Dimerization of Firing Factors for Replication Origin Activation in Eukaryotes: A Crucial Process for Simultaneous Assembly of Bidirectional Replication Forks?
Abstract
:Simple Summary
Abstract
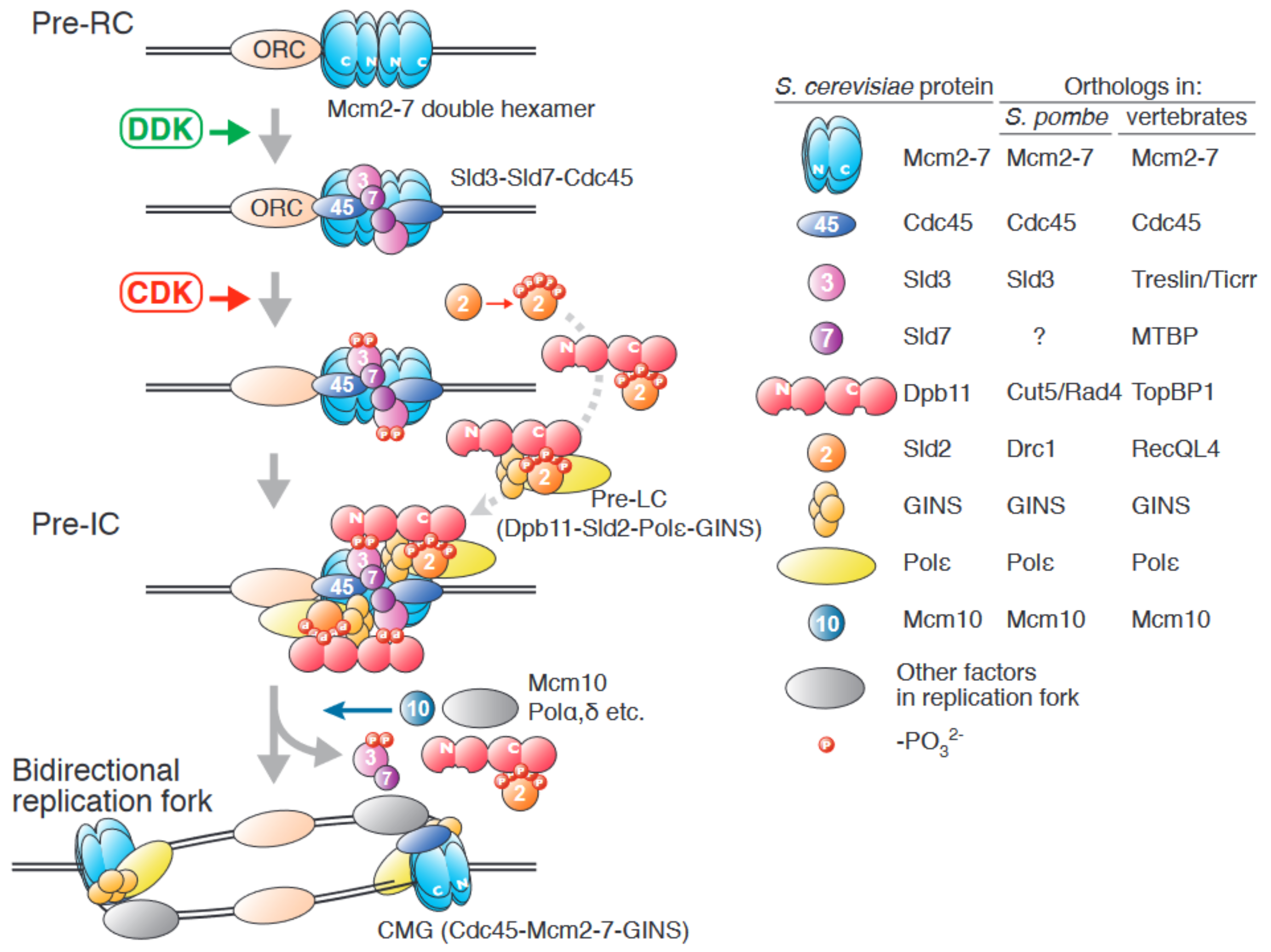
1. An Overview of Origin Firing in Eukaryotes
2. The Role of the Sld3/Treslin/Ticrr-Sld7/MTBP Complex in Origin Firing
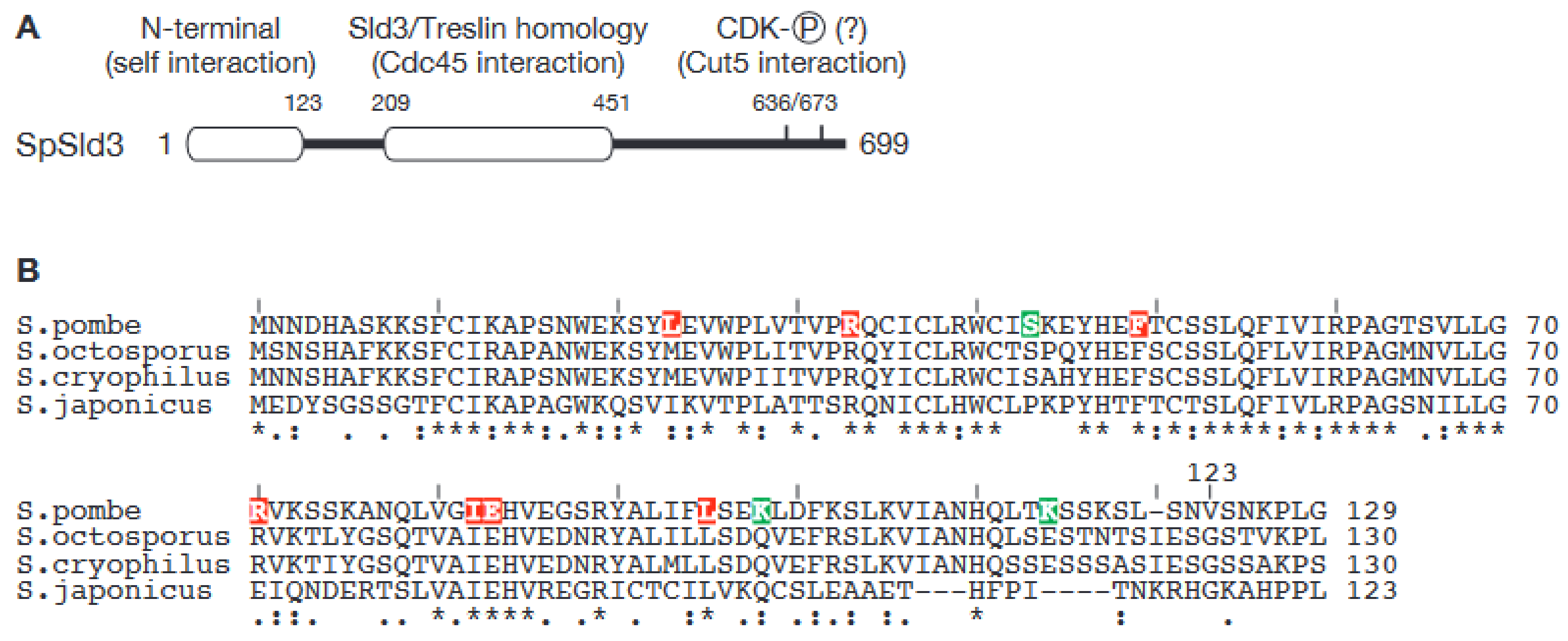
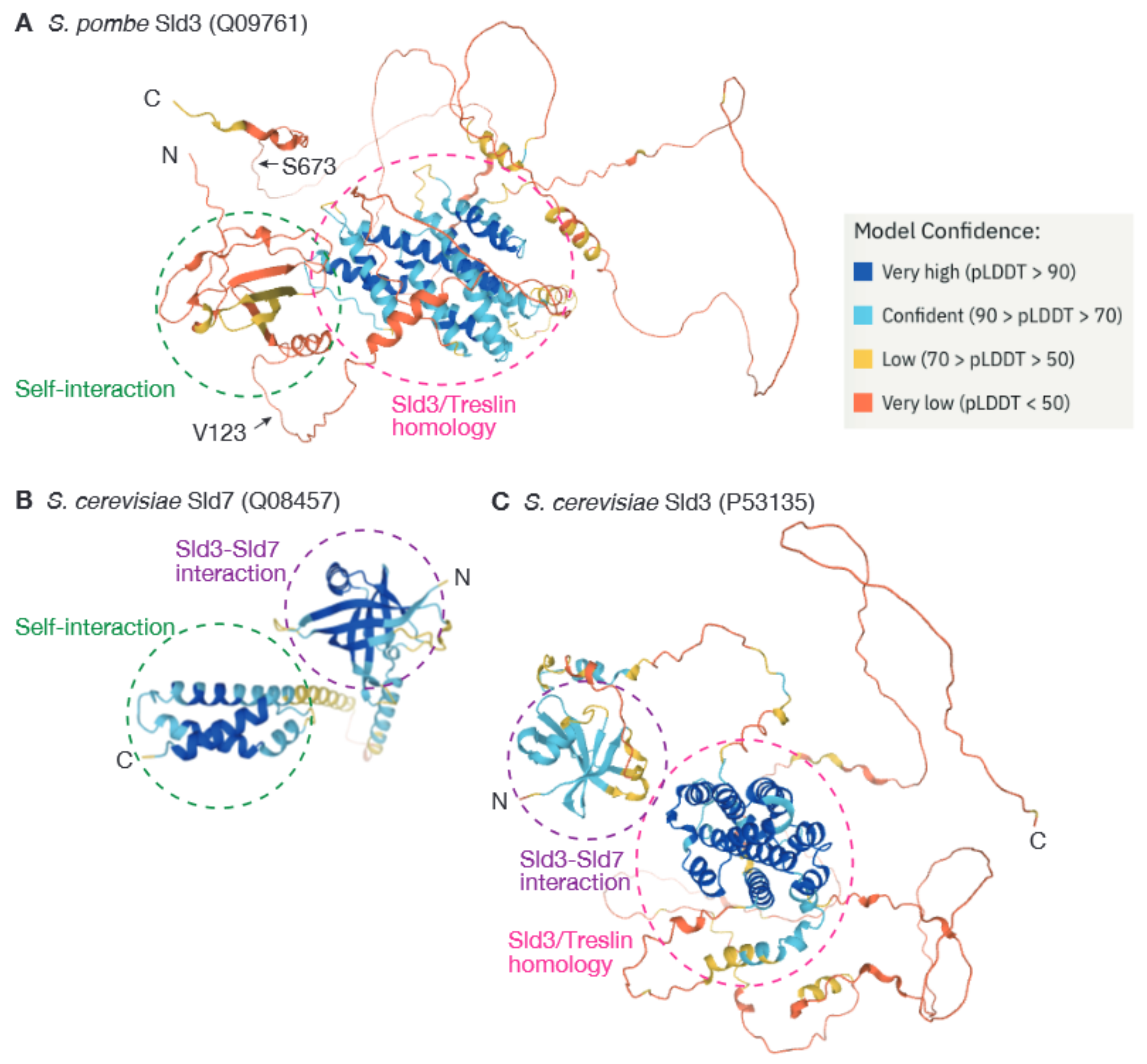
3. Dimerization of Sld3 in the Fission Yeast Schizosaccharomyces pombe Represents a Novel Mechanism for Regulation of Origin Firing
4. Dimerization of Firing Factors May Ensure the Biological Robustness of the Firing Reaction
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Remus, D.; Beuron, F.; Tolun, G.; Griffith, J.D.; Morris, E.P.; Diffley, J.F. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009, 139, 719–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, S.P.; Kaguni, J.M. Helicase loading at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, A.; Diffley, J.F.X. The Initiation of Eukaryotic DNA Replication. Annu. Rev. Biochem. 2022. [CrossRef] [PubMed]
- Moyer, S.E.; Lewis, P.W.; Botchan, M.R. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 2006, 103, 10236–10241. [Google Scholar] [CrossRef] [Green Version]
- Gambus, A.; Jones, R.C.; Sanchez-Diaz, A.; Kanemaki, M.; van Deursen, F.; Edmondson, R.D.; Labib, K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006, 8, 358–366. [Google Scholar] [CrossRef]
- Tanaka, S.; Araki, H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010371. [Google Scholar] [CrossRef] [Green Version]
- Miller, T.C.R.; Locke, J.; Greiwe, J.F.; Diffley, J.F.X.; Costa, A. Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. Nature 2019, 575, 704–710. [Google Scholar] [CrossRef]
- Coster, G.; Diffley, J.F.X. Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science 2017, 357, 314–318. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Friedman, L.J.; Gelles, J.; Bell, S.P. A helicase-tethered ORC flip enables bidirectional helicase loading. eLife 2021, 10, e74282. [Google Scholar] [CrossRef]
- Deegan, T.D.; Yeeles, J.T.; Diffley, J.F. Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. Embo. J. 2016, 35, 961–973. [Google Scholar] [CrossRef]
