Understanding the Role of PIN Auxin Carrier Genes under Biotic and Abiotic Stresses in Olea europaea L.
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
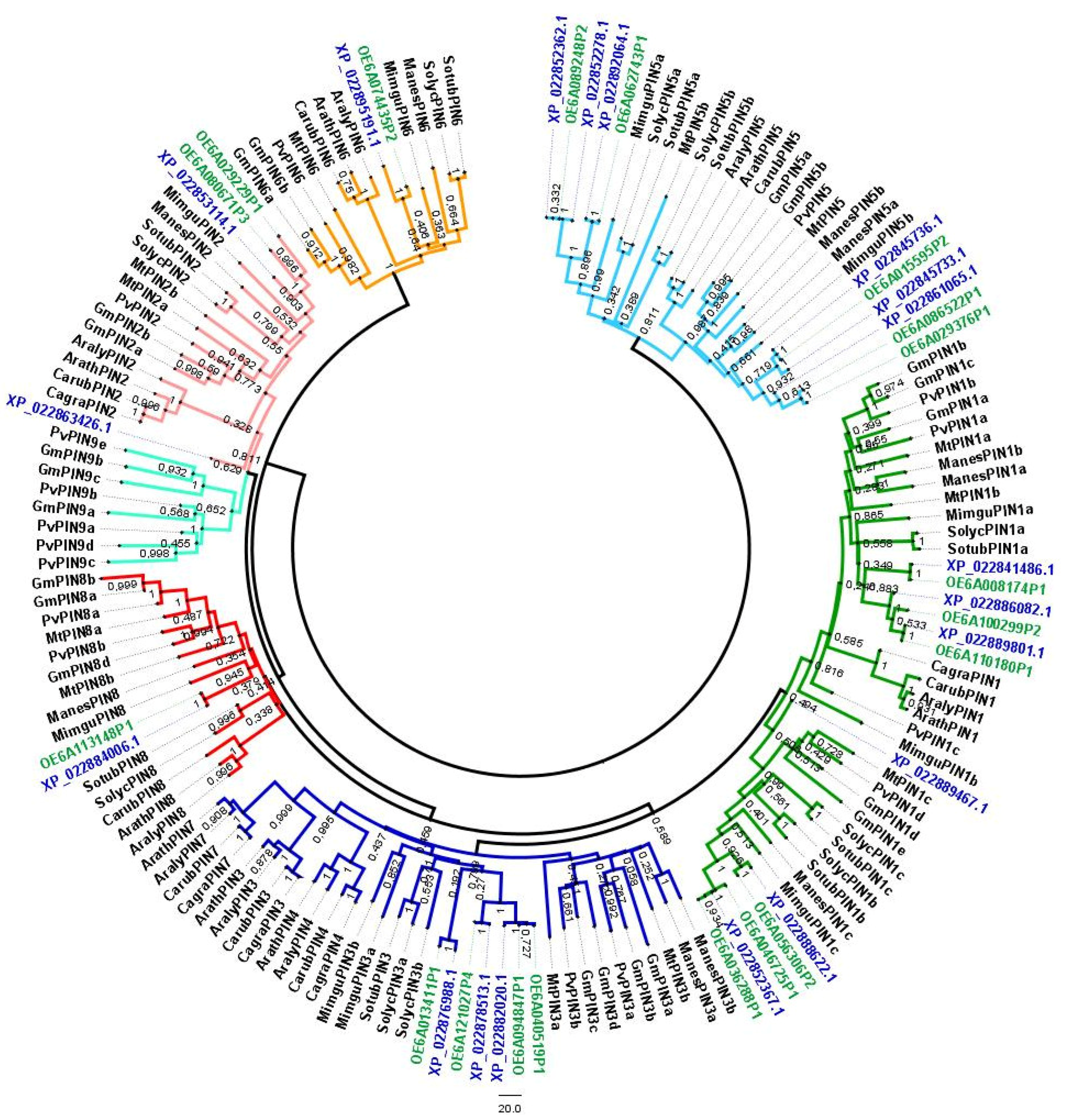
2.1. Identification of OePIN Genes and Phylogenetic Analysis
2.2. In Silico Analysis of PIN Structure and Identification of Regulatory Elements within Genic Regions
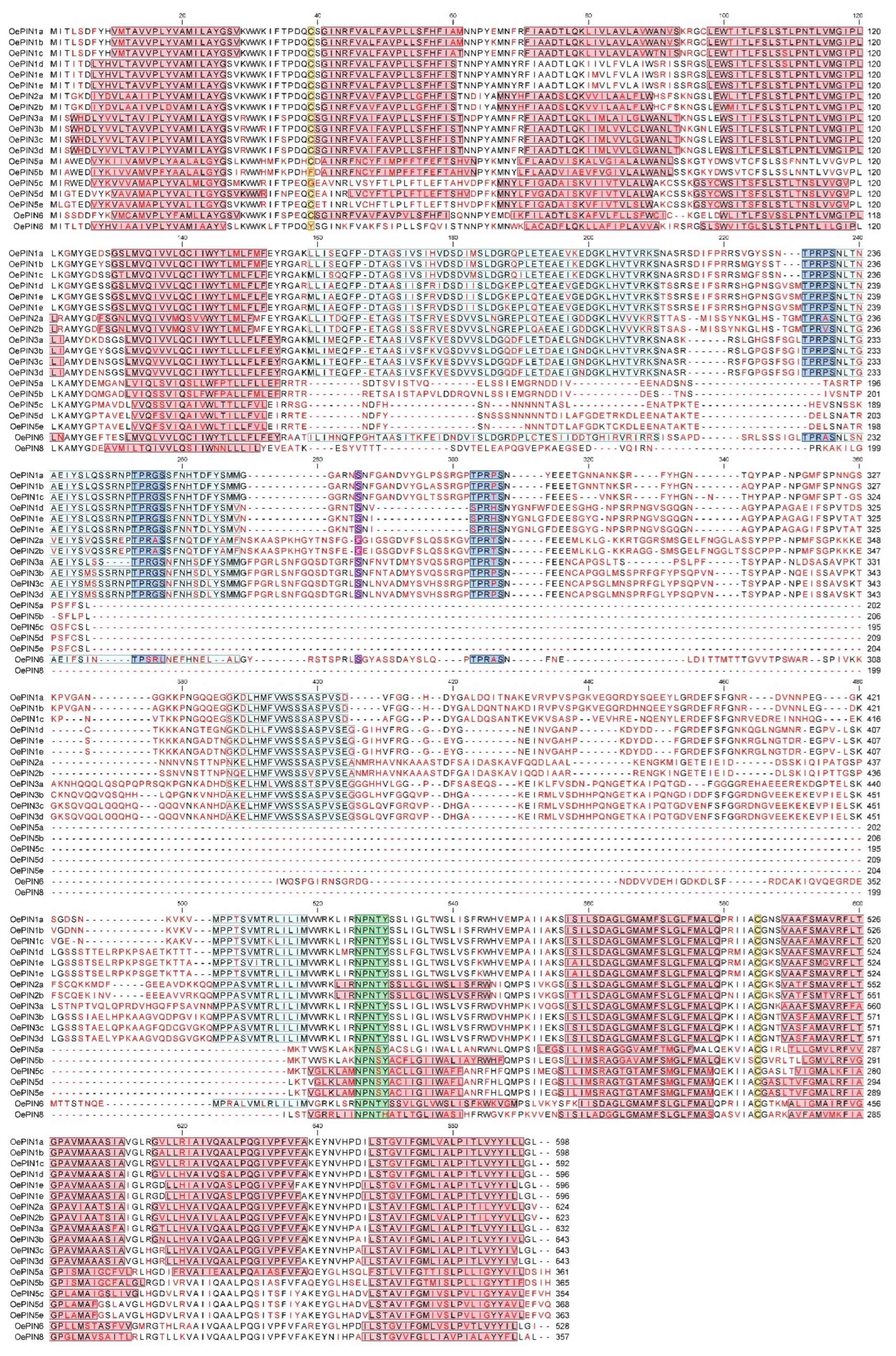
2.3. Identification of PIN Conserved Protein Structural Features
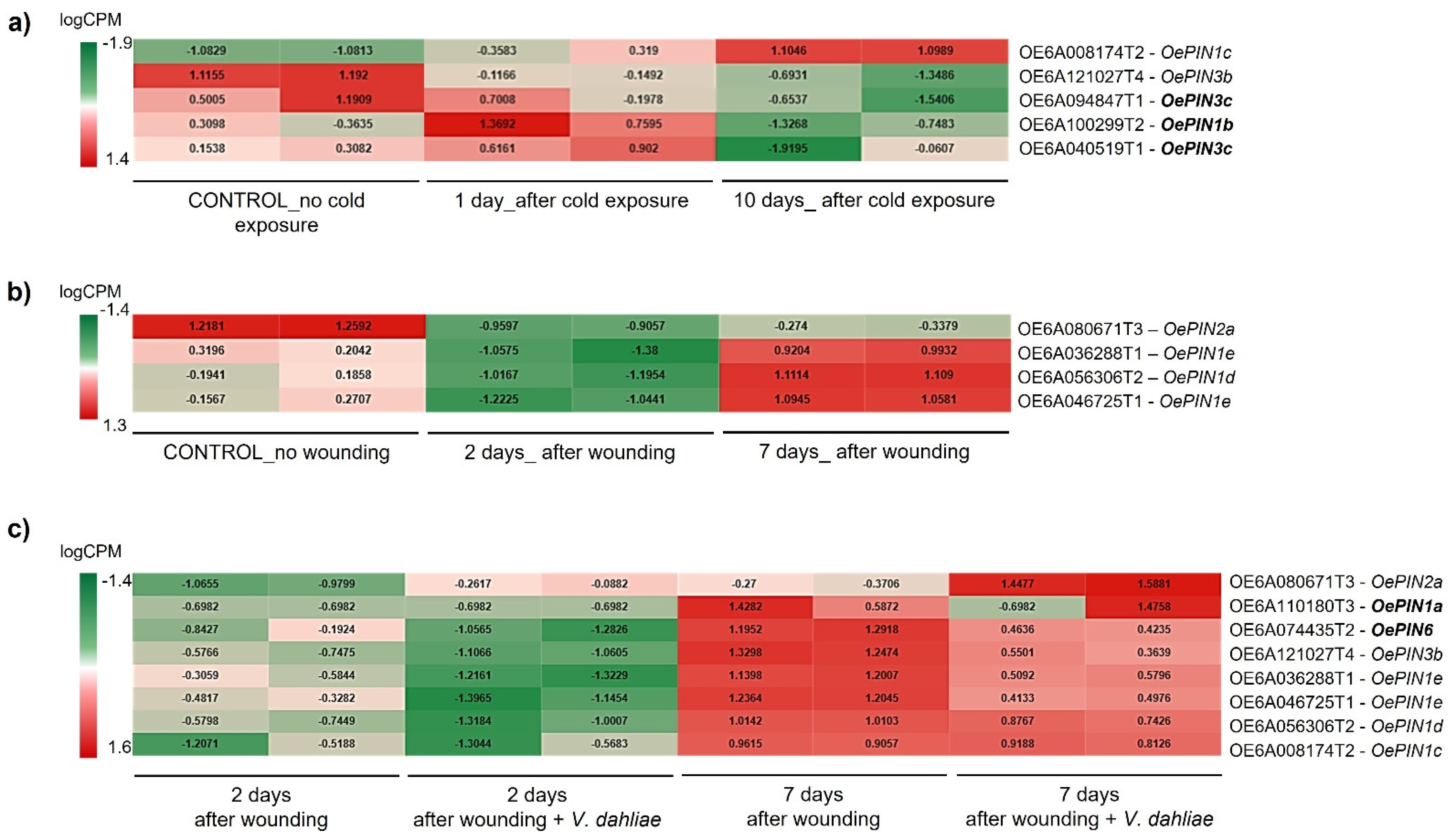
2.4. Mapping Procedures and Identification of Differentially Expressed Genes (DEGs)
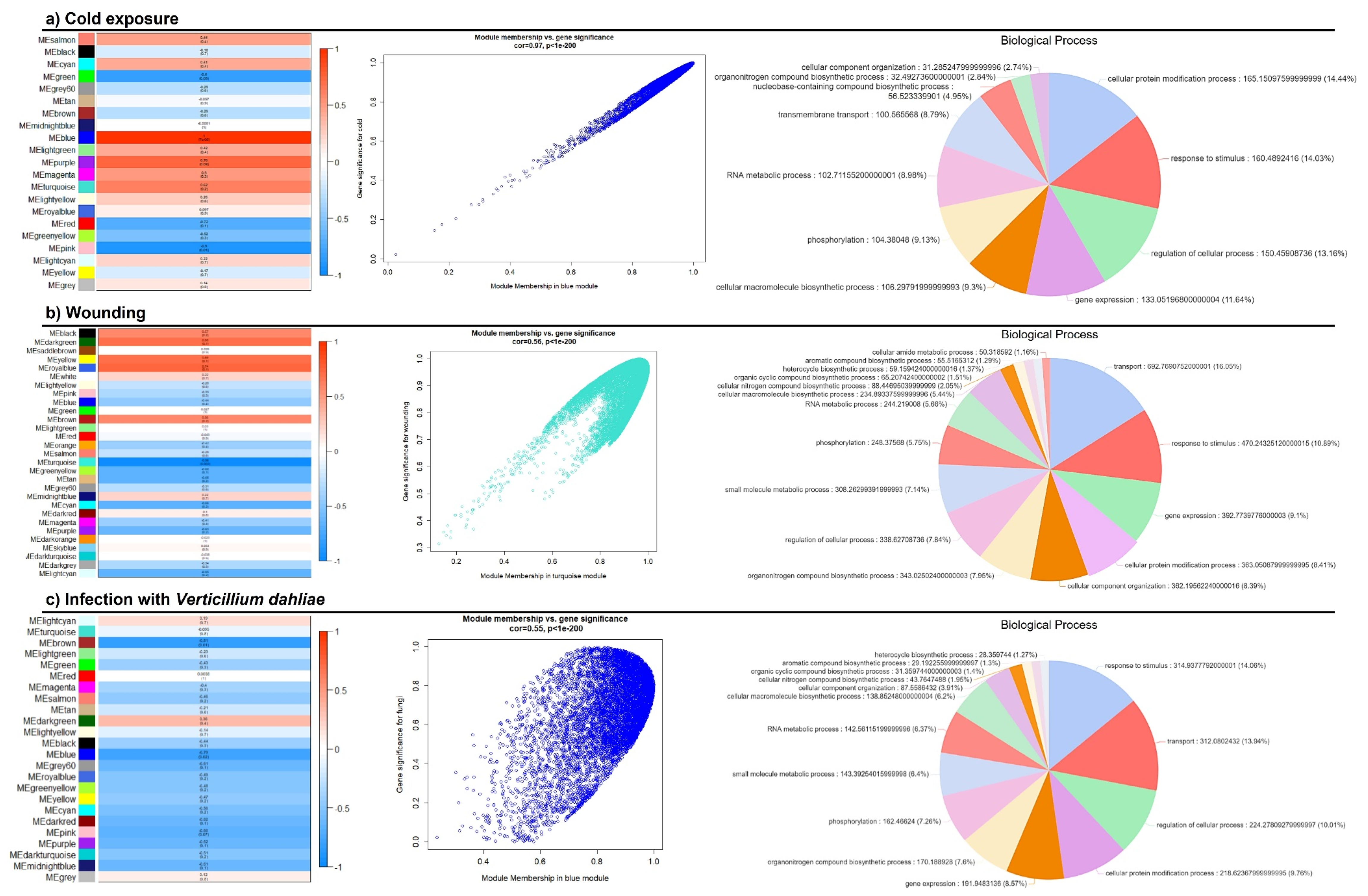
2.5. Construction of Gene Co-Expression Networks
3. Results
3.1. Identification of OePIN Genes—Chromosomal Distribution, Gene Structure and Phylogeny Analysis
3.2. Protein Sequence Analysis
3.3. High-Throughput Expression Analysis of OePINs
3.4. Weighted Gene Co-Expression Network Analysis (WGCNA)
4. Discussion
4.1. OePIN Gene Family Reveals High Diversity and a Genotype-Specific Radiation Pattern
4.2. Variability of OePIN Gene Structure Resulting from Mechanisms Acting at Genomic and Transcriptomic Level
4.3. OePINs Are Co-Expressed with Members Belonging to Diverse Stress Signaling Pathways and Oxidative Stress Homeostasis upon Biotic and Abiotic Stresses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brackmann, K.; Qi, J.; Gebert, M.; Jouannet, V.; Schlamp, T.; Grünwald, K.; Wallner, E.-S.; Novikova, D.D.; Levitsky, V.G.; Agustí, J.; et al. Spatial Specificity of Auxin Responses Coordinates Wood Formation. Nat. Commun. 2018, 9, 875. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Villalon, A. Wiring a Plant: Genetic Networks for Phloem Formation in Arabidopsis thaliana Roots. New Phytol. 2016, 210, 45–50. [Google Scholar] [CrossRef] [PubMed]
- An, J.; LIU, X.; LI, H.; YOU, C.; SHU, J.; WANG, X.; HAO, Y. Molecular Cloning and Functional Characterization of MdPIN1 in Apple. J. Integr. Agric. 2017, 16, 1103–1111. [Google Scholar] [CrossRef]
- Gou, J.; Strauss, S.H.; Tsai, C.J.; Fang, K.; Chen, Y.; Jiang, X.; Busov, V.B. Gibberellins Regulate Lateral Root Formation in Populus through Interactions with Auxin and Other Hormones. Plant Cell 2010, 22, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Palovaara, J.; Hallberg, H.; Stasolla, C.; Luit, B.; Hakman, I. Expression of a Gymnosperm PIN Homologous Gene Correlates with Auxin Immunolocalization Pattern at Cotyledon Formation and in Demarcation of the Procambium during Picea Abies Somatic Embryo Development and in Seedling Tissues. Tree Physiol. 2010, 30, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Pahari, S.; Cormark, R.D.; Blackshaw, M.T.; Liu, C.; Erickson, J.L.; Schultz, E.A. Arabidopsis UNHINGED Encodes a VPS51 Homolog and Reveals a Role for the GARP Complex in Leaf Shape and Vein Patterning. Development 2014, 141, 1894–1905. [Google Scholar] [CrossRef]
- Hakman, I.; Hallberg, H.; Palovaara, J. The Polar Auxin Transport Inhibitor NPA Impairs Embryo Morphology and Increases the Expression of an Auxin Efflux Facilitator Protein PIN during Picea Abies Somatic Embryo Development. Tree Physiol. 2009, 29, 483–496. [Google Scholar] [CrossRef]
