Functional and Seasonal Changes in the Structure of Microbiome Inhabiting Bottom Sediments of a Pond Intended for Ecological King Carp Farming
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Description of the Breeding Pond (Cardinal Pond in Ślesin)
2.2. Bottom Sediment Sampling
2.3. DNA Extraction and NGS Analysis
2.4. Bioinformatic, Functional, and Statistical Analyses
2.5. Community-Level Physiological Profiling (CLPP)
3. Results
3.1. Sequencing Data Quality and Diversity Indices
3.2. Seasonal Changes in the Microbiome Structure—Phylum and Class Taxonomic Level
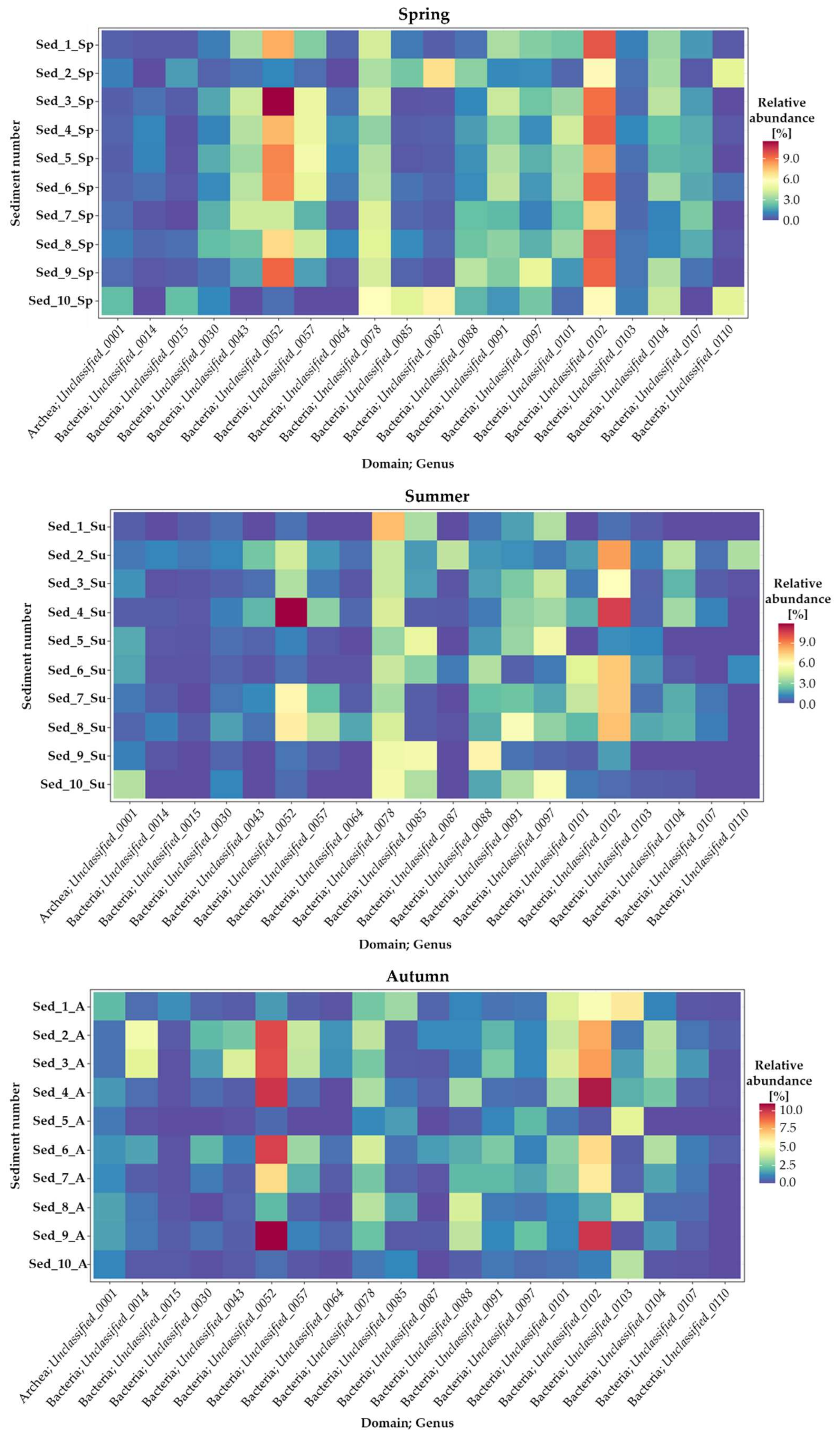
3.3. Seasonal Changes in the Microbiome Structure—Genus Taxonomic Level
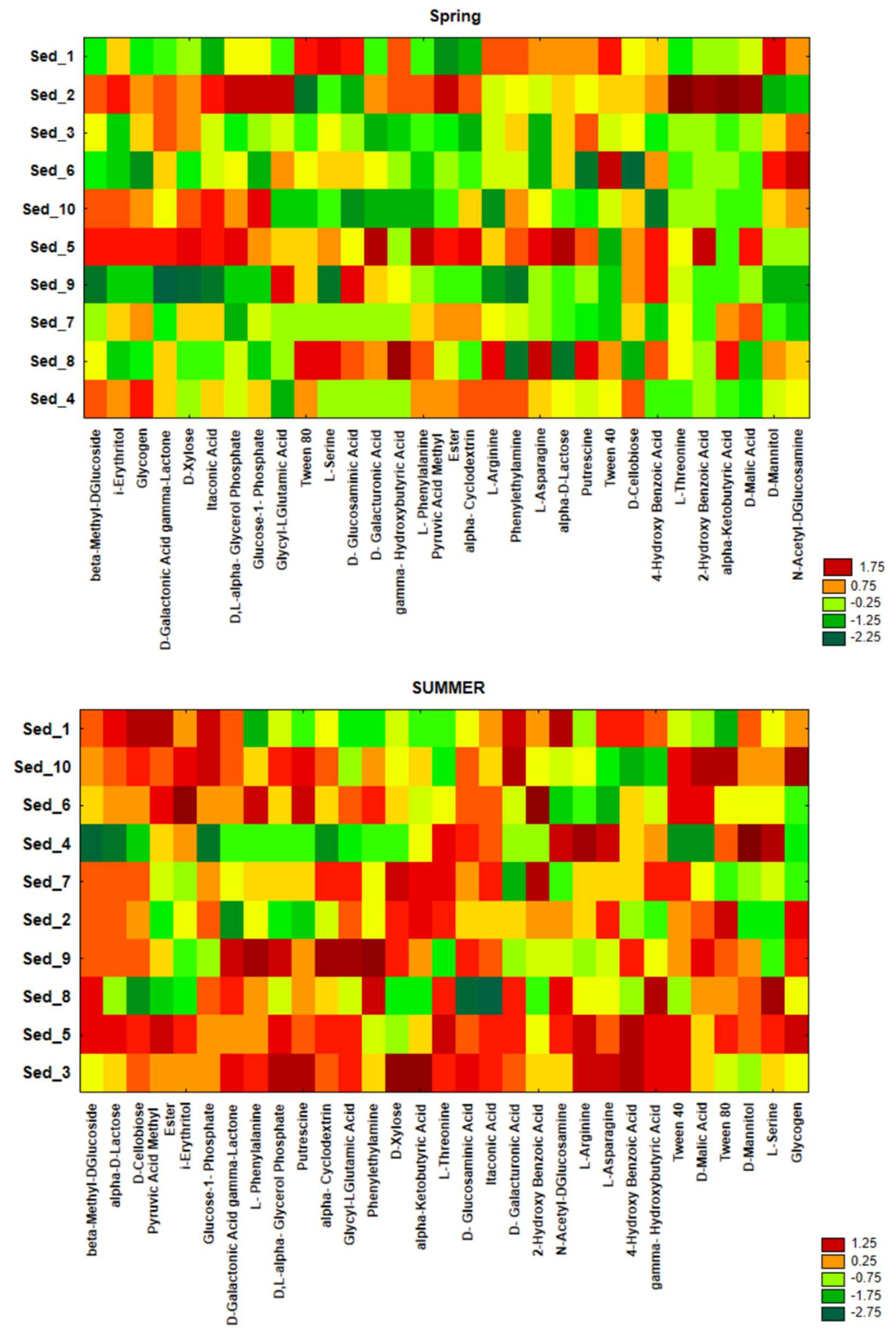
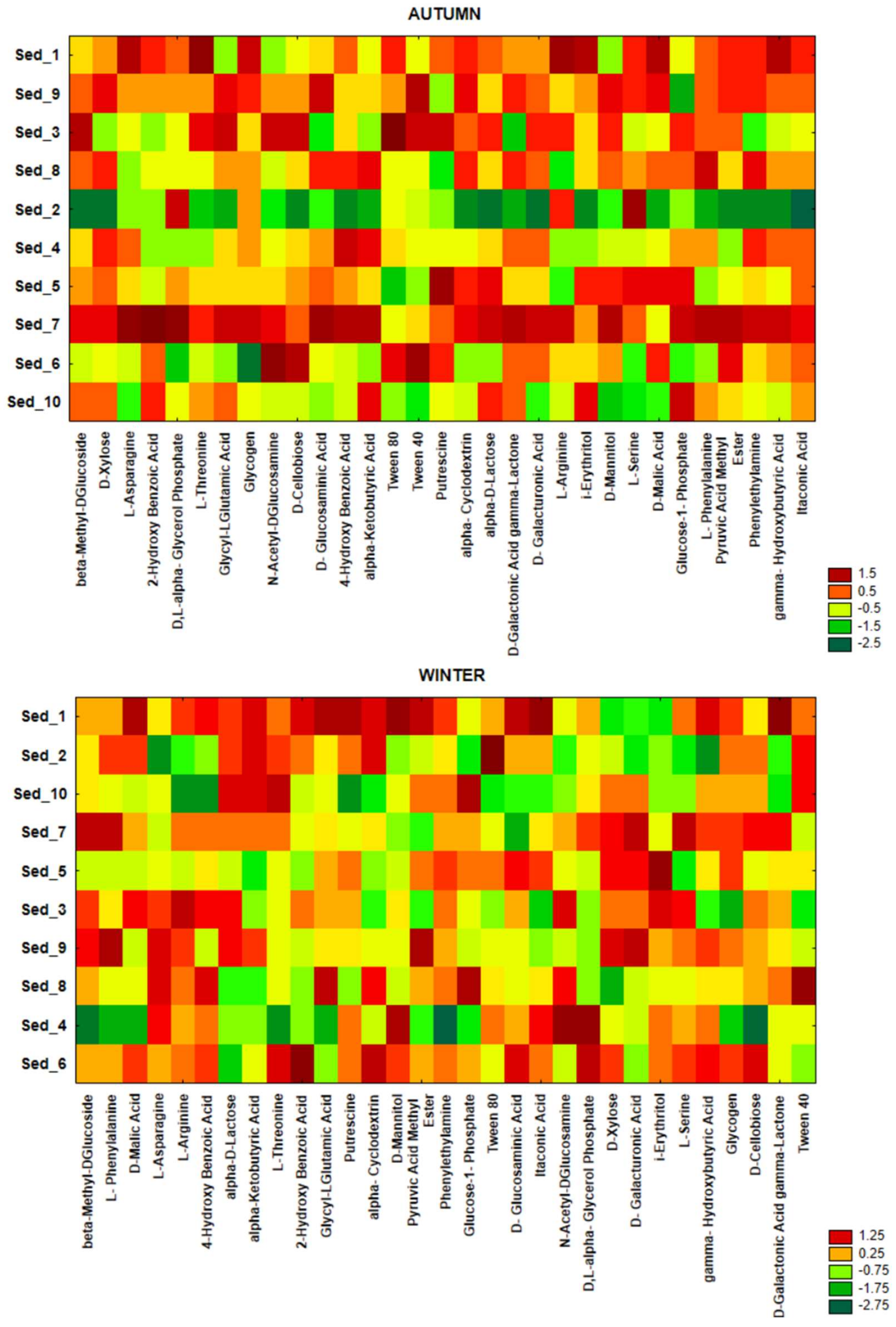
3.4. Seasonal Changes in the Functional Activity of Bacteria Inhabiting Bottom Sediments
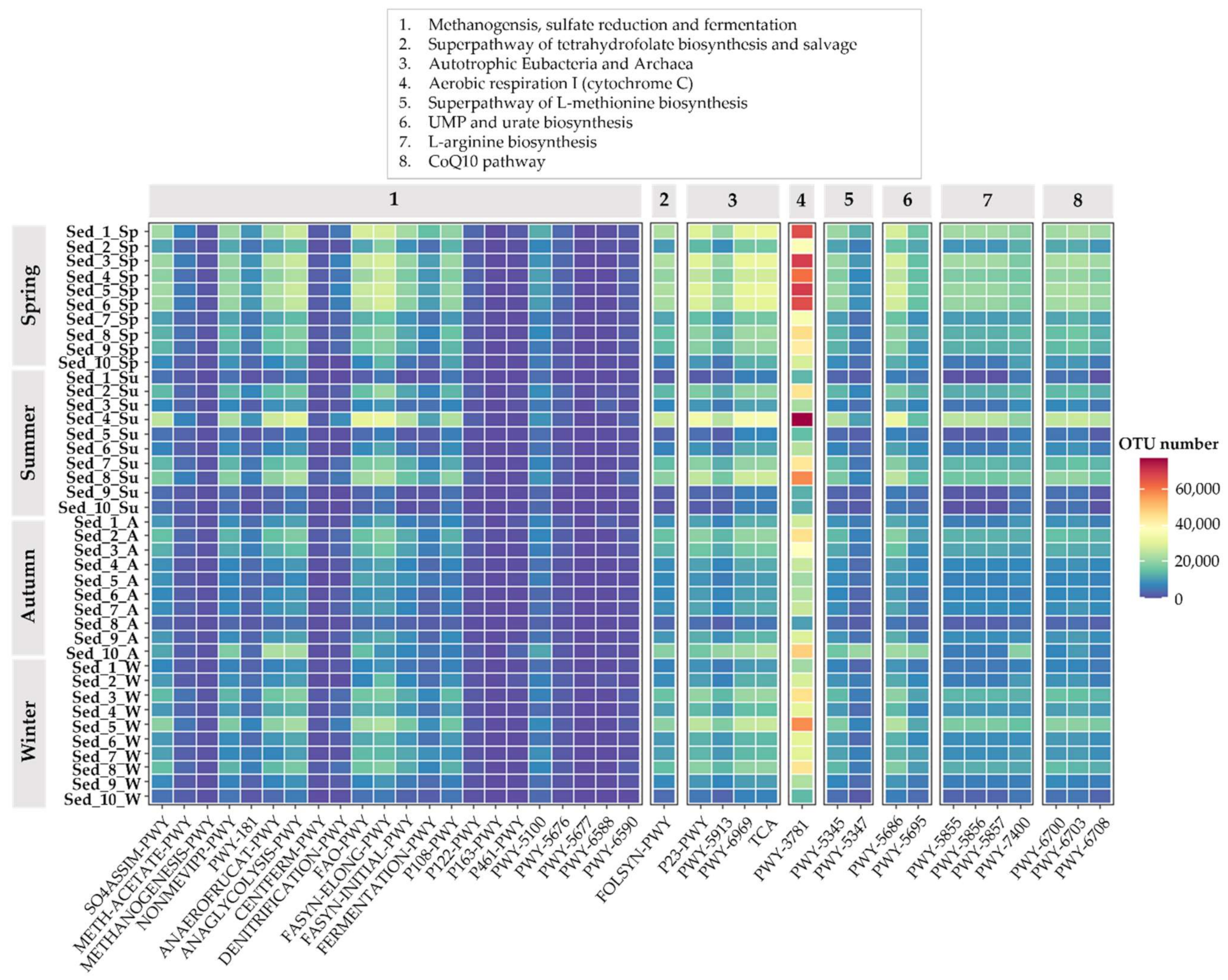
3.5. Bacterial Functional Annotation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

| Sample | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Genus | SPRING | |||||||||
| Thiobacillus | + | + | + | + | + | + | + | + | + | |
| Serratia | + | |||||||||
| Methylobacter | + | |||||||||
| Thermoanaerobaculum | + | + | + | |||||||
| Geobacter | + | |||||||||
| Gp2 | + | |||||||||
| Gp3 | + | + | + | + | + | + | + | |||
| Gp13 | + | + | + | |||||||
| Gp16 | + | + | ||||||||
| Gp18 | + | + | + | + | + | |||||
| SUMMER | ||||||||||
| Thiobacillus | + | + | + | + | + | + | ||||
| Paenibacillus | + | + | ||||||||
| Geobacter | + | + | + | |||||||
| Trichococcus | + | |||||||||
| Thermoanaerobaculum | + | + | + | + | ||||||
| Pseudoarcobacter | + | |||||||||
| Phenylobacterium | + | |||||||||
| Methylobacter | + | |||||||||
| Malikia | + | |||||||||
| Gp3 | + | + | + | + | + | + | + | |||
| Gp13 | + | + | + | + | + | + | + | |||
| Gp18 | + | + | + | + | + | + | + | + | ||
| AUTUMN | ||||||||||
| Aeromonas | + | + | ||||||||
| Arthrobacter | + | |||||||||
| Acinetobacter | + | |||||||||
| Pseudomonas | + | + | ||||||||
| Thiobacillus | + | + | + | + | + | + | + | + | ||
| Sulfuricurvum | + | + | ||||||||