- Francis, L.I.; Randell, J.C.; Takara, T.J.; Uchima, L.; Bell, S.P. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009, 23, 643–654. [Google Scholar] [CrossRef] [Green Version]
- Heller, R.C.; Kang, S.; Lam, W.M.; Chen, S.; Chan, C.S.; Bell, S.P. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 2011, 146, 80–91. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, S.; Nakato, R.; Katou, Y.; Shirahige, K.; Araki, H. Origin association of sld3, sld7, and cdc45 proteins is a key step for determination of origin-firing timing. Curr. Biol. 2011, 21, 2055–2063. [Google Scholar] [CrossRef] [Green Version]
- Yeeles, J.T.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef] [Green Version]
- Randell, J.C.; Fan, A.; Chan, C.; Francis, L.I.; Heller, R.C.; Galani, K.; Bell, S.P. Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol. Cell. 2010, 40, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Kamimura, Y.; Tak, Y.S.; Sugino, A.; Araki, H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. Embo. J. 2001, 20, 2097–2107. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Umemori, T.; Endo, S.; Muramatsu, S.; Kanemaki, M.; Kamimura, Y.; Obuse, C.; Araki, H. Sld7, an Sld3-associated protein required for efficient chromosomal DNA replication in budding yeast. Embo. J. 2011, 30, 2019–2030. [Google Scholar] [CrossRef]
- Tanaka, S.; Umemori, T.; Hirai, K.; Muramatsu, S.; Kamimura, Y.; Araki, H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 2007, 445, 328–332. [Google Scholar] [CrossRef]
- Zegerman, P.; Diffley, J.F. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007, 445, 281–285. [Google Scholar] [CrossRef]
- Masumoto, H.; Muramatsu, S.; Kamimura, Y.; Araki, H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 2002, 415, 651–655. [Google Scholar] [CrossRef]
- Tak, Y.S.; Tanaka, Y.; Endo, S.; Kamimura, Y.; Araki, H. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. Embo. J. 2006, 25, 1987–1996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muramatsu, S.; Hirai, K.; Tak, Y.S.; Kamimura, Y.; Araki, H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev. 2010, 24, 602–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, L.; Stillman, B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science 1998, 280, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa-Onami, M.; Araki, H.; Tanaka, S. Pre-initiation complex assembly functions as a molecular switch that splits the Mcm2-7 double hexamer. EMBO Rep. 2017, 18, 1752–1761. [Google Scholar] [CrossRef]
