Temperature- and Nutrients-Induced Phenotypic Changes of Antarctic Green Snow Bacteria Probed by High-Throughput FTIR Spectroscopy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Antarctic Green Snow Bacterial Isolates
2.2. Cultivation of Bacteria
2.3. FTIR Spectroscopy Analysis
2.4. Data Analysis
3. Results
3.1. Growth Ability under Different Nutrient and Temperature Conditions
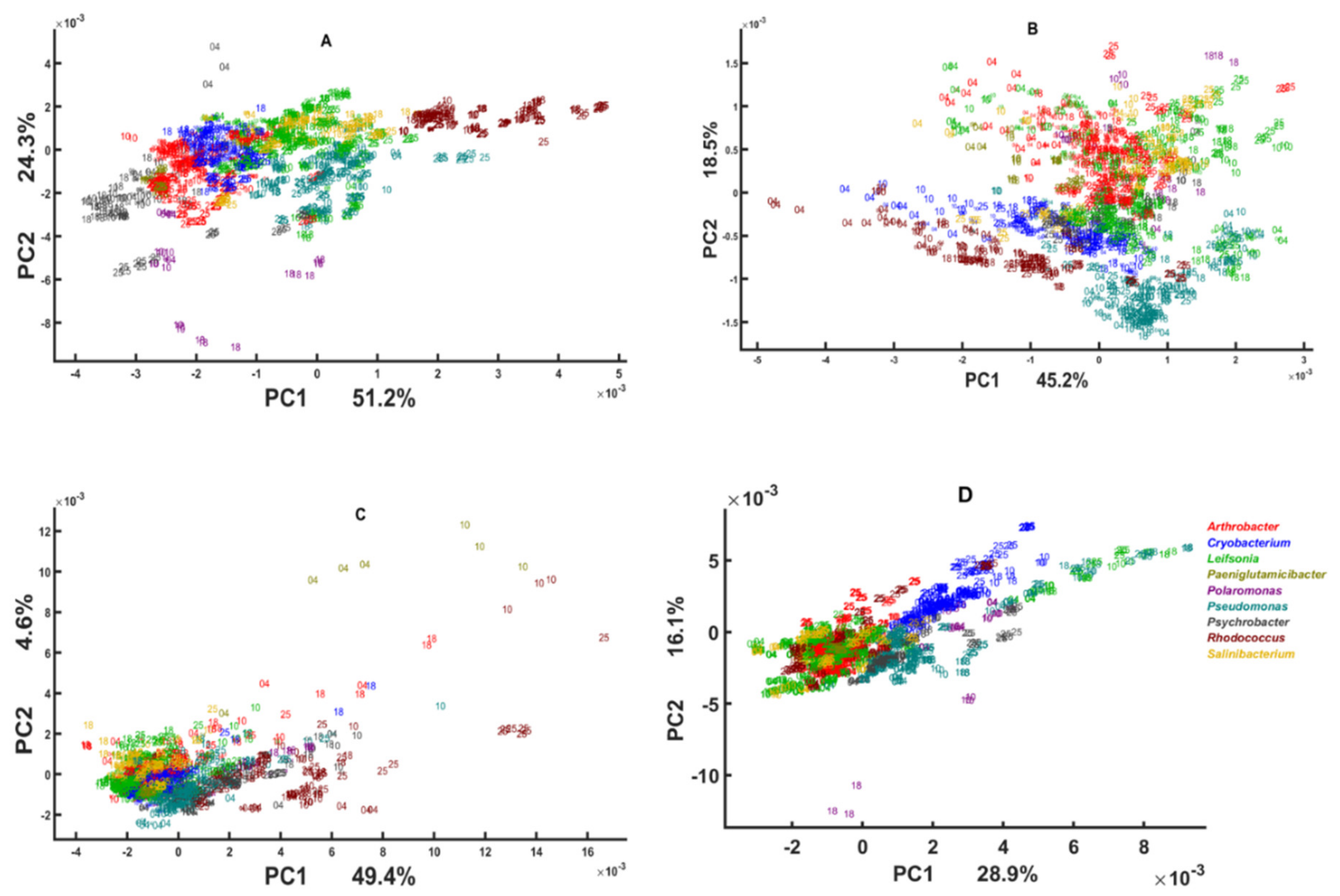
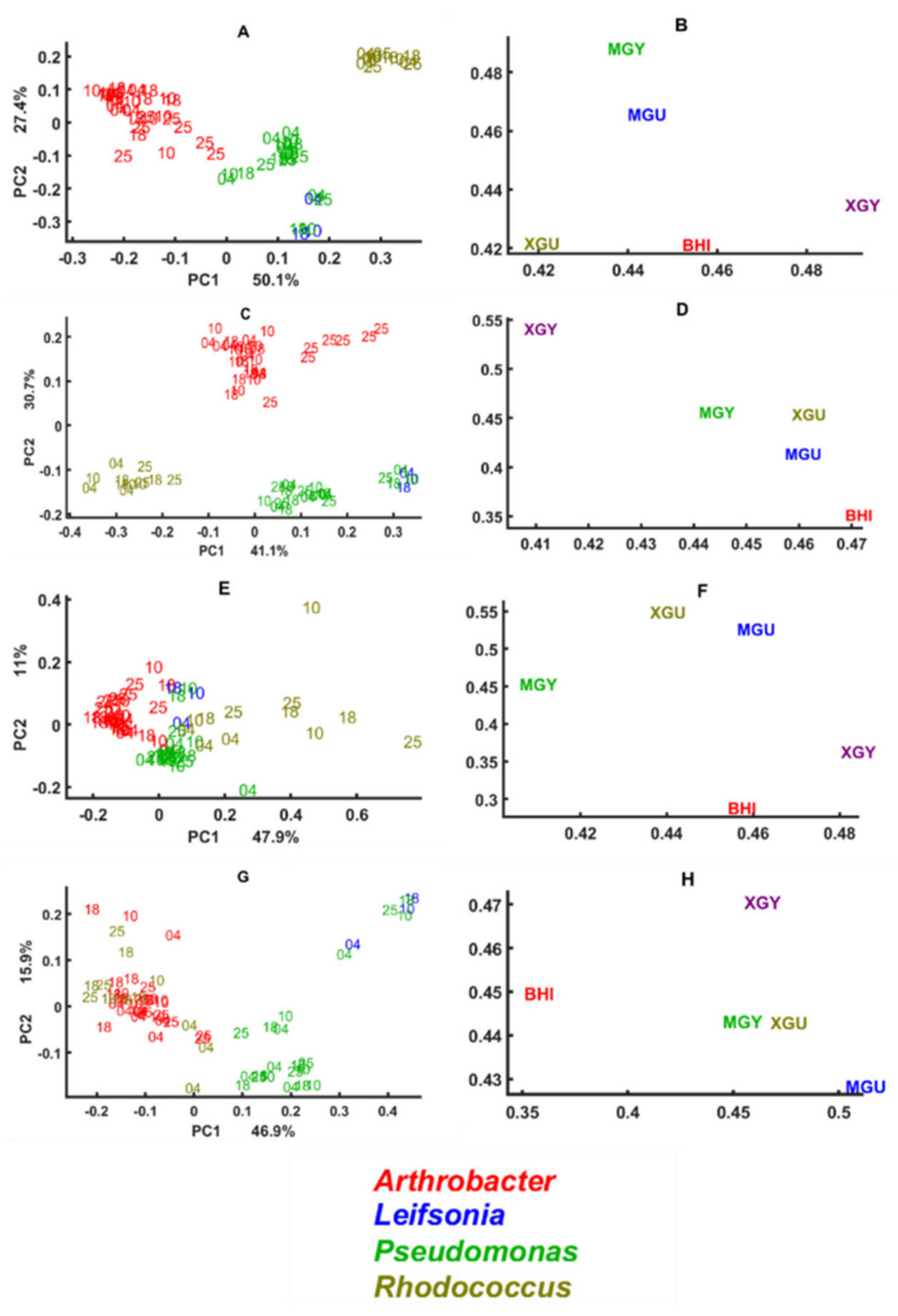
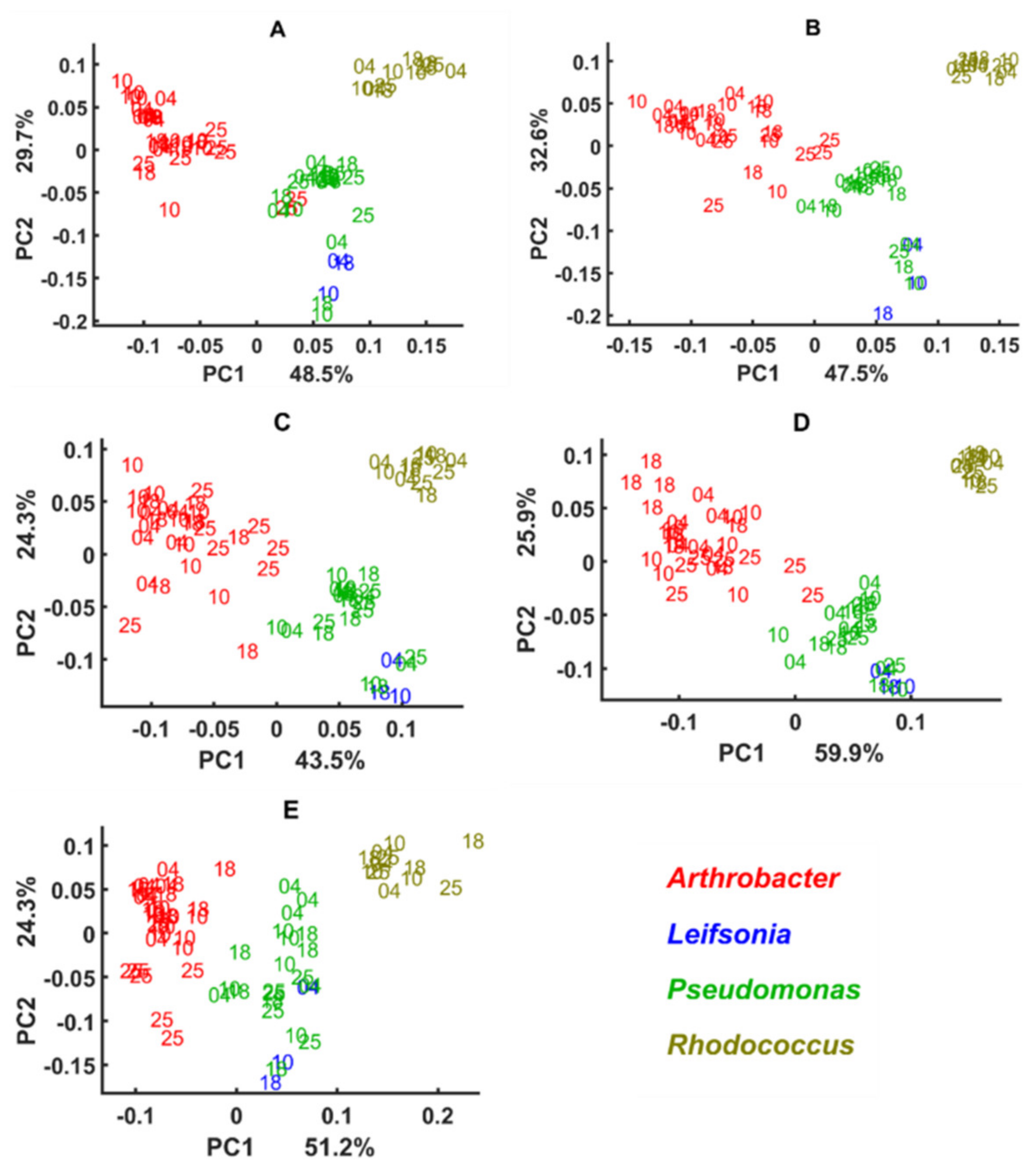
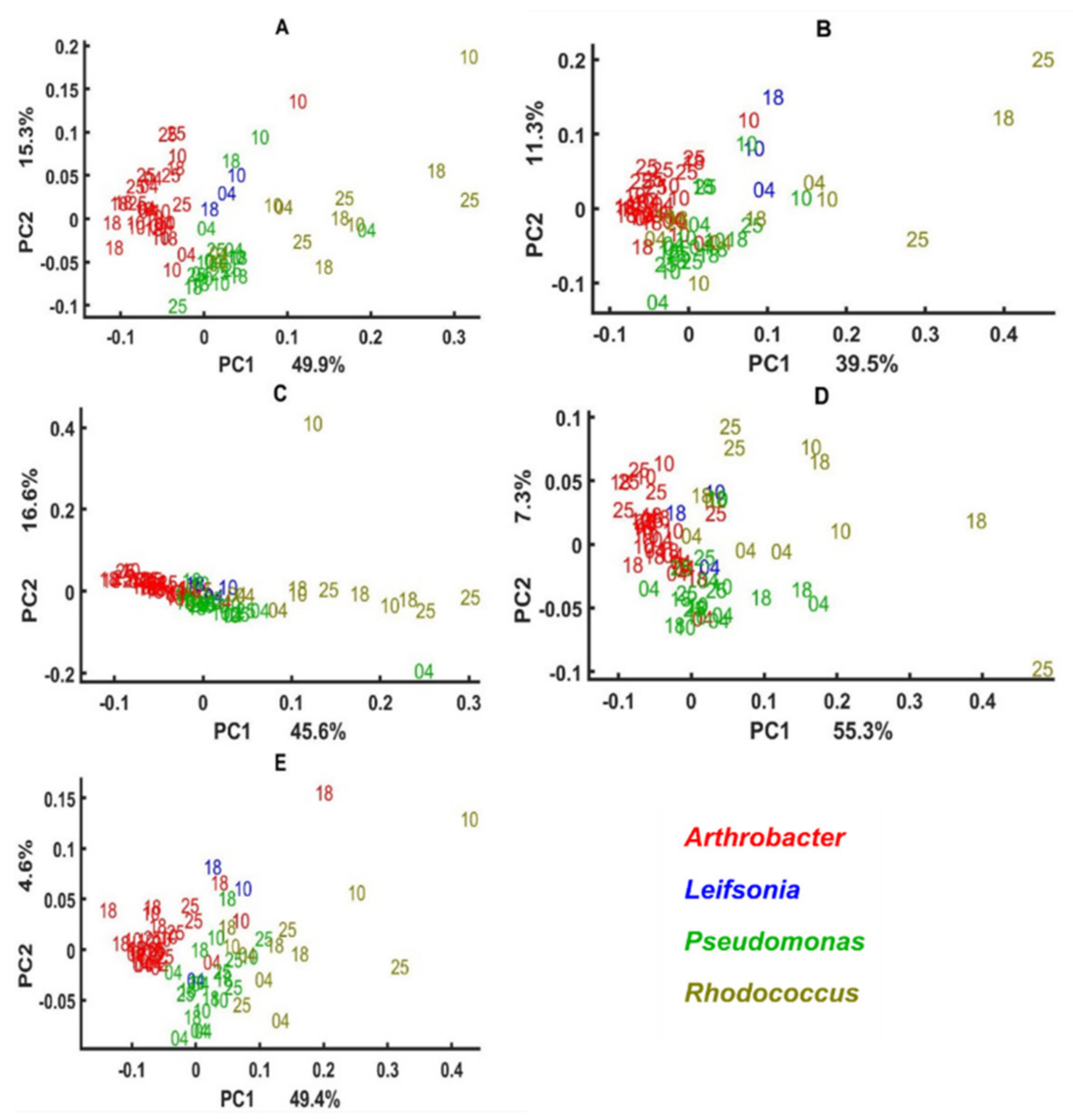
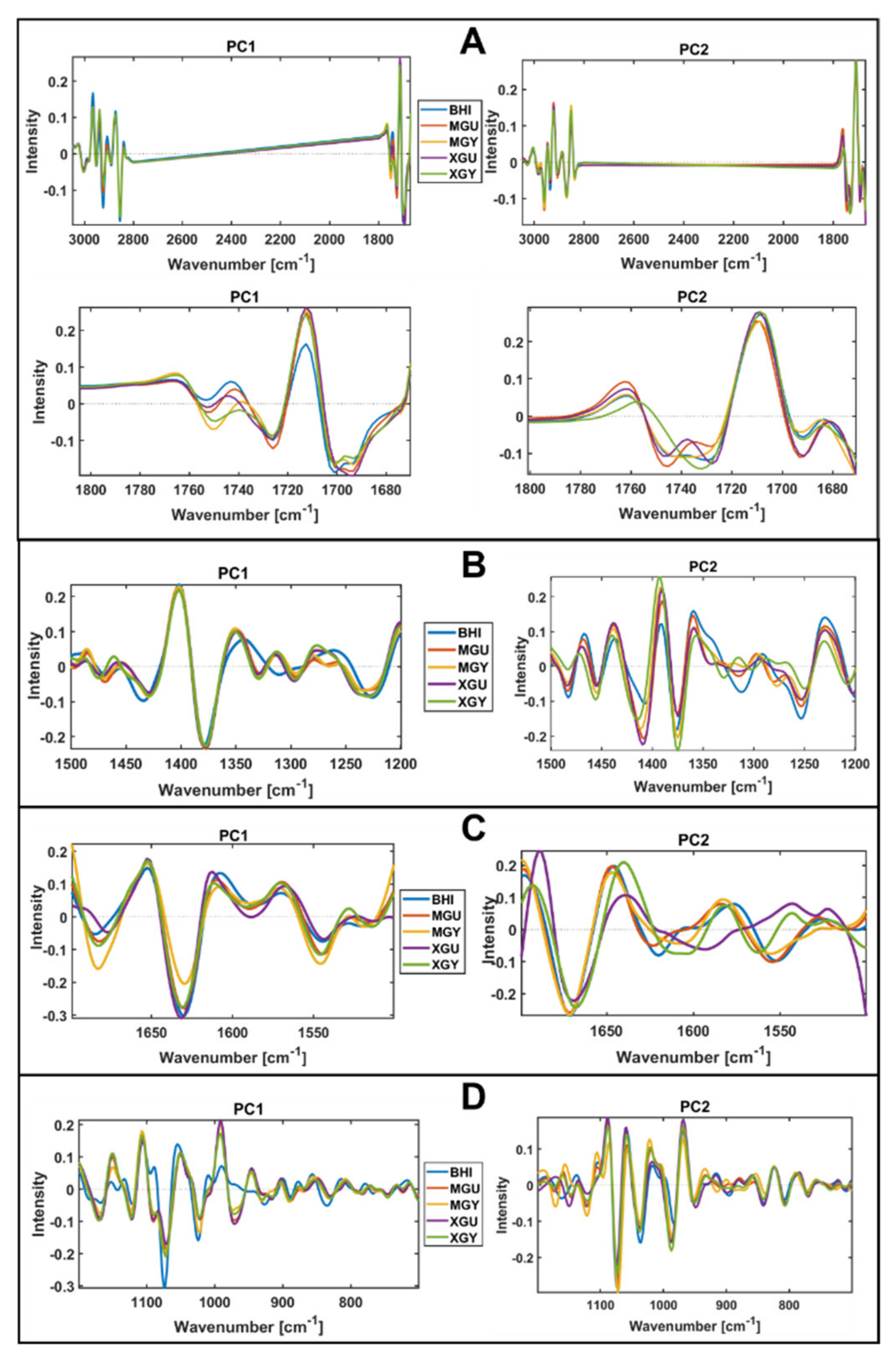
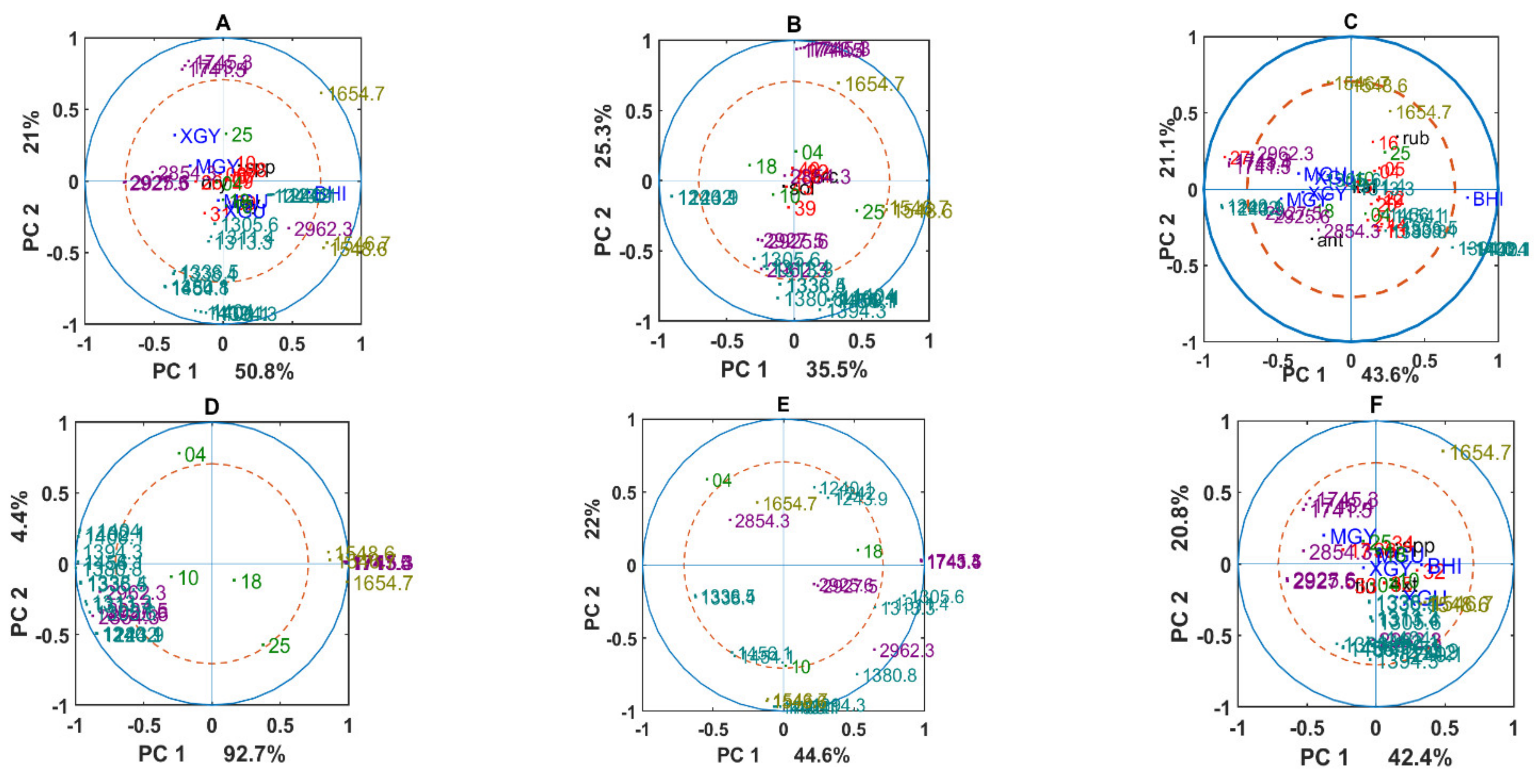
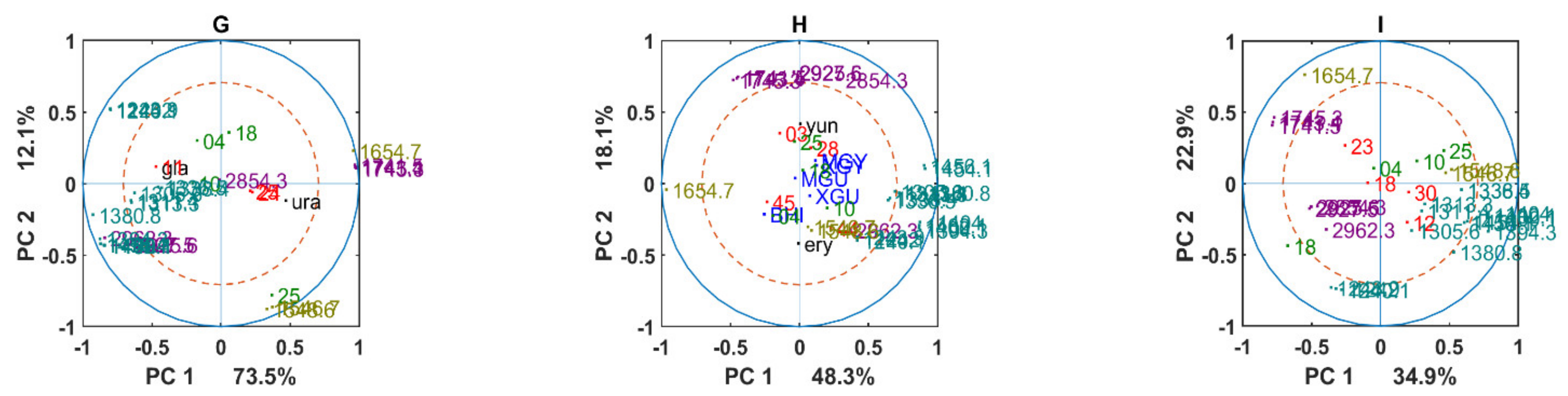
3.2. Chemical Profile Changes under Different Temperatures and Media
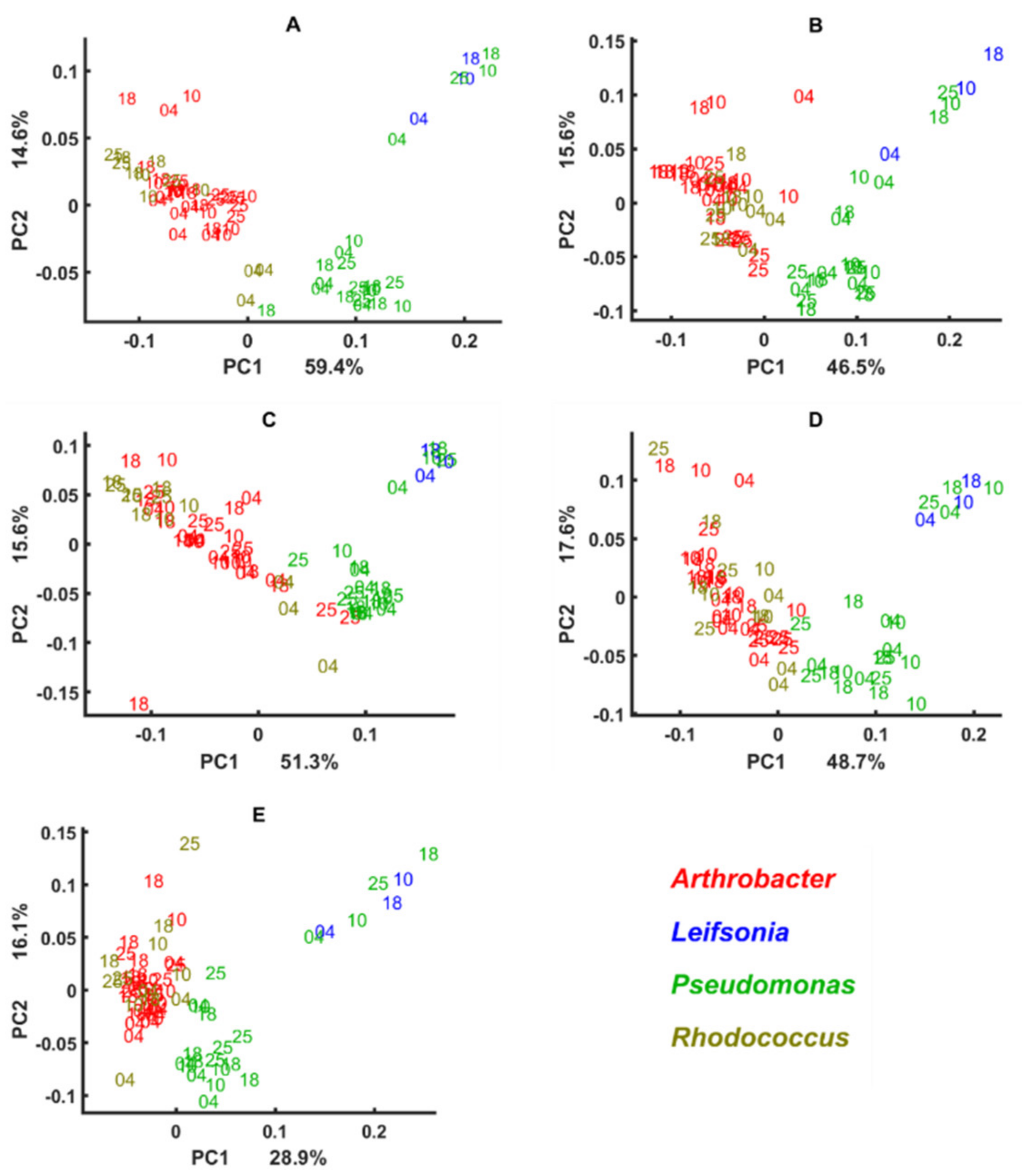
3.3. Genus Specific Responses to Fluctuations in Temperature and Nutrient Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Llano, G.A. The flora of Antarctica. Antarctica 1965, 13, 331–350. [Google Scholar]
- Allen, M.; Babiker, M.; Chen, Y.; de Coninck, H.; Conors, S. Global warming of 1.5 C. IPCC Spec. Rep. 2018, 26, 1–36. [Google Scholar]
- Hoegh-Guldberg, O.; Jacob, D.; Taylor, M.; Bolaños, T.G.; Bindi, M.; Brown, S.; Camilloni, I.A.; Diedhiou, A.; Djalante, R.; Ebi, K. The human imperative of stabilizing global climate change at 1.5 C. Science 2019, 365, eaaw697. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.P.; Norman, L.; Sterk, P.; Huete-Ortega, M.; Bunbury, F.; Loh, B.K.W.; Stockton, S.; Peck, L.S.; Convey, P.; Newsham, K.K. Snow algae communities in Antarctica: Metabolic and taxonomic composition. New Phytol. 2019, 222, 1242–1255. [Google Scholar] [CrossRef]
- Hodson, A.; Nowak, A.; Cook, J.; Sabacka, M.; Wharfe, E.; Pearce, D.; Convey, P.; Vieira, G. Microbes influence the biogeochemical and optical properties of maritime Antarctic snow. J. Geophys. Res. Biogeosci. 2017, 122, 1456–1470. [Google Scholar] [CrossRef]
- Dierssen, H.M.; Smith, R.C.; Vernet, M. Glacial meltwater dynamics in coastal waters west of the Antarctic peninsula. Proc. Natl. Acad. Sci. USA 2002, 99, 1790–1795. [Google Scholar] [CrossRef]
- Hodson, A.; Anesio, A.M.; Tranter, M.; Fountain, A.; Osborn, M.; Priscu, J.; Laybourn-Parry, J.; Sattler, B. Glacial ecosystems. Ecol. Monogr. 2008, 78, 41–67. [Google Scholar] [CrossRef]
- Boetius, A.; Anesio, A.M.; Deming, J.W.; Mikucki, J.A.; Rapp, J.Z. Microbial ecology of the cryosphere: Sea ice and glacial habitats. Nat. Rev. Microbiol. 2015, 13, 677–690. [Google Scholar] [CrossRef]
- Smirnova, M.; Miamin, U.; Kohler, A.; Valentovich, L.; Akhremchuk, A.; Sidarenka, A.; Dolgikh, A.; Shapaval, V. Isolation and characterization of fast-growing green snow bacteria from coastal East Antarctica. MicrobiologyOpen 2020, 10, e1152. [Google Scholar] [CrossRef]
- Russell, N.J. Molecular adaptations in psychrophilic bacteria: Potential for biotechnological applications. Biotechnol. Extrem. 1998, 61, 1–21. [Google Scholar]
- Pucciarelli, S.; Devaraj, R.R.; Mancini, A.; Ballarini, P.; Castelli, M.; Schrallhammer, M.; Petroni, G.; Miceli, C. Microbial consortium associated with the Antarctic marine ciliate Euplotes focardii: An investigation from genomic sequences. Microb. Ecol. 2015, 70, 484–497. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Yadav, A.N.; Rajawat, M.; Kaushik, R.; Kumar, R.; Kumar, M.; Prasanna, R.; Shukla, L. Microbial diversity of extreme regions: An unseen heritage and wealth. Indian J. Plant Genet. Resour. 2016, 29, 246–248. [Google Scholar] [CrossRef]