| Paenibacillus | + | |||||||||
| Polaromonas | + | + | + | + | ||||||
| Pseudoarcobacter | + | |||||||||
| Thermoanaerobaculum | + | + | + | + | + | |||||
| Rheinheimera | + | + | + | |||||||
| Methylobacter | + | |||||||||
| Masillia | + | |||||||||
| Luetolibacter | + | |||||||||
| Hydrogenophaga | + | |||||||||
| Flavobacterium | + | + | + | + | + | |||||
| Deefgea | + | |||||||||
| Gp3 | + | + | + | + | + | + | + | + | ||
| Gp13 | + | + | + | + | + | |||||
| Gp18 | + | + | + | + | + | |||||
| WINTER | ||||||||||
| Flavobacterium | + | + | + | + | + | |||||
| Shewanella | + | |||||||||
| Thiobacillus | + | + | + | + | + | + | + | + | ||
| Pseudomonas | + | + | ||||||||
| Paenibacillus | + | |||||||||
| Thermoanaerobaculum | + | |||||||||
| Polaromonas | + | + | ||||||||
| Luteolibacter | + | |||||||||
| Iodobacter | + | |||||||||
| Hydrogenophaga | + | + | ||||||||
| Gp3 | + | + | + | + | + | |||||
| Gp13 | + | + | + | |||||||
| Gp16 | + | |||||||||
| Gp18 | + | + | + | |||||||
References
- Sehnal, L.; Brammer-Robbins, E.; Wormington, A.M.; Blaha, L.; Bisesi, J.; Larkin, I.; Martyniuk, C.M.; Simonin, M.; Adamovsky, O. Microbiome composition and function in aquatic vertebrates: Small organisms making big impacts on aquatic animal health. Front. Microbiol. 2021, 12, 567408. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and function of the global ocean microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, K.; Huaivan, L.; Zilian, Z.; Weidong, C.; Bin, W.; Fulong, P.; Fanfan, H.; Fanli, H.; Wenging, L. Depth profiles of geochemical features, geochemical activities and biodiversity of microbial communities in marine sediments from the Shenhu area, the northern South China Sea. Sci. Total Environ. 2021, 779, 146233. [Google Scholar] [CrossRef]
- McKenney, E.A.; Koelle, K.; Dunn, R.R.; Yoder, A.D. The ecosystem services of animal micobiomes. Mol. Ecol. 2018, 27, 2164–2172. [Google Scholar] [CrossRef]
- Zhuang, G.-C.; Heuer, V.B.; Lazar, C.; Goldhammer, T.; Wendt, J.; Samarkin, V.A.; Elvert, M.; Teske, A.P.; Joye, S.B.; Hinrichs, K.-U. Relative importance of methylotrophic methanogenesis in sediments of the Western Mediterranean Sea. Geochim. Cosmochim. Acta 2018, 224, 171–186. [Google Scholar] [CrossRef]