- Petrásek, J.; Friml, J. Auxin Transport Routes in Plant Development. Dev. Camb. Engl. 2009, 136, 2675–2688. [Google Scholar] [CrossRef]
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef]
- Salazar, R.; Pollmann, S.; Morales-Quintana, L.; Herrera, R.; Caparrós-Ruiz, D.; Ramos, P. In Seedlings of Pinus radiata, Jasmonic Acid and Auxin Are Differentially Distributed on Opposite Sides of Tilted Stems Affecting Lignin Monomer Biosynthesis and Composition. Plant Physiol. Biochem. 2019, 135, 215–223. [Google Scholar] [CrossRef]
- Campos, M.D.; Nogales, A.; Cardoso, H.G.; Kumar, S.R.; Nobre, T.; Sathishkumar, R.; Arnholdt-Schmitt, B. Stress-Induced Accumulation of DcAOX1 and DcAOX2a Transcripts Coincides with Critical Time Point for Structural Biomass Prediction in Carrot Primary Cultures (Daucus carota L.). Front. Genet. 2016, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Velada, I.; Grzebelus, D.; Lousa, D.; Soares, C.M.; Santos Macedo, E.; Peixe, A.; Arnholdt-Schmitt, B.; Cardoso, H.G. AOX1-Subfamily Gene Members in Olea europaea Cv. “Galega Vulgar”—Gene Characterization and Expression of Transcripts during IBA-Induced in Vitro Adventitious Rooting. Int. J. Mol. Sci. 2018, 19, 597. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, H.G.; Campos, M.C.; Pais, M.S.; Peixe, A. Use of Morphometric Parameters for Tracking Ovule and Microspore Evolution in Grapevine (Vitis vinifera L., Cv. “Aragonez”) and Evaluation of Their Potential to Improve in Vitro Somatic Embryogenesis Efficiency from Gametophyte Tissues. In Vitro Cell. Dev. Biol. Plant 2010, 46, 499–508. [Google Scholar] [CrossRef][Green Version]
- Pires, R.; Cardoso, H.; Ribeiro, A.; Peixe, A.; Cordeiro, A. Somatic Embryogenesis from Mature Embryos of Olea europaea L. Cv. ‘Galega Vulgar’ and Long-Term Management of Calli Morphogenic Capacity. Plants 2020, 9, 758. [Google Scholar] [CrossRef]
- Cardoso, H.G.; Arnholdt-Schmitt, B. Functional Marker Development Across Species in Selected Traits. In Diagnostics in Plant Breeding; Lübberstedt, T., Varshney, R.K., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 467–515. ISBN 978-94-007-5687-8. [Google Scholar]
- Arnholdt-Schmitt, B.; Ragonezi, C.; Cardoso, H. Do Mitochondria Play a Central Role in Stress-Induced Somatic Embryogenesis? Methods Mol. Biol. Clifton NJ 2016, 1359, 87–100. [Google Scholar] [CrossRef]
- Korver, R.A.; Koevoets, I.T.; Testerink, C. Out of Shape During Stress: A Key Role for Auxin. Trends Plant Sci. 2018, 23, 783–793. [Google Scholar] [CrossRef]
- Blakeslee, J.J.; Spatola Rossi, T.; Kriechbaumer, V. Auxin Biosynthesis: Spatial Regulation and Adaptation to Stress. J. Exp. Bot. 2019, 70, 5041–5049. [Google Scholar] [CrossRef]
- Casanova-Sáez, R.; Mateo-Bonmatí, E.; Ljung, K. Auxin Metabolism in Plants. In Auxin Signaling: From Synthesis to Systems Biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2022; pp. 27–48. [Google Scholar]
- Finet, C.; Jaillais, Y. AUXOLOGY: When Auxin Meets Plant Evo-Devo. Dev. Biol. 2012, 369, 19–31. [Google Scholar] [CrossRef]
- Balzan, S.; Johal, G.S.; Carraro, N. The Role of Auxin Transporters in Monocots Development. Front. Plant Sci. 2014, 5, 393. [Google Scholar] [CrossRef]