- Yadav, A.N.; Sachan, S.G.; Verma, P.; Kaushik, R.; Saxena, A.K. Cold active hydrolytic enzymes production by psychrotrophic Bacilli isolated from three sub-glacial lakes of NW Indian Himalayas. J. Basic Microbiol. 2016, 56, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Brakstad, O.G.; Lofthus, S.; Ribicic, D.; Netzer, R. Biodegradation of petroleum oil in cold marine environments. In Psychrophiles: From Biodiversity to Biotechnology; Springer: Berlin/Heidelberg, Germany, 2017; pp. 613–644. [Google Scholar]
- Si-Zhong, Y.; Hui-Jun, J.; Zhi, W.; Rui-Xia, H.; Yan-Jun, J.; Xiu-Mei, L.; Shao-Peng, Y. Bioremediation of oil spills in cold environments: A review. Pedosphere 2009, 19, 371–381. [Google Scholar]
- Gerday, C.; Aittaleb, M.; Bentahir, M.; Chessa, J.-P.; Claverie, P.; Collins, T.; D’Amico, S.; Dumont, J.; Garsoux, G.; Georlette, D. Cold-adapted enzymes: From fundamentals to biotechnology. Trends Biotechnol. 2000, 18, 103–107. [Google Scholar] [CrossRef]
- Huston, A.L.; Krieger-Brockett, B.B.; Deming, J.W. Remarkably low temperature optima for extracellular enzyme activity from Arctic bacteria and sea ice. Environ. Microbiol. 2000, 2, 383–388. [Google Scholar] [CrossRef]
- Groudieva, T.; Kambourova, M.; Yusef, H.; Royter, M.; Grote, R.; Trinks, H.; Antranikian, G. Diversity and cold-active hydrolytic enzymes of culturable bacteria associated with Arctic sea ice, Spitzbergen. Extremophiles 2004, 8, 475–488. [Google Scholar] [CrossRef]
- Hassan, N.; Anesio, A.M.; Rafiq, M.; Holtvoeth, J.; Bull, I.; Haleem, A.; Shah, A.A.; Hasan, F. Temperature driven membrane lipid adaptation in glacial psychrophilic bacteria. Front. Microbiol. 2020, 11, 824. [Google Scholar] [CrossRef]
- Yumoto, I.; Hirota, K.; Iwata, H.; Akutsu, M.; Kusumoto, K.; Morita, N.; Ezura, Y.; Okuyama, H.; Matsuyama, H. Temperature and nutrient availability control growth rate and fatty acid composition of facultatively psychrophilic Cobetia marina strain L-2. Arch. Microbiol. 2004, 181, 345–351. [Google Scholar] [CrossRef]
- Fong, N.; Burgess, M.; Barrow, K.; Glenn, D. Carotenoid accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl. Microbiol. Biotechnol. 2001, 56, 750–756. [Google Scholar] [CrossRef]
- Han, S.J.; Park, H.; Kim, S.; Kim, D.; Park, H.J.; Yim, J.H. Enhanced production of protease by Pseudoalteromonas arctica PAMC 21717 via statistical optimization of mineral components and fed-batch fermentation. Prep. Biochem. Biotechnol. 2016, 46, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Movasaghi, Z.; Rehman, S.; ur Rehman, D.I. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Martin-Hirsch, P.L.; Martin, F.L. ATR-FTIR spectroscopy tools for medical diagnosis and disease investigation. In Nanotechnology Characterization Tools for Biosensing and Medical Diagnosis; Springer: Berlin/Heidelberg, Germany, 2018; pp. 163–211. [Google Scholar]
- Ollesch, J.; Drees, S.L.; Heise, H.M.; Behrens, T.; Brüning, T.; Gerwert, K. FTIR spectroscopy of biofluids revisited: An automated approach to spectral biomarker identification. Analyst 2013, 138, 4092–4102. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Hassan, M.; Essa, S.; Tartor, Y. Use of Fourier transform infrared spectroscopy (FTIR) spectroscopy for rapid and accurate identification of Yeasts isolated from human and animals. Int. J. Vet. Sci. Med. 2013, 1, 15–20. [Google Scholar] [CrossRef]