- Saunois, M.; Bousquet, P.; Poulter, B.; Peregon, A.; Ciais, P.; Canadell, J.G.; Dlugokencky, E.J.; Etiope, G.; Bastviken, D.; Houweling, S.; et al. The global methane budget 2000–2012. Earth Syst. Sci. Data 2016, 8, 697–751. [Google Scholar] [CrossRef] [Green Version]
- Beulig, F.; Røy, H.; McGlynn, S.E.; Jørgensen, B.B. Cryptic CH4 cycling in the sulfate-methane transition of marine sediments apparently mediated by ANME-1 archaea. ISME J. 2019, 13, 250–262. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.; Hyun, J.-H.; You, O.-R.; Kim, M.; Kim, S.-H.; Choi, D.-L.; Green, S.; Kostka, J.E. Microbial community structure associated with biogeochemical processes in the sulfate-methane transition (SMTZ) of gas-hydrate-bearing sediment of the Ulleung Basin, East Sea. Geomicrobiol. J. 2017, 34, 207–219. [Google Scholar] [CrossRef]
- Alfiansah, Y.R.; Hassenruck, C.; Kunzmann, A.; Taslihan, A.; Harder, J.; Gardes, A. Bacterial abundance and community composition in pond water from shrimp aquaculture systems with different stocking densities. Front. Microbiol. 2018, 9, 2457. [Google Scholar] [CrossRef] [Green Version]
- Lastauskiene, E.; Valskys, V.; Stankeviciute, J.; Kalciene, V.; Gegzna, V.; Kavoliunas, J.; Ruzauskas, M.; Armalyte, J. The impact of intensive fish farming in pond sediment microbiome and antibiotic resistance gene composition. Front. Vet. Sci. 2021, 8, 673756. [Google Scholar] [CrossRef]
- Cramm, M.A.; Neves, B.D.M.; Manning, C.C.; Oldenburg, T.B.; Archambault, P.; Chakraborty, A.; Cyr-Parent, A.; Edinger, E.N.; Jaggi, A.; Mort, A.; et al. Characterization of marine microbial communities around an Arctic seabed hydrocarbon seep at Scott Inlet, Balfin Bay. Sci. Total Environ. 2021, 762, 143961. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Heo, Y.M.; Kwon, S.L.; Yoo, Y.; Kim, D.; Lee, J.; Kwon, B.O.; Khim, J.S.; Kim, J.J. Environmental drivers affecting the bacterial community of intertidal sediments in the Yellow Sea. Sci. Total Environ. 