- Grones, P.; Friml, J. Auxin Transporters and Binding Proteins at a Glance. J. Cell Sci. 2015, 128, 1–7. [Google Scholar] [CrossRef]
- Zhou, J.-J.; Luo, J. The PIN-FORMED Auxin Efflux Carriers in Plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef] [PubMed]
- Hille, S.; Akhmanova, M.; Glanc, M.; Johnson, A.; Friml, J. Relative Contribution of PIN-Containing Secretory Vesicles and Plasma Membrane PINs to the Directed Auxin Transport: Theoretical Estimation. Int. J. Mol. Sci. 2018, 19, 3566. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Benková, E.; Blilou, I.; Wisniewska, J.; Hamann, T.; Ljung, K.; Woody, S.; Sandberg, G.; Scheres, B.; Jürgens, G.; et al. AtPIN4 Mediates Sink-Driven Auxin Gradients and Root Patterning in Arabidopsis. Cell 2002, 108, 661–673. [Google Scholar] [CrossRef]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-Dependent Auxin Gradients Establish the Apical-Basal Axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef]
- Mravec, J.; Skůpa, P.; Bailly, A.; Hoyerová, K.; Krecek, P.; Bielach, A.; Petrásek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.-D.; et al. Subcellular Homeostasis of Phytohormone Auxin Is Mediated by the ER-Localized PIN5 Transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Paponov, I.A.; Teale, W.D.; Trebar, M.; Blilou, I.; Palme, K. The PIN Auxin Efflux Facilitators: Evolutionary and Functional Perspectives. Trends Plant Sci. 2005, 10, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chai, C.; Valliyodan, B.; Maupin, C.; Annen, B.; Nguyen, H.T. Genome-Wide Analysis and Expression Profiling of the PIN Auxin Transporter Gene Family in Soybean (Glycine max). BMC Genom. 2015, 16, 951. [Google Scholar] [CrossRef]
- Forestan, C.; Farinati, S.; Varotto, S. The Maize PIN Gene Family of Auxin Transporters. Front. Plant Sci. 2012, 3, 16. [Google Scholar] [CrossRef]
- Roumeliotis, E.; Kloosterman, B.; Oortwijn, M.; Visser, R.; Bachem, C. The PIN Family of Proteins in Potato and Their Putative Role in Tuberization. Front. Plant Sci. 2013, 4, 524. [Google Scholar] [CrossRef]
- Zhang, Y.; He, P.; Yang, Z.; Huang, G.; Wang, L.; Pang, C.; Xiao, H.; Zhao, P.; Yu, J.; Xiao, G. A Genome-Scale Analysis of the PIN Gene Family Reveals Its Functions in Cotton Fiber Development. Front. Plant Sci. 2017, 8, 461. [Google Scholar] [CrossRef]
- Hou, M.; Luo, F.; Wu, D.; Zhang, X.; Lou, M.; Shen, D.; Yan, M.; Mao, C.; Fan, X.; Xu, G.; et al. OsPIN9, an Auxin Efflux Carrier, Is Required for the Regulation of Rice Tiller Bud Outgrowth by Ammonium. New Phytol. 2021, 229, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Bai, X.; Guo, T.; Xie, Z.; Laimer, M.; Du, D.; Gbokie, T.; Zhang, Z.; He, C.; Lu, Y.; et al. Genome-Wide Analysis of the PIN Auxin Efflux Carrier Gene Family in Coffee. Plants 2020, 9, 1061. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Chen, L.; Wang, C.; Zhang, S.; Yang, Y.; Liu, J.; Li, D.; Song, J.; Wang, R. Characterization of the Auxin Efflux Transporter PIN Proteins in Pear. Plants 2020, 9, 349. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Friml, J. PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef]