- Figoli, C.B.; Garcea, M.; Bisioli, C.; Tafintseva, V.; Shapaval, V.; Peña, M.G.; Gibbons, L.; Althabe, F.; Yantorno, O.M.; Horton, M. A robust metabolomics approach for the evaluation of human embryos from in vitro fertilization. Analyst 2021, 146, 6156–6169. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, V.; Nippolainen, E.; Shaikh, R.; Afara, I.O.; Töyräs, J.; Solheim, J.; Tafintseva, V.; Zimmermann, B.; Kohler, A.; Saarakkala, S. Infrared Fiber-Optic Spectroscopy Detects Bovine Articular Cartilage Degeneration. Cartilage 2021, 13, 285S–294S. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, A.C.S.; De, G.C.R. Traceability of active compounds of essential oils in antimicrobial food packaging using a chemometric method by ATR-FTIR. Am. J. Anal. Chem. 2017, 8, 726. [Google Scholar] [CrossRef]
- Wang, Y.-D.; Li, X.-L.; Liu, Z.-X.; Zhang, X.-X.; Hu, J.; Lü, J.-H. Discrimination of foodborne pathogenic bacteria using synchrotron FTIR microspectroscopy. Nucl. Sci. Tech. 2017, 28, 49. [Google Scholar] [CrossRef]
- Zeng, W.; Guo, L.; Xu, S.; Chen, J.; Zhou, J. High-throughput screening technology in industrial biotechnology. Trends Biotechnol. 2020, 38, 888–906. [Google Scholar] [CrossRef]
- Brown, C.; Martin, A.P.; Grof, C.P. The application of Fourier transform mid-infrared (FTIR) spectroscopy to identify variation in cell wall composition of Setaria italica ecotypes. J. Integr. Agric. 2017, 16, 1256–1267. [Google Scholar] [CrossRef]
- Kosa, G.; Shapaval, V.; Kohler, A.; Zimmermann, B. FTIR spectroscopy as a unified method for simultaneous analysis of intra-and extracellular metabolites in high-throughput screening of microbial bioprocesses. Microb. Cell Factories 2017, 16, 195. [Google Scholar] [CrossRef] [PubMed]
- Magazù, S.; Calabrò, E.; Caccamo, M. Experimental study of thermal restraint in bio-protectant disaccharides by FTIR spectroscopy. Open Biotechnol. J. 2018, 12, 123–133. [Google Scholar] [CrossRef]
- Lin, M.; Al-Holy, M.; Chang, S.-S.; Huang, Y.; Cavinato, A.G.; Kang, D.-H.; Rasco, B.A. Rapid discrimination of Alicyclobacillus strains in apple juice by Fourier transform infrared spectroscopy. Int. J. Food Microbiol. 2005, 105, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Al-Qadiri, H.M.; Lin, M.; Cavinato, A.G.; Rasco, B.A. Fourier transform infrared spectroscopy, detection and identification of Escherichia coli O157: H7 and Alicyclobacillus strains in apple juice. Int. J. Food Microbiol. 2006, 111, 73–80. [Google Scholar] [CrossRef]
- Harz, M.; Rösch, P.; Popp, J. Vibrational spectroscopy—A powerful tool for the rapid identification of microbial cells at the single-cell level. Cytom. Part A J. Int. Soc. Anal. Cytol. 2009, 75, 104–113. [Google Scholar] [CrossRef]
- Shapaval, V.; Møretrø, T.; Suso, H.P.; Åsli, A.W.; Schmitt, J.; Lillehaug, D.; Martens, H.; Böcker, U.; Kohler, A. A high-throughput microcultivation protocol for FTIR spectroscopic characterization and identification of fungi. J. Biophotonics 2010, 3, 512–521. [Google Scholar] [CrossRef]
- Amamcharla, J.K.; Panigrahi, S.; Logue, C.M.; Marchello, M.; Sherwood, J.S. Fourier transform infrared spectroscopy (FTIR) as a tool for discriminating Salmonella typhimurium contaminated beef. Sens. Instrum. Food Qual. Saf. 2010, 4, 1–12. [Google Scholar] [CrossRef]
- Kiefer, J.; Ebel, N.; Schlücker, E.; Leipertz, A. Characterization of Escherichia coli suspensions using UV/Vis/NIR absorption spectroscopy. Anal. Methods 2010, 2, 123–128. [Google Scholar] [CrossRef]
- Shapaval, V.; Walczak, B.; Gognies, S.; Møretrø, T.; Suso, H.; Åsli, A.W.; Belarbi, A.; Kohler, A. FTIR spectroscopic characterization of differently cultivated food related yeasts. Analyst 2013, 138, 4129–4138. [Google Scholar] [CrossRef]
- Shapaval, V.; Schmitt, J.; Møretrø, T.; Suso, H.; Skaar, I.; Åsli, A.; Lillehaug, D.; Kohler, A. Characterization of food spoilage fungi by FTIR spectroscopy. J. Appl. Microbiol. 2013, 114, 788–796. [Google Scholar] [CrossRef]