2021, 755, 142726. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.; Yu, H.; Sheng, H.; Chen, C.; Tang, Y.; Liang, Z. Seasonal variations and co-occurrence networks of bacterial communities in the water and sediment of artificial habitat in Laoshan Bay, China. Peer J. 2022, 10, e12705. [Google Scholar] [CrossRef] [PubMed]
- Oertli, B.; Parris, K.M. Review: Toward management of urban ponds for freshwater biodiversity. Ecosphere 2019, 10, e02810. [Google Scholar] [CrossRef] [Green Version]
- Harper, L.R.; Buxton, A.S.; Rees, H.C.; Bruce, K.; Brys, R.; Halfmaerten, D.; Read, D.S.; Watson, H.V.; Sayer, C.D.; Jones, E.P.; et al. Prospects and challenges of environmental DNA (eDNA) monitoring in freshwater ponds. Hydrobiologia 2019, 826, 25–41. [Google Scholar] [CrossRef] [Green Version]
- Hill, M.J.; Hassall, C.; Oertli, B.; Fahrig, L.; Robson, B.J.; Biggs, J.; Samways, M.J.; Usio, N.; Takamura, N.; Krishnaswamy, J.; et al. New policy directions for global pond conservation. Cons. Lett. 2018, 142, e12447. [Google Scholar] [CrossRef] [Green Version]
- Wolińska, A.; Kuźniar, A.; Gałązka, A. Biodiversity in the rhizosphere of selected winter wheat (Triticum aestivum L.) cultivars—genetic and catabolic fingerprinting. Agronomy 2020, 10, 953. [Google Scholar] [CrossRef]
- Wolińska, A.; Kruczyńska, A.; Podlewski, J.; Słomczewski, A.; Grządziel, J.; Gałązka, A.; Kuźniar, A. Does the use of an intercropping mixyure really improve the biology of monocultural soils?—A search for bacterial indicators of sensitivity and resistance to long-term maize monoculture. Agronomy 2022, 12, 613. [Google Scholar] [CrossRef]
- Kuźniar, A.; Włodarczyk, K.; Grządziel, J.; Goraj, W.; Gałązka, A.; Wolińska, A. Culture-independent analysis of an endophytic core microbiome in two species of wheat: Triticum aestivum L. (cv. ‘Hondia’) and the first report of microbiota in Triticum spelta L. (cv. ‘Rokosz’). Syst. Appl. Microbiol. 2020, 43, 126025. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Meth. 