- Van Deursen, F.; Sengupta, S.; De Piccoli, G.; Sanchez-Diaz, A.; Labib, K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. Embo. J. 2012, 31, 2195–2206. [Google Scholar] [CrossRef] [Green Version]
- Watase, G.; Takisawa, H.; Kanemaki, M.T. Mcm10 Plays a Role in Functioning of the Eukaryotic Replicative DNA Helicase, Cdc45-Mcm-GINS. Curr. Biol. 2012, 22, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Langston, L.D.; Mayle, R.; Schauer, G.D.; Yurieva, O.; Zhang, D.; Yao, N.Y.; Georgescu, R.E.; O’Donnell, M.E. Mcm10 promotes rapid isomerization of CMG-DNA for replisome bypass of lagging strand DNA blocks. eLife 2017, 6, e29118. [Google Scholar] [CrossRef]
- Looke, M.; Maloney, M.F.; Bell, S.P. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 2017, 31, 291–305. [Google Scholar] [CrossRef] [Green Version]
- Douglas, M.E.; Ali, F.A.; Costa, A.; Diffley, J.F.X. The mechanism of eukaryotic CMG helicase activation. Nature 2018, 555, 265–268. [Google Scholar] [CrossRef]
- Fukuura, M.; Nagao, K.; Obuse, C.; Takahashi, T.S.; Nakagawa, T.; Masukata, H. CDK promotes interactions of Sld3 and Drc1 with Cut5 for initiation of DNA replication in fission yeast. Mol. Biol. Cell 2011, 22, 2620–2633. [Google Scholar] [CrossRef]
- Noguchi, E.; Shanahan, P.; Noguchi, C.; Russell, P. CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr. Biol. 2002, 12, 599–605. [Google Scholar] [CrossRef] [Green Version]
- Matsuno, K.; Kumano, M.; Kubota, Y.; Hashimoto, Y.; Takisawa, H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol. Cell Biol. 2006, 26, 4843–4852. [Google Scholar] [CrossRef] [Green Version]
- Sangrithi, M.N.; Bernal, J.A.; Madine, M.; Philpott, A.; Lee, J.; Dunphy, W.G.; Venkitaraman, A.R. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 2005, 121, 887–898. [Google Scholar] [CrossRef] [Green Version]