- Shapaval, V.; Afseth, N.K.; Vogt, G.; Kohler, A. Fourier transform infrared spectroscopy for the prediction of fatty acid profiles in Mucor fungi grown in media with different carbon sources. Microb. Cell Factories 2014, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Böcker, U.; Shapaval, V.; Forsmark, A.; Andersson, M.; Warringer, J.; Martens, H.; Omholt, S.W.; Blomberg, A. High-throughput biochemical fingerprinting of Saccharomyces cerevisiae by Fourier transform infrared spectroscopy. PLoS ONE 2015, 10, e0118052. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shapaval, V.; Møretrø, T.; Wold Åsli, A.; Suso, H.-P.; Schmitt, J.; Lillehaug, D.; Kohler, A. A novel library-independent approach based on high-throughput cultivation in Bioscreen and fingerprinting by FTIR spectroscopy for microbial source tracking in food industry. Lett. Appl. Microbiol. 2017, 64, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Colabella, C.; Corte, L.; Roscini, L.; Shapaval, V.; Kohler, A.; Tafintseva, V.; Tascini, C.; Cardinali, G. Merging FT-IR and NGS for simultaneous phenotypic and genotypic identification of pathogenic Candida species. PLoS ONE 2017, 12, e0188104. [Google Scholar] [CrossRef] [PubMed]
- Kosa, G.; Zimmermann, B.; Kohler, A.; Ekeberg, D.; Afseth, N.K.; Mounier, J.; Shapaval, V. High-throughput screening of Mucoromycota fungi for production of low-and high-value lipids. Biotechnol. Biofuels 2018, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Shapaval, V.; Brandenburg, J.; Blomqvist, J.; Tafintseva, V.; Passoth, V.; Sandgren, M.; Kohler, A. Biochemical profiling, prediction of total lipid content and fatty acid profile in oleaginous yeasts by FTIR spectroscopy. Biotechnol. Biofuels 2019, 12, 1–12. [Google Scholar] [CrossRef]
- Dzurendova, S.; Zimmermann, B.; Kohler, A.; Tafintseva, V.; Slany, O.; Certik, M.; Shapaval, V. Microcultivation and FTIR spectroscopy-based screening revealed a nutrient-induced co-production of high-value metabolites in oleaginous Mucoromycota fungi. PLoS ONE 2020, 15, e0234870. [Google Scholar] [CrossRef]
- Liland, K.H.; Kohler, A.; Shapaval, V. Hot PLS—A framework for hierarchically ordered taxonomic classification by partial least squares. Chemom. Intell. Lab. Syst. 2014, 138, 41–47. [Google Scholar] [CrossRef]
- Li, J.; Shapaval, V.; Kohler, A.; Talintyre, R.; Schmitt, J.; Stone, R.; Gallant, A.J.; Zeze, D.A. A modular liquid sample handling robot for high-throughput Fourier transform infrared spectroscopy. In Advances in Reconfigurable Mechanisms and Robots II; Springer: Berlin/Heidelberg, Germany, 2016; pp. 769–778. [Google Scholar]
- Xiong, Y.; Shapaval, V.; Kohler, A.; Li, J.; From, P.J. A fully automated robot for the preparation of fungal samples for FTIR spectroscopy using deep learning. IEEE Access 2019, 7, 132763–132774. [Google Scholar] [CrossRef]
- Xiong, Y.; Shapaval, V.; Kohler, A.; From, P.J. A laboratory-built fully automated ultrasonication robot for filamentous fungi homogenization. SLAS Technol. Transl. Life Sci. Innov. 2019, 24, 583–595. [Google Scholar] [CrossRef]
- Bioquest, A. M9 Minimal Salts Preparation. Available online: https://www.aatbio.com/resources/buffer-preparations-and-recipes/m9-salts (accessed on 9 March 2022).
- Tafintseva, V.; Shapaval, V.; Smirnova, M.; Kohler, A. Extended multiplicative signal correction for FTIR spectral quality test and pre-processing of infrared imaging data. J. Biophotonics 2020, 13, e201960112. [Google Scholar] [CrossRef] [PubMed]
- Tafintseva, V.; Shapaval, V.; Blazhko, U.; Kohler, A. Correcting replicate variation in spectroscopic data by machine learning and model-based pre-processing. Chemom. Intell. Lab. Syst. 2021, 215, 104350. [Google Scholar] [CrossRef]