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Wright, E.S. Using DECIPHER v2.0 to analyze biological sequence data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. Microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Hou, J.; Deng, M.; Liu, Q.; Wu, C.; Ji, Y.; He, X. Bacterial abundance and diversity in pond water supplied with different feeds. Sci. Rep. 2016, 6, 35232. [Google Scholar] [CrossRef] [Green Version]
- Bergkessel, M.; Delavaine, L. Diversity in starvation survival strategies and outcomes among heterotrophic Proteobacteria. Microb. Physiol. 2021, 31, 146–162. [Google Scholar] [CrossRef]
- Hedrich, S.; Schlomann, M.; Johnson, B. The iron-oxidizing Proteobacteria. Microbiol. Soc. 2011, 157, 1551–1564. [Google Scholar] [CrossRef] [Green Version]
- Buijs, Y.; Bech, P.; Vazquez-Albacete, D.; Bentzon-Tilia, M.; Sonnenschein, E.; Gram, L.; Zhang, S.D. Marine Proteobacteria as a source of natural products: Advances in molecular tools and strategies. Nat. Prod. Rep. 2019, 36, 1333–1350. [Google Scholar] [CrossRef]
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.Z.; El-Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent understanding of soil Acidobacteria and their ecological significance: A critical review. Front. Microbiol. 2020, 30, 580024. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Izak, D.; Szafranek-Nakonieczna, A.; Banach, A.; Błaszczyk, M. Bacteroidetes as a sensitive biological indicator of agricultural soil usage revealed by culture independent approach. Appl. Soil Ecol. 2017, 119, 128–137. [Google Scholar] [CrossRef]
- Larsbrink, J.; McKee, L. Bacteroidetes bacteria in the soil: Glycan acquisition, enzyme secretion, and gilding motility. Adv. Appl. Microbiol. 2020, 110, 63–98. [Google Scholar] [CrossRef]
- Obbels, D.; Verleyen, E.; Mano, M.-J.; Namsaraev, Z.; Sweetlove, M.; Tytgat, B.; Fernandez-Carazo, R.; de Wever, A.; D’Hondt, S.; Ertz, D.; et al. Bacterial and eukaryotic biodiversity patterns in terrestrial and aquatic habitats in the SørRondane Mountains, Dronning Maud Land, East Antarctica. FEMS Microbiol. Ecol. 2016, 92, fiw041. [Google Scholar] [CrossRef] [PubMed]
