operon smeU1-V-W-U2-X to multidrug resistance of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2011, 55, 5826–5833. [Google Scholar] [CrossRef]
- Lin, C.W.; Huang, Y.W.; Hu, R.M.; Yang, T.C. SmeOP-tolCsm efflux pump contributes to the multidrug resistance of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2014, 58, 2405–2408. [Google Scholar] [CrossRef]
- Li, L.H.; Zhang, M.S.; Wu, C.J.; Lin, Y.T.; Yang, T.C. Overexpression of SmeGH contributes to the acquired MDR of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2019, 74, 2225–2229. [Google Scholar] [CrossRef]
- Al-Hamad, A.; Upton, M.; Burnie, J. Molecular cloning and characterization of SmrA, a novel ABC multidrug efflux pump from Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2009, 64, 731–734. [Google Scholar] [CrossRef][Green Version]
- Lin, Y.T.; Huang, Y.W.; Liou, R.S.; Chang, Y.C.; Yang, T.C. MacABCsm, an ABC-type tripartite efflux pump of Stenotrophomonas maltophilia involved in drug resistance, oxidative and envelope stress tolerances and biofilm formation. J. Antimicrob. Chemother. 2014, 69, 3221–3226. [Google Scholar] [CrossRef]
- Dulyayangkul, P.; Calvopiña, K.; Kate, J.; Heesom, K.J.; Avison, M.B. Novel mechanisms of efflux-mediated levofloxacin resistance and reduced amikacin susceptibility in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2021, 65, e01284-e20. [Google Scholar] [CrossRef]
- Huang, Y.W.; Hu, R.M.; Chu, F.Y.; Lin, H.R.; Yang, T.C. Characterization of a major facilitator superfamily (MFS) tripartite efflux pump EmrCABsm from Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2013, 68, 2498–2505. [Google Scholar] [CrossRef]
- Hu, R.M.; Liao, S.T.; Huang, C.C.; Huang, Y.W.; Yang, T.C. An inducible fusaric acid tripartite efflux pump contributes to the fusaric acid resistance in Stenotrophomonas maltophilia. PLoS ONE 2012, 7, e51053. [Google Scholar] [CrossRef]
- Gil-Gil, T.; Martinez, J.L.; Blanco, P. Mechanisms of antimicrobial resistance in Stenotrophomonas maltophilia: A review of current knowledge. Expert Rev. Anti. Infect. Ther. 2020, 18, 335–347. [Google Scholar] [CrossRef]
- Garcia-Leon, G.; Salgado, F.; Oliveros, J.C.; Sanchez, M.B.; Martinez, J.L. Interplay between intrinsic and acquired resistance to quinolones in Stenotrophomonas maltophilia. Environ. Microbiol. 2014, 16, 1282–1296. [Google Scholar] [CrossRef]
- Garcia-Leon, G.; Hernandez, A.; Hernando-Amado, S.; Alavi, P.; Berg, G.; Martinez, J.L. A function of SmeDEF, the major quinolone resistance determinant of Stenotrophomonas maltophilia, is the colonization of plant roots. Appl. Environ. Microbiol. 2014, 80, 4559–4565. [Google Scholar] [CrossRef]
- Sanchez, P.; Alonso, A.; Martinez, J.L. Cloning and characterization of SmeT, a repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. Antimicrob. Agents Chemother. 2002, 46, 3386–3393. [Google Scholar] [CrossRef]
- Sanchez, M.B.; Martinez, J.L. Overexpression of the efflux pumps SmeVWX and SmeDEF is a major cause of resistance to co-trimoxazole in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2018, 62, e00301–e00318. [Google Scholar] [CrossRef]
- Li, X.Z.; Zhang, L.; Poole, K. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2002, 46, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Blanco, P.; Corona, F.; Martinez, J.L. Mechanisms and phenotypic consequences of acquisition of tigecycline resistance by Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2019, 74, 3221–3230. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-W.; Liou, R.-S.; Lin, Y.-T.; Huang, H.-H.; Yang, T.-C. A linkage between SmeIJK efflux pump, cell envelope integrity, and σE-mediated envelope stress response in Stenotrophomonas maltophilia. PLoS ONE 2014, 9, e111784. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.B.; Martinez, J.L. SmQnr contributes to intrinsic resistance to quinolones in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2010, 54, 580–581. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.J.; Lu, H.F.; Lin, Y.T.; Zhang, M.S.; Li, L.H.; Yang, T.C. Substantial contribution of SmeDEF, SmeVWX, SmQnr and heat shock response to fluoroquinolone resistance in clinical isolates of Stenotrophomonas maltophilia. Front. Microbiol. 2019, 10, 822. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Leon, G.; Ruiz de Alegria Puig, C.; Garcia de la Fuente, C.; Martinez-Martinez, L.; Martinez, J.L.; Sanchez, M.B. High-level quinolone resistance is associated with the overexpression of smeVWX in Stenotrophomonas maltophilia clinical isolates. Clin. Microbiol. Infect. 2015, 21, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard, Document M07-A9, 9th ed.; Clinical and Laboratory Standards Institute CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Liofilchem Inc. Liofilchem MTS—MIC Test Strip Guide. 2020. Available online: https://www.liofilchem.com/solutions/clinical/arm/mts-mic-test-strip.html (accessed on 18 September 2021).