- Tafintseva, V.; Vigneau, E.; Shapaval, V.; Cariou, V.; Qannari, E.M.; Kohler, A. Hierarchical classification of microorganisms based on high-dimensional phenotypic data. J. Biophotonics 2018, 11, e201700047. [Google Scholar] [CrossRef] [PubMed]
- Wold, S.; Hellberg, S.; Lundstedt, T.; Sjostrom, M.; Wold, H. Theory and application. In Proceedings of the Symposium on PLS Model Building, Frankfurt am Main, Germany, 23–25 September 1987. [Google Scholar]
- Hanafi, M.; Kohler, A.; Qannari, E.M. Shedding new light on hierarchical principal component analysis. J. Chemom. 2010, 24, 703–709. [Google Scholar] [CrossRef]
- Diehn, S.; Zimmermann, B.; Tafintseva, V.; Seifert, S.; Bağcıoğlu, M.; Ohlson, M.; Weidner, S.; Fjellheim, S.; Kohler, A.; Kneipp, J. Combining chemical information from grass pollen in multimodal characterization. Front. Plant Sci. 2020, 10, 1788. [Google Scholar] [CrossRef]
- Blazhko, U.; Shapaval, V.; Kovalev, V.; Kohler, A. Comparison of augmentation and pre-processing for deep learning and chemometric classification of infrared spectra. Chemom. Intell. Lab. Syst. 2021, 215, 104367. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Zimmermann, B.; Kohler, A. Optimizing Savitzky-Golay parameters for improving spectral resolution and quantification in infrared spectroscopy. Appl. Spectrosc. 2013, 67, 892–902. [Google Scholar] [CrossRef]
- Martens, H.; Stark, E. Extended multiplicative signal correction and spectral interference subtraction: New preprocessing methods for near infrared spectroscopy. J. Pharm. Biomed. Anal. 1991, 9, 625–635. [Google Scholar] [CrossRef]
- Kohler, A.; Solheim, J.H.; Tafintseva, V.; Zimmermann, B.; Shapaval, V. Model-based pre-processing in vibrational spectroscopy. In Comprehensive Chemometrics: Chemical and Biochemical Data Analysis, 2nd ed.; Brown, S., Tauler, R., Walczak, B., Eds.; Elsevier: New York, NY, USA, 2020; pp. 83–100. [Google Scholar]
- Naumann, D. Infrared spectroscopy in microbiology. Encycl. Anal. Chem. 2000, 102, 131. [Google Scholar]
- Busse, H.J.; Wieser, M. Paeniglutamicibacter. In Bergey’s Manual of Systematic Bacteriology; Wiley: Hoboken, NJ, USA, 2015; pp. 1–9. [Google Scholar]
- Han, S.K.; Nedashkovskaya, O.I.; Mikhailov, V.V.; Kim, S.B.; Bae, K.S. Salinibacterium amurskyense gen. nov., sp. nov., a novel genus of the family Microbacteriaceae from the marine environment. Int. J. Syst. Evol. Microbiol. 2003, 53, 2061–2066. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.B.; Nedashkovskaya, O.I. Salinibacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds.; Wiley: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Kourti, T.; MacGregor, J.F. Analysis of multiblock and hierarchical PCA and PLS models. J. Chemom. J. Chemom. Soc. 1998, 12, 301–321. [Google Scholar] [CrossRef]
- Faghihzadeh, F.; Anaya, N.M.; Schifman, L.A.; Oyanedel-Craver, V. Fourier transform infrared spectroscopy to assess molecular-level changes in microorganisms exposed to nanoparticles. Nanotechnol. Environ. Eng. 2016, 1, 1. [Google Scholar] [CrossRef]
- Bajerski, F.; Ganzert, L.; Mangelsdorf, K.; Lipski, A.; Wagner, D. Cryobacterium arcticum sp. nov., a psychrotolerant bacterium from an Arctic soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 1849–1853. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P.; Cavanagh, J.; Austin, J.J.; Sanderson, K. Novel Psychrobacter species from Antarctic ornithogenic soils. Int. J. Syst. Evol. Microbiol. 1996, 46, 841–848. [Google Scholar] [CrossRef]