- Ferro, P.; Vaz-Moreira, I.; Manaia, C. Betaproteobacteria are predominant in drinking water: Are there reasons for concern? Crit. Rev. Microbiol. 2019, 45, 649–667. [Google Scholar] [CrossRef] [PubMed]
- Evans, F.; Egan, S.; Kjelleberg, S. Ecology of type II secretion in marine Gammaproteobacteria. Environ. Microbiol. 2008, 10, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Dyksma, S.; Bischof, K.; Fuchs, B.; Hoffmann, K.; Meier, D.; Meyerdierks, A.; Pjevac, P.; Probandt, D.; Richter, M.; Stepanauskas, R.; et al. Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J. 2016, 10, 1939–1953. [Google Scholar] [CrossRef] [Green Version]
- Wee, S.; Burns, J.; DiChristina, T. Identification of a molecular signature unique to metal-reducing Gammaproteobacteria. FEMS Microbiol. Lett. 2014, 350, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Haggblom, M. Genome-guided identification of organohalide-respiring Deltaproteobacteria from the marine environment. mBio 2018, 9, e02471-18. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.H.; Stoven, K.; Haneklaus, S.; Singh, B.R.; Schung, E. Elemental sulfur oxidation by Thiobacillus spp. and aerobic heterotrophic sulfur-oxidizing bacteria. Pedosphere 2010, 20, 71–79. [Google Scholar] [CrossRef]
- Darcy, J.L.; Lynch, R.C.; King, A.J.; Robeson, M.S.; Schmidt, S.K. Global distribution of Polaromonas phylityoes—Evidence for a highly successful dispersal capacity. PLoS ONE 2011, 6, e23742. [Google Scholar] [CrossRef] [Green Version]
- Sizova, M.; Panikov, N. Polaromonas hydrogenivorans sp. nov., a psychrotolerant hydrogen-oxidizing bacterium from Alaskan soil. Int. J. Syst. Evol. Microbiol 2007, 57, 616–619. [Google Scholar] [CrossRef] [Green Version]
- Osborne, T.H.; E Jamieson, H.; A Hudson-Edwards, K.; Nordstrom, D.K.; Walker, S.R.; A Ward, S.; Santini, J.M. Microbial oxidation of arsenite in a subarctic environment: Diversity of arsenite oxidase genes and identification of a psychrotolerant arsenite oxidiser. BMC Microbiol. 2010, 10, 205. [Google Scholar] [CrossRef] [Green Version]