- Claussin, C.; Vazquez, J.; Whitehouse, I. Single-molecule mapping of replisome progression. Mol. Cell 2022, 82, 1372–1382.e1374. [Google Scholar] [CrossRef]
- Itou, H.; Shirakihara, Y.; Araki, H. The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3. Acta Crystallographica. Sect. D Biol. Crystallogr. 2015, 71, 1649–1656. [Google Scholar] [CrossRef]
- Kumagai, A.; Shevchenko, A.; Dunphy, W.G. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 2010, 140, 349–359. [Google Scholar] [CrossRef] [Green Version]
- Sansam, C.L.; Cruz, N.M.; Danielian, P.S.; Amsterdam, A.; Lau, M.L.; Hopkins, N.; Lees, J.A. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 2010, 24, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Pulido, L.; Diffley, J.F.; Ponting, C.P. Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr. Biol. 2010, 20, R509–R510. [Google Scholar] [CrossRef] [Green Version]
- Boos, D.; Yekezare, M.; Diffley, J.F. Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science 2013, 340, 981–984. [Google Scholar] [CrossRef]
- Kohler, K.; Sanchez-Pulido, L.; Hofer, V.; Marko, A.; Ponting, C.P.; Snijders, A.P.; Feederle, R.; Schepers, A.; Boos, D. The Cdk8/19-cyclin C transcription regulator functions in genome replication through metazoan Sld7. PLoS Biol. 2019, 17, e2006767. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, P.; Sanchez-Pulido, L.; Marko, A.; Ponting, C.P.; Boos, D. Refining the domain architecture model of the replication origin firing factor Treslin/TICRR. Life Sci. Alliance 2022, 5, e202101088. [Google Scholar] [CrossRef] [PubMed]
- Zaffar, E.; Ferreira, F.; Sanchez-Pulido, L.; Boos, D. The Role of MTBP as a Replication Origin Firing Factor. Biology 2022, 11, 827. [Google Scholar] [CrossRef]
- Tanaka, S. An efficient method for the isolation of interaction-null/impaired mutants using the yeast two-hybrid technique. Genes Cells 2019, 24, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Rhind, N.; Gilbert, D.M. DNA replication timing. Cold Spring Harb. Perspect. Biol. 2013, 5, a010132. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, S.; Araki, H. Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma 2010, 119, 565–574. [Google Scholar] [CrossRef]