- Amaresan, N.; Kumar, M.S.; Annapurna, K.; Kumar, K.; Sankaranarayanan, A. Beneficial Microbes in Agro-Ecology: Bacteria and Fungi; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- López, N.I.; Pettinari, M.J.; Stackebrandt, E.; Tribelli, P.M.; Põtter, M.; Steinbüchel, A.; Méndez, B.S. Pseudomonas extremaustralis sp. nov., a poly (3-hydroxybutyrate) producer isolated from an Antarctic environment. Curr. Microbiol. 2009, 59, 514–519. [Google Scholar] [CrossRef]
- Zhang, Y.-Q.; Li, W.-J.; Kroppenstedt, R.M.; Kim, C.-J.; Chen, G.-Z.; Park, D.-J.; Xu, L.-H.; Jiang, C.-L. Rhodococcus yunnanensis sp. nov., a mesophilic actinobacterium isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2005, 55, 1133–1137. [Google Scholar] [CrossRef]
- Kutalik, Z.; Razaz, M.; Baranyi, J. Connection between stochastic and deterministic modelling of microbial growth. J. Theor. Biol. 2005, 232, 285–299. [Google Scholar] [CrossRef]
- Palleroni, N.J. The genus Pseudomonas. In Practical Handbook of Microbiology; CRC Press: Boca Raton, FL, USA, 2008; pp. 251–262. [Google Scholar]
- Palleroni, N.J. Pseudomonas. In Bergey’s Manual of Systematic Bacteriology; Wiley: Hoboken, NJ, USA, 2015; Volume 1. [Google Scholar]
- Singh, R.N.; Gaba, S.; Yadav, A.N.; Gaur, P.; Gulati, S.; Kaushik, R.; Saxena, A.K. First high quality draft genome sequence of a plant growth promoting and cold active enzyme producing psychrotrophic Arthrobacter agilis strain L77. Stand. Genom. Sci. 2016, 11, 54. [Google Scholar] [CrossRef]
- Ding, L.; Hirose, T.; Yokota, A. Four novel Arthrobacter species isolated from filtration substrate. Int. J. Syst. Evol. Microbiol. 2009, 59, 856–862. [Google Scholar] [CrossRef][Green Version]
- Bakermans, C.; Nealson, K.H. Relationship of critical temperature to macromolecular synthesis and growth yield in Psychrobacter cryopegella. J. Bacteriol. 2004, 186, 2340–2345. [Google Scholar] [CrossRef] [PubMed]
- Goordial, J.; Raymond-Bouchard, I.; Zolotarov, Y.; de Bethencourt, L.; Ronholm, J.; Shapiro, N.; Woyke, T.; Stromvik, M.; Greer, C.W.; Bakermans, C. Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol. Ecol. 2016, 92, fiv154. [Google Scholar] [PubMed]
- Evtushenko, L.I. Leifsonia. In Bergey’s Manual of Systematic Bacteriology; Wiley: Hoboken, NJ, USA, 2015; pp. 1–32. [Google Scholar]
- Simonescu, C.M. Application of FTIR spectroscopy in environmental studies. Adv. Asp. Spectrosc. 2012, 29, 77–86. [Google Scholar]
- Alvarez-Ordonez, A.; Mouwen, D.; Lopez, M.; Prieto, M. Fourier transform infrared spectroscopy as a tool to characterize molecular composition and stress response in foodborne pathogenic bacteria. J. Microbiol. Methods 2011, 84, 369–378. [Google Scholar] [CrossRef]
- Busse, H.-J. Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov., Paenarthrobacter gen. nov. and Pseudarthrobacter gen. nov., and emended description of Arthrobacter roseus. Int. J. Syst. Evol. Microbiol. 2016, 66, 9–37. [Google Scholar]
- Guckert, J.B.; Ringelberg, D.B.; White, D.C.; Hanson, R.S.; Bratina, B.J. Membrane fatty acids as phenotypic markers in the polyphasic taxonomy of methylotrophs within the Proteobacteria. Microbiology 1991, 137, 2631–2641. [Google Scholar] [CrossRef]
- Zahran, H.; Abdel-Fattah, M.; Ahmad, M.; Zaky, A. Polyphasic taxonomy of symbiotic rhizobia from wild leguminous plants growing in Egypt. Folia Microbiol. 2003, 48, 510–520. [Google Scholar] [CrossRef]
- Abdulrasheed, M.; Zakaria, N.N.; Roslee, A.F.A.; Shukor, M.Y.; Zulkharnain, A.; Napis, S.; Convey, P.; Alias, S.A.; Gonzalez-Rocha, G.; Ahmad, S.A. Biodegradation of diesel oil by cold-adapted bacterial strains of Arthrobacter spp. from Antarctica. Antarct. Sci. 2020, 32, 341–353. [Google Scholar] [CrossRef]