- Abdollahi Sisi, N.; Růžička, K. ER-Localized PIN Carriers: Regulators of Intracellular Auxin Homeostasis. Plants 2020, 9, 1527. [Google Scholar] [CrossRef] [PubMed]
- Kühn, N.; Serrano, A.; Abello, C.; Arce, A.; Espinoza, C.; Gouthu, S.; Deluc, L.; Arce-Johnson, P. Regulation of Polar Auxin Transport in Grapevine Fruitlets (Vitis vinifera L.) and the Proposed Role of Auxin Homeostasis during Fruit Abscission. BMC Plant Biol. 2016, 16, 234. [Google Scholar] [CrossRef]
- Song, C.; Zhang, D.; Zhang, J.; Zheng, L.; Zhao, C.; Ma, J.; An, N.; Han, M. Expression Analysis of Key Auxin Synthesis, Transport, and Metabolism Genes in Different Young Dwarfing Apple Trees. Acta Physiol. Plant. 2016, 38, 43. [Google Scholar] [CrossRef]
- Carraro, N.; Tisdale-Orr, T.E.; Clouse, R.M.; Knöller, A.S.; Spicer, R. Diversification and Expression of the PIN, AUX/LAX, and ABCB Families of Putative Auxin Transporters in Populus. Front. Plant Sci. 2012, 3, 17. [Google Scholar] [CrossRef]
- Grunewald, W.; Cannoot, B.; Friml, J.; Gheysen, G. Parasitic Nematodes Modulate PIN-Mediated Auxin Transport to Facilitate Infection. PLOS Pathog. 2009, 5, e1000266. [Google Scholar] [CrossRef]
- Pasternak, T.; Rudas, V.; Potters, G.; Jansen, M.A.K. Morphogenic Effects of Abiotic Stress: Reorientation of Growth in Arabidopsis thaliana Seedlings. Environ. Exp. Bot. 2005, 53, 299–314. [Google Scholar] [CrossRef]
- Shibasaki, K.; Uemura, M.; Tsurumi, S.; Rahman, A. Auxin Response in Arabidopsis under Cold Stress: Underlying Molecular Mechanisms. Plant Cell 2009, 21, 3823–3838. [Google Scholar] [CrossRef] [PubMed]
- Ruedell, C.M.; de Almeida, M.R.; Fett-Neto, A.G. Concerted Transcription of Auxin and Carbohydrate Homeostasis-Related Genes Underlies Improved Adventitious Rooting of Microcuttings Derived from Far-Red Treated Eucalyptus globulus Labill Mother Plants. Plant Physiol. Biochem. 2015, 97, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Velada, I.; Cardoso, H.; Porfirio, S.; Peixe, A. Expression Profile of PIN-Formed Auxin Efflux Carrier Genes during IBA-Induced In Vitro Adventitious Rooting in Olea europaea L. Plants 2020, 9, 185. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for Inferring Very Large Phylogenies by Using the Neighbor-Joining Method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v1.3.1; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2010. [Google Scholar]
- Kohany, O.; Gentles, A.J.; Hankus, L.; Jurka, J. Annotation, Submission and Screening of Repetitive Elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 2006, 7, 474. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 95–98. [Google Scholar]
- Barghini, E.; Natali, L.; Giordani, T.; Cossu, R.M.; Scalabrin, S.; Cattonaro, F.; Šimková, H.; Vrána, J.; Doležel, J.; Morgante, M.; et al. LTR Retrotransposon Dynamics in the Evolution of the Olive (Olea europaea) Genome. DNA Res. 2015, 22, 91–100. [Google Scholar] [CrossRef]
- Nakai, K.; Kanehisa, M. Expert System for Predicting Protein Localization Sites in Gram-Negative Bacteria. Proteins 1991, 11, 95–110. [Google Scholar] [CrossRef]