- Polz, M.F.; Hunt, D.E.; Preheim, S.P.; Weinreich, D.M. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Phil. Trans. R. Soc. B 2006, 361, 2009–2021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaud, L.; Caruso, C.; Mangano, S.; Interdonato, F.; Bruni, V.; Lo Giudice, A. Predominance of Flavobacterium, Psedudomonas, and Polaromonas within the prokaryotic community of freshwater shallow lakes in the northern Victoria Land, East Antarctica. FEMS Microbiol. Ecol. 2012, 82, 391–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Chaves, M.G.; Silva, G.G.Z.; Rossetto, R.; Edwards, R.A.; Tsai, S.M.; Navarette, A.A. Acidobacteria subgroups and their metabolic potential for carbon degradation in sugarcane soil amended with vinasse and nitrogen fertilizers. Front. Microbiol. 2019, 10, 1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Xie, F.; Zhang, F.; Zhou, K.; Sun, H.; Yang, Q. Analysis of bacterial community functional diversity in late-stage shrimp (Litopenaeus vannamei) ponds using Biolog EcoPlates and PICRUSt2. Aquaqulture 2022, 546, 737288. [Google Scholar] [CrossRef]
- Deng, H.; Ge, L.; Xu, T.; Zhang, M.; Wang, X.; Zhang, Y.; Peng, H. Analysis of the metabolic utilization of carbon sources and potential functional diversity of the bacterial community in lab-scale horizontal subsurface-flow constructed wetlands. J. Environ. Qual. 2011, 40, 1730–1736. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Zhang, M.; Ding, W.; Lan, L.; Liu, Z.; Miao, L.; Hou, J. Microbial carbon metabolic functions in sediments influenced by resuspension event. Water 2020, 13, 7. [Google Scholar] [CrossRef]
- Oest, A.; Alsaffar, A.; Fenner, M.; Azzopardi, D.; Tiquia-Arashiro, S.M. Patterns of change in metabolic capabilities of sediment microbial communities in river and lake ecosystems. Int. J. Microbiol. 2018, 2018, 6234931. [Google Scholar] [CrossRef]
- Hamada, M.; Toyofuku, M.; Miyano, T.; Nomura, N. cbb3-type cytochrome C oxidases, aerobic respiratory enzymes impact the anaerobic life of Pseudomonas aeruginosa PAO1. J. Bacteriol. 2014, 196, 3881–3889. [Google Scholar] [CrossRef] [Green Version]