- Ibrahim, S.; Abdul Khalil, K.; Zahri, K.N.M.; Gomez-Fuentes, C.; Convey, P.; Zulkharnain, A.; Sabri, S.; Alias, S.A.; González-Rocha, G.; Ahmad, S.A. Biosurfactant production and growth kinetics studies of the waste canola oil-degrading bacterium Rhodococcus erythropolis AQ5-07 from Antarctica. Molecules 2020, 25, 3878. [Google Scholar] [CrossRef]
- Roslee, A.F.A.; Zakaria, N.N.; Convey, P.; Zulkharnain, A.; Lee, G.L.Y.; Gomez-Fuentes, C.; Ahmad, S.A. Statistical optimisation of growth conditions and diesel degradation by the Antarctic bacterium, Rhodococcus sp. strain AQ5–07. Extremophiles 2020, 24, 277–291. [Google Scholar] [CrossRef]
- Sinensky, M. Homeoviscous adaptation—A homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. USA 1974, 71, 522–525. [Google Scholar] [CrossRef] [PubMed]
- Georlette, D.; Blaise, V.; Collins, T.; D’Amico, S.; Gratia, E.; Hoyoux, A.; Marx, J.-C.; Sonan, G.; Feller, G.; Gerday, C. Some like it cold: Biocatalysis at low temperatures. FEMS Microbiol. Rev. 2004, 28, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S.; Williams, T.J.; Wilkins, D.; Yau, S.; Allen, M.A.; Brown, M.V.; Lauro, F.M.; Cavicchioli, R. Psychrophiles. Annu. Rev. Earth Planet. Sci. 2013, 41, 87–115. [Google Scholar] [CrossRef]
- Fukunaga, N.; Russell, N.J. Membrane lipid composition and glucose uptake in two psychrotolerant bacteria from Antarctica. Microbiology 1990, 136, 1669–1673. [Google Scholar] [CrossRef]


| Bacterial Isolate Name | Collection No/Samples Number [9] |
|---|---|
| Arthrobacter cryoconiti | BIM B-1627/G.S.9 |
| Arthrobacter oryzae | BIM B-1663/G.S.26 |
| Arthrobacter sp. | BIM B-1624/G.S.6 |
| Arthrobacter sp. | BIM B-1625/G.S.7 |
| Arthrobacter sp. | BIM B-1626/G.S.8 |
| Arthrobacter sp. | BIM B-1628/G.S.10 |
| Arthrobacter sp. | BIM B-1664/G.S.29 |
| Arthrobacter sp. | BIM B-1666/G.S.31 |
| Arthrobacter sp. | BIM B-1656/G.S.37 |
| Cryobacterium arcticum | BIM B-1619/G.S.1 |
| Cryobacterium arcticum | BIM B-1620/G.S.2 |
| Cryobacterium soli | BIM B-1658/G.S.39 |
| Cryobacterium soli | BIM B-1659/G.S.40 |
| Cryobacterium soli | BIM B-1677/G.S.41 |
| Cryobacterium soli | BIM B-1675/G.S.43 |
| Leifsonia antarctica | BIM B-1631/G.S.13 |
| Leifsonia antarctica | BIM B-1632/G.S.14 |
| Leifsonia antarctica | BIM B-1637/G.S.19 |
| Leifsonia antarctica | BIM B-1638/G.S.20 |
| Leifsonia antarctica | BIM B-1639/G.S.21 |
| Leifsonia antarctica | BIM B-1669/G.S.22 |
| Leifsonia antarctica | BIM B-1671/G.S.27 |
| Leifsonia kafniensis | BIM B-1633/G.S.15 |
| Leifsonia rubra | BIM B-1622/G.S.4 |
| Leifsonia rubra | BIM B-1623/G.S.5 |
| Leifsonia rubra | BIM B-1634/G.S.16 |
| Paeniglutamicibacter antarcticus | BIM B-1657/G.S.38 |
| Polaromonas sp. | BIM B-1676/G.S.42 |
| Pseudomonas extremaustralis | BIM B-1672/G.S.35 |
| Pseudomonas fluorescens | BIM B-1668/G.S.33 |
| Pseudomonas sp. | BIM B-1635/G.S.17 |
| Pseudomonas sp. | BIM B-1667/G.S.32 |
| Pseudomonas sp. | BIM B-1673/G.S.34 |
| Pseudomonas versuta | BIM B-1674/G.S.36 |
| Psychrobacter glacinicola | BIM B-1629/G.S.11 |
| Psychrobacter urativorans | BIM B-1655/G.S.24 |
| Psychrobacter urativorans | BIM B-1662/G.S.25 |
| Rhodococcus yunnanensis | BIM B-1621/G.S.3 |
| Rhodococcus yunnanensis | BIM B-1670/G.S.28 |
| Rhodococcus erythropolis | BIM B-1660/G.S.44 |
| Rhodococcus erythropolis | BIM B-1661/G.S.45 |
| Salinibacterium sp. | BIM B-1630/G.S.12 |
| Salinibacterium sp. | BIM B-1636/G.S.18 |
| Salinibacterium sp. | BIM B-1654/G.S.23 |
| Salinibacterium sp. | BIM B-1665/G.S.30 |
| Composition of Salt Solution (g/L) | MGU | MGY | XGU | XGY | BHI |
|---|---|---|---|---|---|
| Na2HPO4 | 24 | 24 | - | - | - |