- Bennett, T.; Brockington, S.F.; Rothfels, C.; Graham, S.W.; Stevenson, D.; Kutchan, T.; Rolf, M.; Thomas, P.; Wong, G.K.-S.; Leyser, O.; et al. Paralogous Radiations of PIN Proteins with Multiple Origins of Noncanonical PIN Structure. Mol. Biol. Evol. 2014, 31, 2042–2060. [Google Scholar] [CrossRef]
- Leyva-Pérez, M.d.l.O.; Valverde-Corredor, A.; Valderrama, R.; Jiménez-Ruiz, J.; Muñoz-Merida, A.; Trelles, O.; Barroso, J.B.; Mercado-Blanco, J.; Luque, F. Early and Delayed Long-Term Transcriptional Changes and Short-Term Transient Responses during Cold Acclimation in Olive Leaves. DNA Res. 2015, 22, 1–11. [Google Scholar] [CrossRef]
- Jiménez-Ruiz, J.; Leyva-Pérez, M.d.l.O.; Schilirò, E.; Barroso, J.B.; Bombarely, A.; Mueller, L.; Mercado-Blanco, J.; Luque, F. Transcriptomic Analysis of Olea europaea L. Roots during the Verticillium dahliae Early Infection Process. Plant Genome 2017, 10, plantgenome2016.07.0060. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Cruz, F.; Julca, I.; Gómez-Garrido, J.; Loska, D.; Marcet-Houben, M.; Cano, E.; Galán, B.; Frias, L.; Ribeca, P.; Derdak, S.; et al. Genome Sequence of the Olive Tree, Olea europaea. GigaScience 2016, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Barak, L.S.; Ménard, L.; Ferguson, S.S.; Colapietro, A.M.; Caron, M.G. The Conserved Seven-Transmembrane Sequence NP(X)2,3Y of the G-Protein-Coupled Receptor Superfamily Regulates Multiple Properties of the Beta 2-Adrenergic Receptor. Biochemistry 1995, 34, 15407–15414. [Google Scholar] [CrossRef]
- Retzer, K.; Lacek, J.; Skokan, R.; Del Genio, C.I.; Vosolsobě, S.; Laňková, M.; Malínská, K.; Konstantinova, N.; Zažímalová, E.; Napier, R.M.; et al. Evolutionary Conserved Cysteines Function as Cis-Acting Regulators of Arabidopsis PIN-FORMED 2 Distribution. Int. J. Mol. Sci. 2017, 18, 2274. [Google Scholar] [CrossRef]
- Julca, I.; Marcet-Houben, M.; Vargas, P.; Gabaldón, T. Phylogenomics of the Olive Tree (Olea europaea) Reveals the Relative Contribution of Ancient Allo- and Autopolyploidization Events. BMC Biol. 2018, 16, 15. [Google Scholar] [CrossRef]
- Mandáková, T.; Li, Z.; Barker, M.S.; Lysak, M.A. Diverse Genome Organization Following 13 Independent Mesopolyploid Events in Brassicaceae Contrasts with Convergent Patterns of Gene Retention. Plant J. 2017, 91, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; DePamphilis, C.W. Patterns of Gene Duplication in the Plant SKP1 Gene Family in Angiosperms: Evidence for Multiple Mechanisms of Rapid Gene Birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.; Zhang, J.; Liu, X.; Lin, C.; Xin, H.; Xue, L.; Wang, C. De Novo Assembly of a New Olea europaea Genome Accession Using Nanopore Sequencing. Hortic. Res. 2021, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, D.; Zhang, C.; Kong, N.; Ma, H.; Chen, Q. Comparative Analysis of the PIN Auxin Transporter Gene Family in Different Plant Species: A Focus on Structural and Expression Profiling of PINs in Solanum tuberosum. Int. J. Mol. Sci. 2019, 20, 3270. [Google Scholar] [CrossRef]
- Velada, I.; Cardoso, H.G.; Ragonezi, C.; Nogales, A.; Ferreira, A.; Valadas, V.; Arnholdt-Schmitt, B. Alternative Oxidase Gene Family in Hypericum perforatum L.: Characterization and Expression at the Post-Germinative Phase. Front. Plant Sci. 2016, 7, 1043. [Google Scholar] [CrossRef]