- Mantiero, D.; Mackenzie, A.; Donaldson, A.; Zegerman, P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. Embo. J. 2011, 30, 4805–4814. [Google Scholar] [CrossRef] [Green Version]
- Kanke, M.; Kodama, Y.; Takahashi, T.S.; Nakagawa, T.; Masukata, H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. Embo. J. 2012, 31, 2182–2194. [Google Scholar] [CrossRef] [Green Version]
- Cook, C.R.; Kung, G.; Peterson, F.C.; Volkman, B.F.; Lei, M. A novel zinc finger is required for Mcm10 homocomplex assembly. J. Biol. Chem. 2003, 278, 36051–36058. [Google Scholar] [CrossRef] [Green Version]
- De Jesus-Kim, L.; Friedman, L.J.; Looke, M.; Ramsoomair, C.K.; Gelles, J.; Bell, S.P. DDK regulates replication initiation by controlling the multiplicity of Cdc45-GINS binding to Mcm2-7. eLife 2021, 10, e65471. [Google Scholar] [CrossRef]
- Greiwe, J.F.; Miller, T.C.R.; Locke, J.; Martino, F.; Howell, S.; Schreiber, A.; Nans, A.; Diffley, J.F.X.; Costa, A. Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase. Nat. Struct. Mol. Biol. 2022, 29, 10–20. [Google Scholar] [CrossRef]
- Cheng, J.; Li, N.; Huo, Y.; Dang, S.; Tye, B.K.; Gao, N.; Zhai, Y. Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase. Nat. Commun. 2022, 13, 1396. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, S.; Ogawa, S. Dimerization of Firing Factors for Replication Origin Activation in Eukaryotes: A Crucial Process for Simultaneous Assembly of Bidirectional Replication Forks? Biology 2022, 11, 928. https://doi.org/10.3390/biology11060928
Tanaka S, Ogawa S. Dimerization of Firing Factors for Replication Origin Activation in Eukaryotes: A Crucial Process for Simultaneous Assembly of Bidirectional Replication Forks? Biology. 2022; 11(6):928. https://doi.org/10.3390/biology11060928
Chicago/Turabian StyleTanaka, Seiji, and Shiho Ogawa. 2022. "Dimerization of Firing Factors for Replication Origin Activation in Eukaryotes: A Crucial Process for Simultaneous Assembly of Bidirectional Replication Forks?" Biology 11, no. 6: 928. https://doi.org/10.3390/biology11060928
APA StyleTanaka, S., & Ogawa, S. (2022). Dimerization of Firing Factors for Replication Origin Activation in Eukaryotes: A Crucial Process for Simultaneous Assembly of Bidirectional Replication Forks? Biology, 11(6), 928. https://doi.org/10.3390/biology11060928