| KH2PO4 | 12 | 12 | 4 | 4 | - |
| K2HPO4 | - | - | 12 | 12 | - |
| NaCl | 2.5 | 2.5 | - | - | - |
| NH4Cl | 5 | 5 | 20 | 20 | - |
| NH4NO3 | - | - | 4 | 4 | - |
| Na2SO4 × 10H2O | - | - | 8 | 8 | - |
| MgSO4 × 7H2O | - | - | 0.4 | 0.4 | - |
| Commercial powder | - | - | - | - | 37 |
| Media composition (mL) | |||||
| Salt solution | 100 | 100 | 100 | 100 | - |
| 20% agar | 300 | 300 | 300 | 300 | 1000 |
| 0.1 M CaCl2 | 4 | 4 | - | - | - |
| 0.1 M MgSO4 | 4 | 4 | - | - | - |
| 20% glucose | 4 | - | 4 | - | - |
| 20% glycerol | - | 4 | - | 4 | - |
| Arthrobacter | ||||||||||||
| 1624 * G.S.6 | 1625 G.S.7 | 1626 G.S.8 | 1627 G.S.9 | 1628 G.S.10 | 1663 G.S.26 | 1664 G.S.29 | 1666 G.S.31 | 1656 G.S.37 | ||||
| BHI | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| XGU | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| XGY | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| MGU | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| MGY | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| Pseudomonas | Rhodococcus | |||||||||||
| 1635 G.S.17 | 1667 G.S.32 | 1668 G.S.33 | 1673 G.S.34 | 1672 G.S.35 | 1674 G.S.36 | 1621 G.S.3 | 1670 G.S.28 | 1660 G.S.44 | 1661 G.S.45 | |||
| BHI | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| XGU | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| XGY | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| MGU | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| MGY | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| Cryobacterium | Salinibacterium | |||||||||||
| 1619 G.S.1 | 1620 G.S.2 | 1658 G.S.39 | 1659 G.S.40 | 1677 G.S.41 | 1675 G.S.43 | 1630 G.S.12 | 1636 G.S.18 | 1654 G.S.23 | 1665 G.S.30 | |||
| BHI | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| XGU XGY MGU MGY | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| Leifsonia | ||||||||||||
| 1622 G.S.4 | 1623 G.S.5 | 1631 G.S.13 | 1632 G.S.14 | 1633 G.S.15 | 1634 G.S.16 | 1637 G.S.19 | 1638 G.S.20 | 1639 G.S.21 | 1669 G.S.22 | 1671 G.S.27 | ||
| BHI | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| XGU XGY MGU MGY | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| Psychrobacter | Paeniglutamicibcter antarcticus 1657 G.S.38 | Polaromonas sp. 1676 G.S.42 | ||||||||||
| 1629 G.S.11 | 1655 G.S.24 | 1662 G.S.25 | ||||||||||
| BHI | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
| XGU XGY MGU MGY | 4 °C | |||||||||||
| 10 °C | ||||||||||||
| 18 °C | ||||||||||||
| 25 °C | ||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smirnova, M.; Tafintseva, V.; Kohler, A.; Miamin, U.; Shapaval, V. Temperature- and Nutrients-Induced Phenotypic Changes of Antarctic Green Snow Bacteria Probed by High-Throughput FTIR Spectroscopy. Biology 2022, 11, 890. https://doi.org/10.3390/biology11060890
Smirnova M, Tafintseva V, Kohler A, Miamin U, Shapaval V. Temperature- and Nutrients-Induced Phenotypic Changes of Antarctic Green Snow Bacteria Probed by High-Throughput FTIR Spectroscopy. Biology. 2022; 11(6):890. https://doi.org/10.3390/biology11060890
Chicago/Turabian StyleSmirnova, Margarita, Valeria Tafintseva, Achim Kohler, Uladzislau Miamin, and Volha Shapaval. 2022. "Temperature- and Nutrients-Induced Phenotypic Changes of Antarctic Green Snow Bacteria Probed by High-Throughput FTIR Spectroscopy" Biology 11, no. 6: 890. https://doi.org/10.3390/biology11060890
APA StyleSmirnova, M., Tafintseva, V., Kohler, A., Miamin, U., & Shapaval, V. (2022). Temperature- and Nutrients-Induced Phenotypic Changes of Antarctic Green Snow Bacteria Probed by High-Throughput FTIR Spectroscopy. Biology, 11(6), 890. https://doi.org/10.3390/biology11060890