- Cardoso, H.G.; Nogales, A.; Frederico, A.M.; Svensson, J.T.; Macedo, E.S.; Valadas, V.; Arnholdt-Schmitt, B. Exploring AOX Gene Diversity. In Alternative Respiratory Pathways in Higher Plants; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015; pp. 239–254. ISBN 978-1-118-78997-1. [Google Scholar]
- Knowles, D.G.; McLysaght, A. High Rate of Recent Intron Gain and Loss in Simultaneously Duplicated Arabidopsis Genes. Mol. Biol. Evol. 2006, 23, 1548–1557. [Google Scholar] [CrossRef]
- Fiume, E.; Christou, P.; Gianì, S.; Breviario, D. Introns Are Key Regulatory Elements of Rice Tubulin Expression. Planta 2004, 218, 693–703. [Google Scholar] [CrossRef]
- Zhang, J.; Gu, H.; Dai, H.; Zhang, Z.; Miao, M. Alternative Polyadenylation of the Stacyose Synthase Gene Mediates Source-Sink Regulation in Cucumber. J. Plant Physiol. 2020, 245, 153111. [Google Scholar] [CrossRef]
- Tian, B.; Manley, J.L. Alternative Polyadenylation of MRNA Precursors. Nat. Rev. Mol. Cell Biol. 2017, 18, 18–30. [Google Scholar] [CrossRef]
- Tu, Z.; Shen, Y.; Wen, S.; Liu, H.; Wei, L.; Li, H. A Tissue-Specific Landscape of Alternative Polyadenylation, LncRNAs, TFs, and Gene Co-Expression Networks in Liriodendron chinense. Front. Plant Sci. 2021, 12, 1506. [Google Scholar] [CrossRef]
- Choi, J.Y.; Lee, Y.C.G. Double-Edged Sword: The Evolutionary Consequences of the Epigenetic Silencing of Transposable Elements. PLoS Genet. 2020, 16, e1008872. [Google Scholar] [CrossRef] [PubMed]
- Negi, P.; Rai, A.N.; Suprasanna, P. Moving through the Stressed Genome: Emerging Regulatory Roles for Transposons in Plant Stress Response. Front. Plant Sci. 2016, 7, 1448. [Google Scholar] [CrossRef] [PubMed]
- Thieme, M.; Bucher, E. Chapter Six—Transposable Elements as Tool for Crop Improvement. In Advances in Botanical Research; Mirouze, M., Bucher, E., Gallusci, P., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 88, pp. 165–202. [Google Scholar]
- Giles, N.M.; Watts, A.B.; Giles, G.I.; Fry, F.H.; Littlechild, J.A.; Jacob, C. Metal and Redox Modulation of Cysteine Protein Function. Chem. Biol. 2003, 10, 677–693. [Google Scholar] [CrossRef]
- Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.-Q. Interplay between Reactive Oxygen Species and Hormones in the Control of Plant Development and Stress Tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef]
- Cardoso, H.; Peixe, A.; Bellini, C.; Porfírio, S.; Druege, U. Editorial: Advances on the Biological Mechanisms Involved in Adventitious Root Formation: From Signaling to Morphogenesis. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Wilkins, K.A.; Matthus, E.; Swarbreck, S.M.; Davies, J.M. Calcium-Mediated Abiotic Stress Signaling in Roots. Front. Plant Sci. 2016, 7, 1296. [Google Scholar] [CrossRef]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.-P. Calcium Signalling in Plant Biotic Interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef]
- Tognetti, V.B.; Mühlenbock, P.; Van Breusegem, F. Stress Homeostasis—The Redox and Auxin Perspective. Plant Cell Environ. 2012, 35, 321–333. [Google Scholar] [CrossRef]