- George, D.M.; Vincent, A.S.; Mackey, H.R. An overview of anoxygenic phototrophic bacteria and their applications in environmental biotechnology for sustainable resource recovery. Biotechnol. Rep. 2020, 19, e00563. [Google Scholar] [CrossRef]

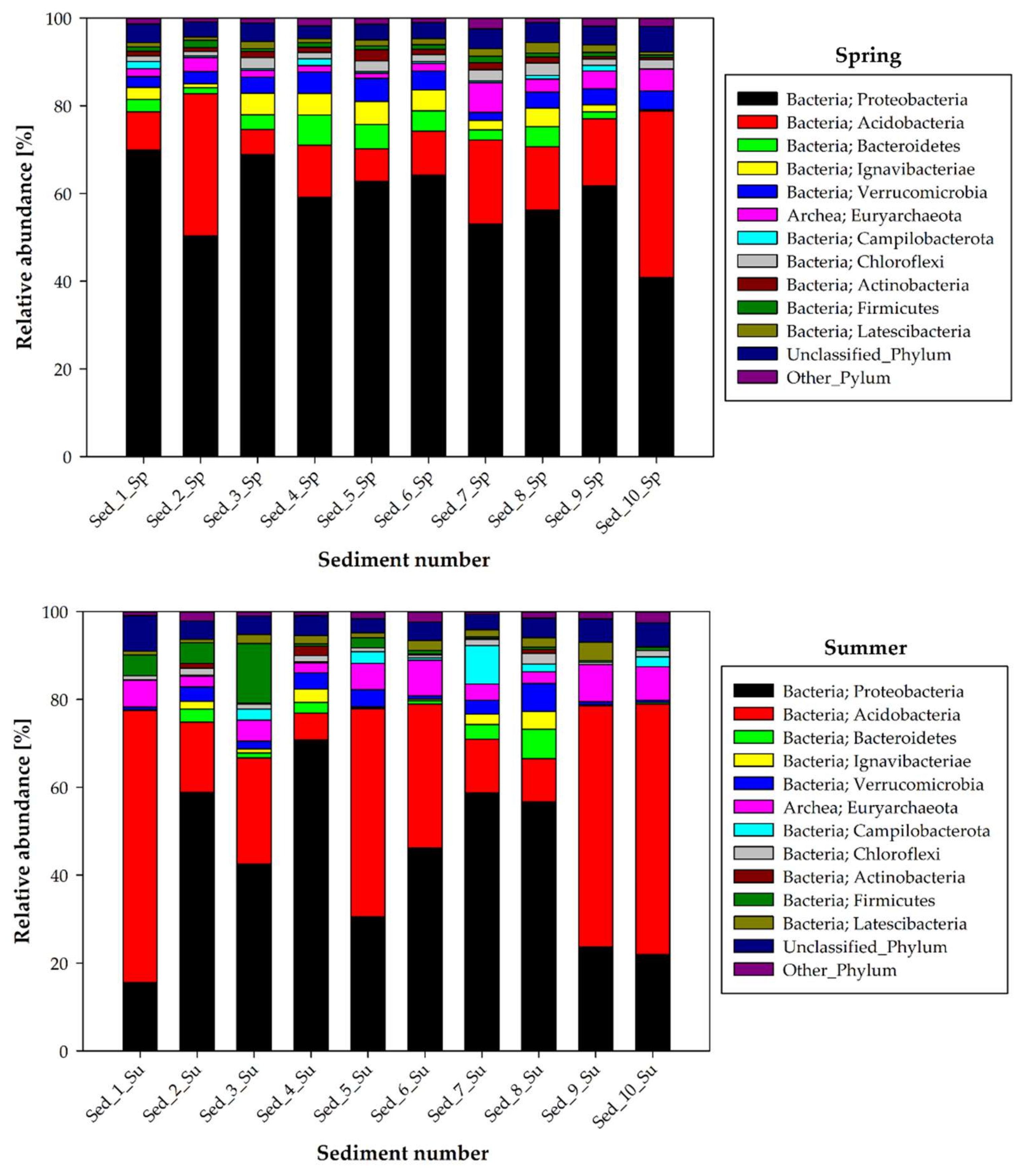

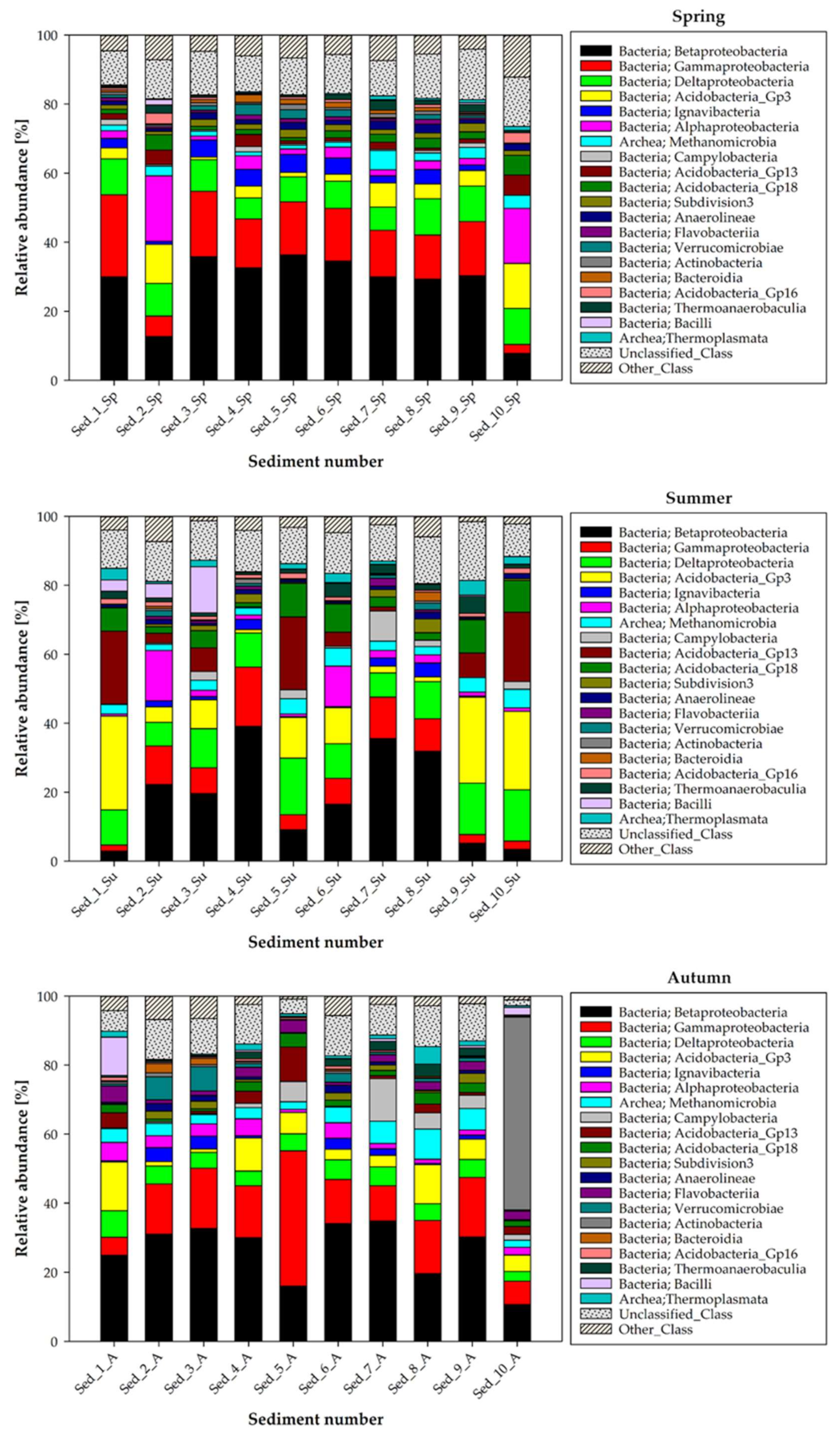

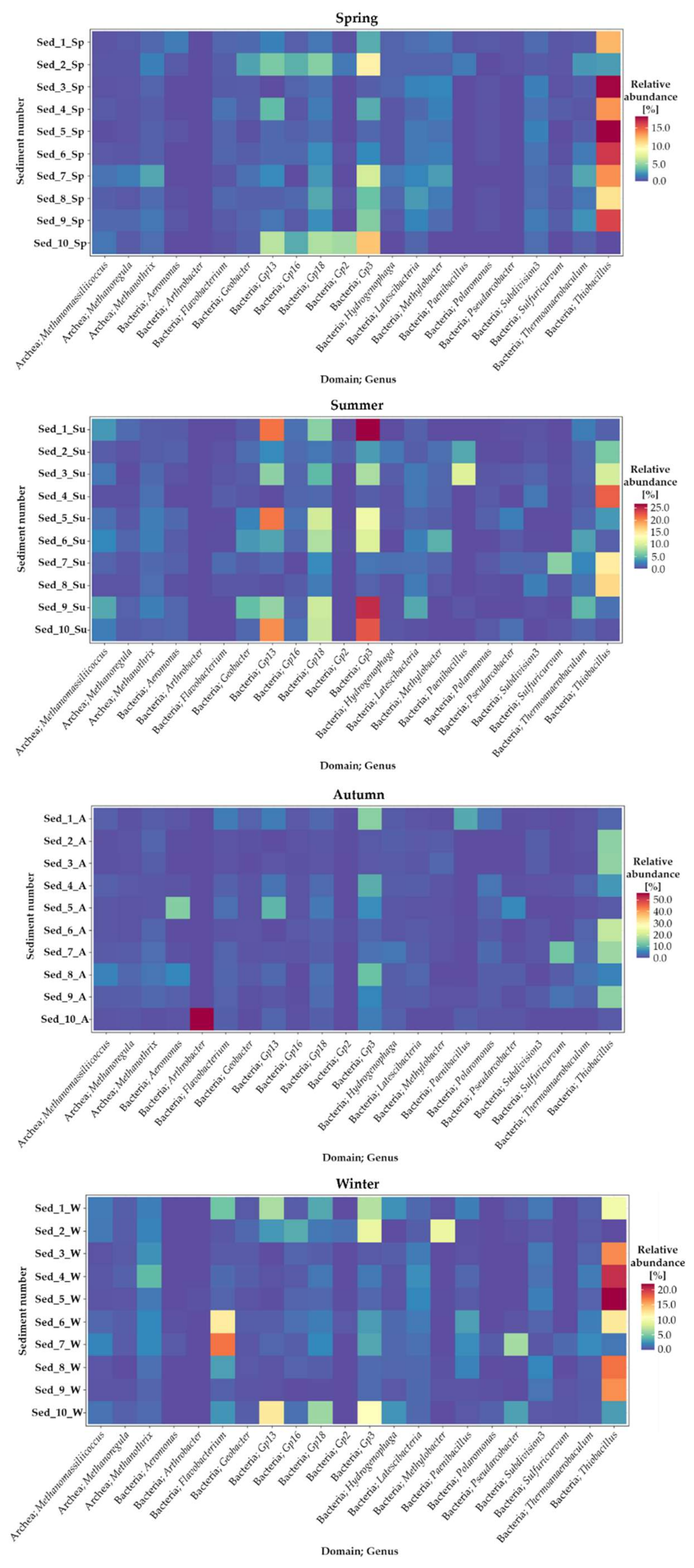
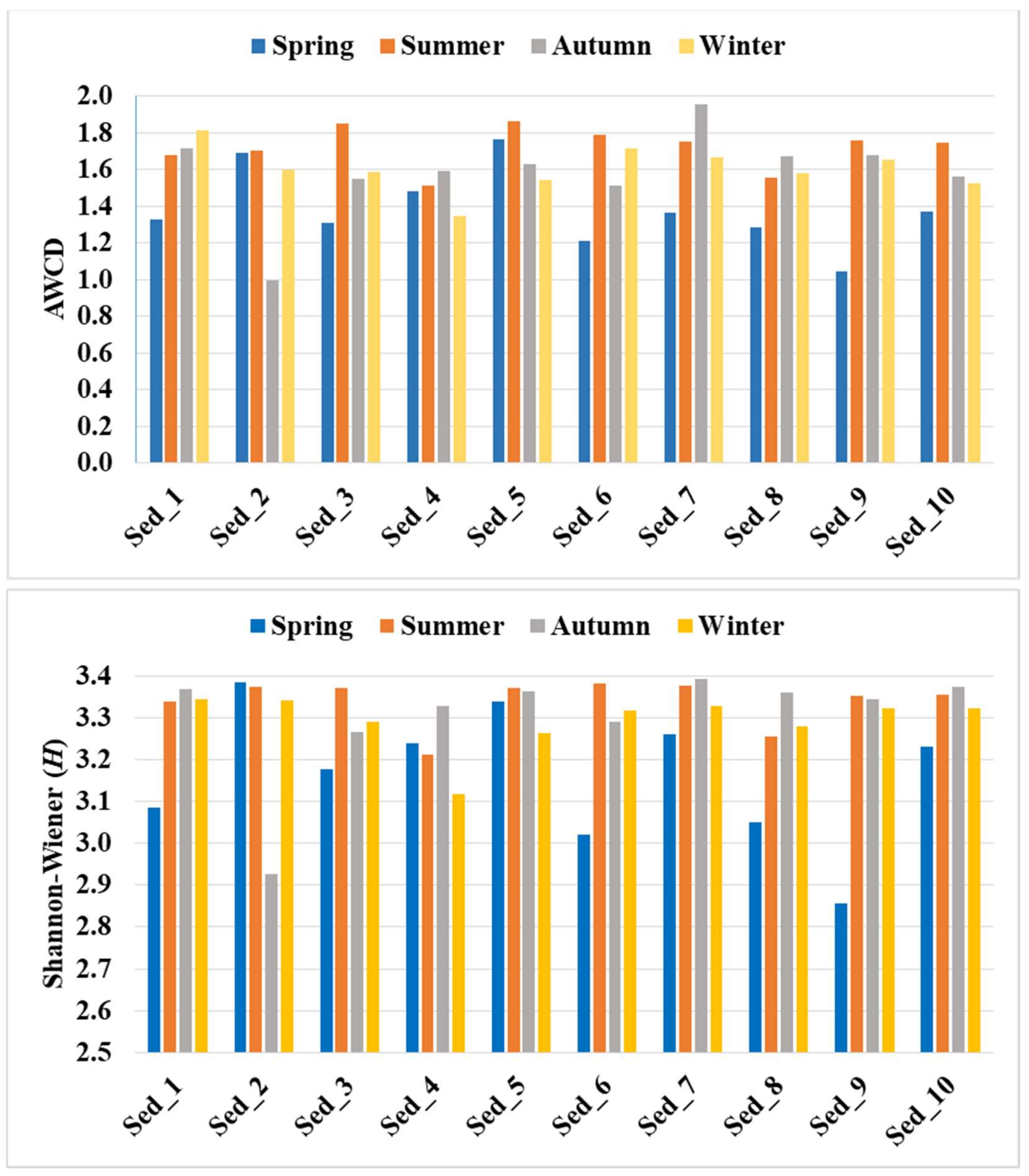

| Taxonomic Affiliation | Seasonal Variation in Abundance | Main Dominants | |||
|---|---|---|---|---|---|
| Spring | Summer | Autumn | Winter | ||
| Phyla | 13 | 14 | 12 | 15 | Proteobacteria Acidobacteria Bacteroidetes Euryarchaeota |
| Classes | 28 | 28 | 22 | 29 | Betaproteobacteria Gammaproteobacteria Deltaproteobacteria Methanobacteria |
| Genera | 28 | 28 | 35 | 39 | Table A1 (Appendix A) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolińska, A.; Kruczyńska, A.; Grządziel, J.; Gałązka, A.; Marzec-Grządziel, A.; Szałaj, K.; Kuźniar, A. Functional and Seasonal Changes in the Structure of Microbiome Inhabiting Bottom Sediments of a Pond Intended for Ecological King Carp Farming. Biology 2022, 11, 913. https://doi.org/10.3390/biology11060913
Wolińska A, Kruczyńska A, Grządziel J, Gałązka A, Marzec-Grządziel A, Szałaj K, Kuźniar A. Functional and Seasonal Changes in the Structure of Microbiome Inhabiting Bottom Sediments of a Pond Intended for Ecological King Carp Farming. Biology. 2022; 11(6):913. https://doi.org/10.3390/biology11060913
Chicago/Turabian StyleWolińska, Agnieszka, Anna Kruczyńska, Jarosław Grządziel, Anna Gałązka, Anna Marzec-Grządziel, Klaudia Szałaj, and Agnieszka Kuźniar. 2022. "Functional and Seasonal Changes in the Structure of Microbiome Inhabiting Bottom Sediments of a Pond Intended for Ecological King Carp Farming" Biology 11, no. 6: 913. https://doi.org/10.3390/biology11060913
APA StyleWolińska, A., Kruczyńska, A., Grządziel, J., Gałązka, A., Marzec-Grządziel, A., Szałaj, K., & Kuźniar, A. (2022). Functional and Seasonal Changes in the Structure of Microbiome Inhabiting Bottom Sediments of a Pond Intended for Ecological King Carp Farming. Biology, 11(6), 913. https://doi.org/10.3390/biology11060913