- Clinical and Laboratory Standards Institute (CLSI). M100: Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI: Wayne, PA, USA, 2022. [Google Scholar]
- Laudy, A.E.; Kulińska, E.; Tyski, S. The impact of efflux pump inhibitors on the activity of selected non-antibiotic medicinal products against Gram-negative bacteria. Molecules 2017, 22, 114. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, A.; Corona, F.; Dias, R.; Sanchez, M.B.; Martinez, J.L. The inactivation of RNase G reduces the Stenotrophomonas maltophilia susceptibility to quinolones by triggering the heat shock response. Front Microbiol. 2015, 6, 1068. [Google Scholar] [CrossRef]
- Valdezate, S.; Vindel, A.; Echeita, A.; Baquero, F.; Canton, R. Topoisomeraze II and IV quinolone resistance-determining regions in Stenotrophomonas maltophilia clinical isolates with different levels of quinolone susceptibility. Antimicrob. Agents Chemother. 2002, 44, 665–671. [Google Scholar] [CrossRef]
- Rhee, J.Y.; Choi, J.Y.; Soo Ko, K. Efflux pump inhibitor carbonyl cyanide-m-chlorophenylhydrazone (CCCP) enhances bacteriostatic activity of trimethoprim-sulfamethoxazole against clinical Stenotrophomonas maltophilia isolates from Korea. J. Bacteriol. Virol. 2016, 46, 185–192. [Google Scholar] [CrossRef]
- Jia, W.; Wang, J.; Xu, H.; Li, G. Resistance of Stenotrophomonas maltophilia to fluoroquinolones: Prevalence in a university hospital and possible mechanisms. Int. J. Environ. Res. Public Health 2015, 12, 5177–5195. [Google Scholar] [CrossRef]
- Lamers, R.P.; Cavallari, J.F.; Burrows, L.L. The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAβN) permeabilizes the outer membrane of gram-negative bacteria. PLoS ONE 2013, 8, e60666. [Google Scholar] [CrossRef]
- Laudy, A.E. Non-antibiotics, efflux pumps and drug resistance of Gram-negative rods. Pol. J. Microbiol. 2018, 67, 129–135. [Google Scholar] [CrossRef]
- Osei Sekyere, J.; Amoako, D.G. Carbonyl cyanide m-chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae. Front. Microbiol. 2017, 8, 228. [Google Scholar] [CrossRef]
- Sanchez, P.; Alonso, A.; Martinez, J.L. Regulatory regions of smeDEF in Stenotrophomonas maltophilia strains expressing different amounts of the multidrug efflux pump SmeDEF. Antimicrob. Agents Chemother. 2004, 48, 2274–2276. [Google Scholar] [CrossRef]
- Sporer, A.J.; Beierschmitt, C.; Bendebury, A.; Zink, K.E.; Price-Whelan, A.; Buzzeo, M.C.; Sanchez, L.M.; Dietrich, L.E.P. Pseudomonas aeruginosa PumA acts on an endogenous phenazine to promote self-resistance. Microbiology 2018, 164, 790–800. [Google Scholar] [CrossRef]
- Wu, C.J.; Huang, Y.W.; Lin, Y.T.; Ning, H.C.; Yang, T.C. Inactivation of SmeSyRy two-component regulatory system inversely regulates the expression of SmeYZ and SmeDEF efflux pumps in Stenotrophomonas maltophilia. PLoS ONE 2016, 11, e0160943. [Google Scholar] [CrossRef]
- Yero, D.; Huedo, P.; Conchillo-Solé, O.; Martínez-Servat, S.; Mamat, U.; Coves, X.; Llanas, F.; Roca, I.; Vila, J.; Schaible, U.E.; et al. Genetic variants of the DSF quorum sensing system in Stenotrophomonas maltophilia influence virulence and resistance phenotypes among genotypically diverse clinical isolates. Front. Microbiol. 2020, 11, 1160. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).