- Grunewald, W.; Friml, J. The March of the PINs: Developmental Plasticity by Dynamic Polar Targeting in Plant Cells. EMBO J. 2010, 29, 2700–2714. [Google Scholar] [CrossRef]
- Campos, M.D.; Campos, C.; Nogales, A.; Cardoso, H. Carrot AOX2a Transcript Profile Responds to Growth and Chilling Exposure. Plants 2021, 10, 2369. [Google Scholar] [CrossRef]
- Fradin, E.F.; Thomma, B.P.H.J. Physiology and Molecular Aspects of Verticillium Wilt Diseases Caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Prieto, P.; Navarro-Raya, C.; Valverde-Corredor, A.; Amyotte, S.G.; Dobinson, K.F.; Mercado-Blanco, J. Colonization Process of Olive Tissues by Verticillium dahliae and Its in Planta Interaction with the Biocontrol Root Endophyte Pseudomonas fluorescens PICF7. Microb. Biotechnol. 2009, 2, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Meents, A.K.; Furch, A.C.U.; Almeida-Trapp, M.; Özyürek, S.; Scholz, S.S.; Kirbis, A.; Lenser, T.; Theißen, G.; Grabe, V.; Hansson, B.; et al. Beneficial and Pathogenic Arabidopsis Root-Interacting Fungi Differently Affect Auxin Levels and Responsive Genes During Early Infection. Front. Microbiol. 2019, 10, 380. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Hentrich, M.; Pollmann, S. Auxin-Oxylipin Crosstalk: Relationship of AntagonistsF. J. Integr. Plant Biol. 2011, 53, 429–445. [Google Scholar] [CrossRef]

| Gene | Locus ID | Gene (bp) | Orf (bp) | Event |
|---|---|---|---|---|
| PIN1 subfamily | ||||
| OePIN1a | OE6A110180P1 | 3287 | 1794 | - |
| OE6A110180P2 | 1731 | APA_Int5 | ||
| OE6A110180P3 | 1677 | APA_Int4 | ||
| OE6A110180P4 | 1530 | APA_Int3 | ||
| OePIN1b | OE6A100299P2 | 3818 | 1794 | - |
| OePIN1c | OE6A008174P1 | 3192 | 1776 | - |
| OE6A008174P2 | 1707 | APA_Int4 | ||
| OePIN1d | OE6A056306P2 | 2853 | 1788 | - |
| OE6A056306P3 | 1404 | APA_Int2 | ||
| OePIN1e | OE6A046725P1 | 2746 | 1788 | - |
| OePIN1e | OE6A036288P1 | 2740 | 1788 | - |
| * | OE6A045718P1 | 1681 | 858 | - |
| * | OE6A004471P1 | 1191 | 1188 | - |
| PIN2 subfamily | ||||
| OePIN2a | OE6A080671P1 | 1557 | APA_Int2 | |
| OE6A080671P2 | 1685 | APA_Int3 | ||
| OE6A080671P3 | 3332 | 1872 | - | |
| OePIN2b | OE6A029229P1 | 9225 | 1869 | - |
| PIN3 subfamily | ||||
| OePIN3a | OE6A013411P1 | 2873 | 1896 | - |
| OE6A013411P2 | 1866 | AS_Int1 | ||
| OePIN3b | OE6A121027P3 | 1866 | APA_Int5 + AS_Int4 | |
| OE6A121027P4 | 2927 | 1929 | - | |
| OePIN3c | OE6A040519P1 | 2897 | 1929 | - |
| OE6A040519P2 | 1905 | AS_Int1 | ||
| OePIN3c | OE6A094847P1 | 2881 | 1929 | - |
| OE6A094847P2 | 1866 | APA_Int5 | ||
| OE6A094847P3 | 1905 | AS_Int1 | ||
| PIN5 subfamily | ||||
| OePIN5a | OE6A089248P1 | 1017 | APA_Int3 | |
| OE6A089248P2 | 1689 | 1149 | - | |
| OePIN5b | OE6A062743P1 | 1567 | 1095 | - |
| OePIN5c | OE6A015595P1 | 1041 | AS_Int1 | |
| OE6A015595P2 | 1681 | 1062 | - | |
| OE6A015595P3 | 1002 | APA_Int3 | ||
| OE6A015595P5 | 1164 | AS_Int2 | ||
| OePIN5d | OE6A086522P1 | 1736 | 1104 | - |
| OePIN5e | OE6A029376P1 | 1713 | 1089 | - |
| PIN6 subfamily | ||||
| OePIN6 | OE6A074435P1 | 1506 | APA_Ex7 | |
| OE6A074435P2 | 5086 | 1584 | ||
| PIN8 subfamily | ||||
| OePIN8 | OE6A113148P1 | 2376 | 1071 | - |
| Gene | Locus ID | Chr. | Position | Gene (bp) | ORF (bp) | Accession | Event |
|---|---|---|---|---|---|---|---|
| PIN1 subfamily | |||||||
| OePIN1a | NC_036248 | 12 | 3,293,149..3,297,089 | 3288 | 1794 | X1: XM_023034033.1 X2: XM_023034034.1 X3: XM_023034035.1 | - |
| 1794 | - | ||||||
| 3308 | 1794 | APA_int5+AS3′utr | |||||
| OePIN1b | NC_036237 | 1 | 58,361..60,794 | 1697 | 1530 | XM_023030314.1 | |
| OePIN1c | NC_036253 | 17 | 15,003,301..15,007,290 | 3187 | 1776 | X1: XM_022985718.1 | |
| 1776 | X2: XM_022985719.1 | ||||||
| OePIN1d | NC_036247 | 11 | 32,656,543..32,660,906 | 3639 | 1800 | XM_023032854.1 | |
| OePIN1e | NW_019229312 | Unk | 244,819..247,974 | 2741 | 1788 | XM_022996599.1 | |
| * | NC_036247 | 11 | 32,669,953..32,671,485 | 1212 | 597 | XM_023033699.1 | |
| PIN2 subfamily | |||||||
| OePIN2a | NC_036238 | 2 | 26,682,952..26,689,244 | 1457 | 1338 | XM_022997346.1 | |
| OePIN2b | NW_019245527 | Unk | 38678..41693 | 2646 | 1101 | XM_023007658.1 | |
| PIN3 subfamily | |||||||
| OePIN3a | NC_036237 | 1 | 14651491..14655627 | 2878 | 1896 | XM_023021220.1 | |
| OePIN3b | NC_036242 | 6 | 28,142,004..28,145,594 | 2924 | 1929 | XM_023022745.1 | |
| OePIN3c | NC_036244 | 8 | 17,824,834..17,828,738 | 3327 | 1929 | XM_023026252.1 | |
| PIN5 subfamily | |||||||
| OePIN5a | NC_036237 | 1 | 3,246,084..3,247,884 | 1623 | 1083 | XM_022996510.1 | |
| OePIN5a | NC_036237 | 1 | 3,254,921..3,256,721 | 1623 | 1083 | XM_022996594.1 | |
| OePIN5b | NC_036248 | 12 | 6,751,323..6,753,009 | 1572 | 1095 | XM_023036296.1 | |
| OePIN5c | NC_036255 | 19 | 6,145,679..6,147,551 | 1681 | 1062 | X1: XM_022989965.1 | - |
| 1041 | X2: XM_022989967.1 | AS_Int1 | |||||
| OePIN5c | NC_036255 | 19 | 6,156,663..6,158,539 | 1681 | 1062 | X1: XM_022989968.1 | - |
| 1041 | X2: XM_022989969.1 | AS_Int1 | |||||
| OePIN5d | NW_019241282 | Unk | 233..1706 | 1449 | 825 | XM_023005297.1 | |
| PIN6 subfamily | |||||||
| OePIN6 | NC_036250 | 14 | 1,655,349..1,660,725 | 5323 | 1539 | XM_023039423.1 | |
| PIN8 subfamily | |||||||
| OePIN8 | NC_036246 | 10 | 1,724,672..1,727,598 | 2377 | 1071 | XM_023028238.1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardoso, H.; Campos, C.; Grzebelus, D.; Egas, C.; Peixe, A. Understanding the Role of PIN Auxin Carrier Genes under Biotic and Abiotic Stresses in Olea europaea L. Biology 2022, 11, 1040. https://doi.org/10.3390/biology11071040
Cardoso H, Campos C, Grzebelus D, Egas C, Peixe A. Understanding the Role of PIN Auxin Carrier Genes under Biotic and Abiotic Stresses in Olea europaea L. Biology. 2022; 11(7):1040. https://doi.org/10.3390/biology11071040
Chicago/Turabian StyleCardoso, Hélia, Catarina Campos, Dariusz Grzebelus, Conceição Egas, and Augusto Peixe. 2022. "Understanding the Role of PIN Auxin Carrier Genes under Biotic and Abiotic Stresses in Olea europaea L." Biology 11, no. 7: 1040. https://doi.org/10.3390/biology11071040
APA StyleCardoso, H., Campos, C., Grzebelus, D., Egas, C., & Peixe, A. (2022). Understanding the Role of PIN Auxin Carrier Genes under Biotic and Abiotic Stresses in Olea europaea L. Biology, 11(7), 1040. https://doi.org/10.3390/biology11071040










