Reversion of MRAP2 Protein Sequence Generates a Functional Novel Pharmacological Modulator for MC4R Signaling
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Multiple Sequence Alignment and TM Prediction
2.2. Plasmids
2.3. Cell Culture and Transfection
2.4. Western Blot and Co-Immunoprecipitation (CoIP)
2.5. Immunofluorescence Assay
2.6. Bimolecular Fluorescent Complimentary (BiFC) Assay
2.7. ELISA Assay
2.8. cAMP Luminescent Assay
2.9. Statistical Analysis
3. Results
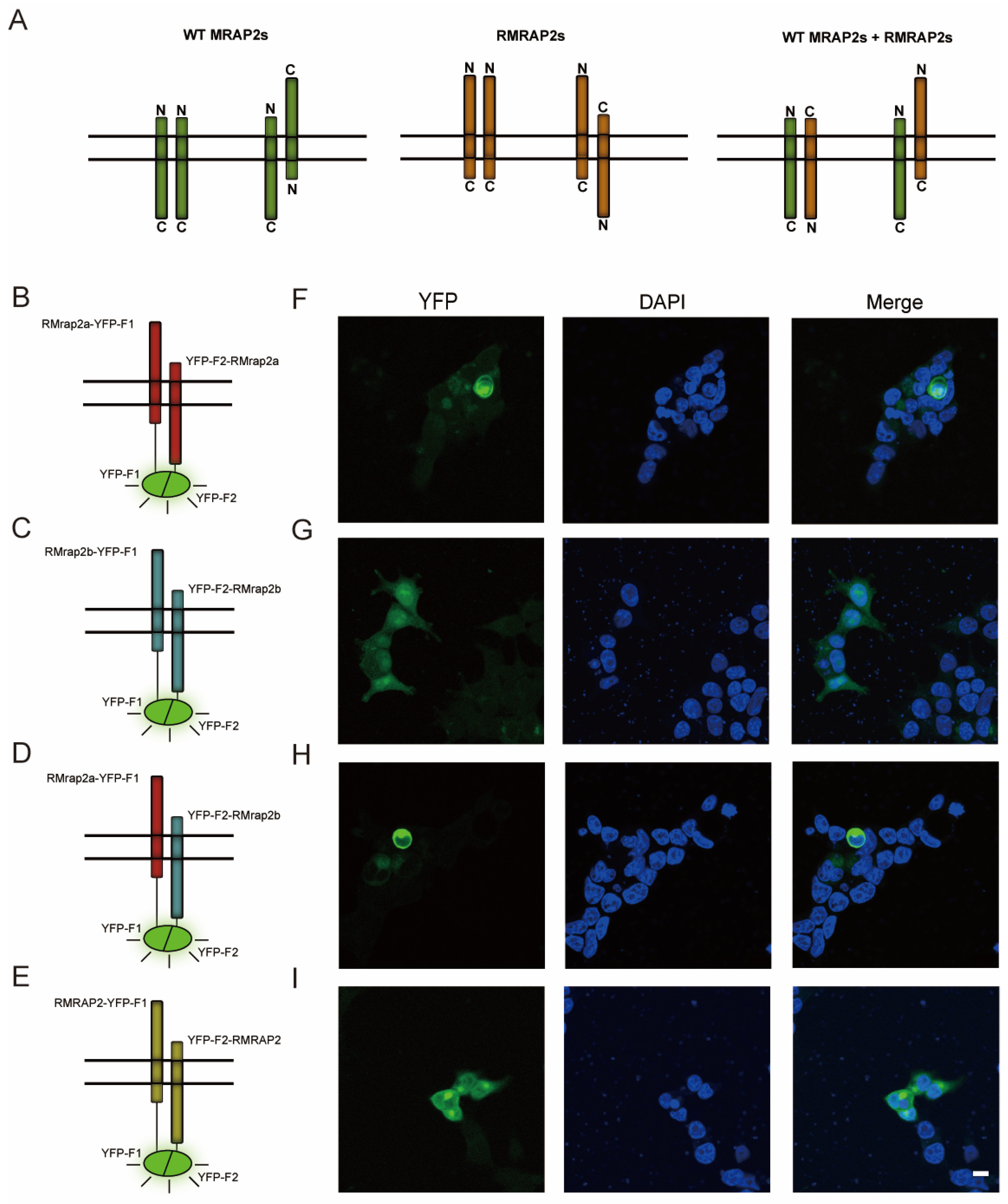
3.1. Interaction of Mouse and Zebrafish RMRAP2s with MC4Rs
3.2. RMRAP2s Could Form Homodimers and Heterodimers
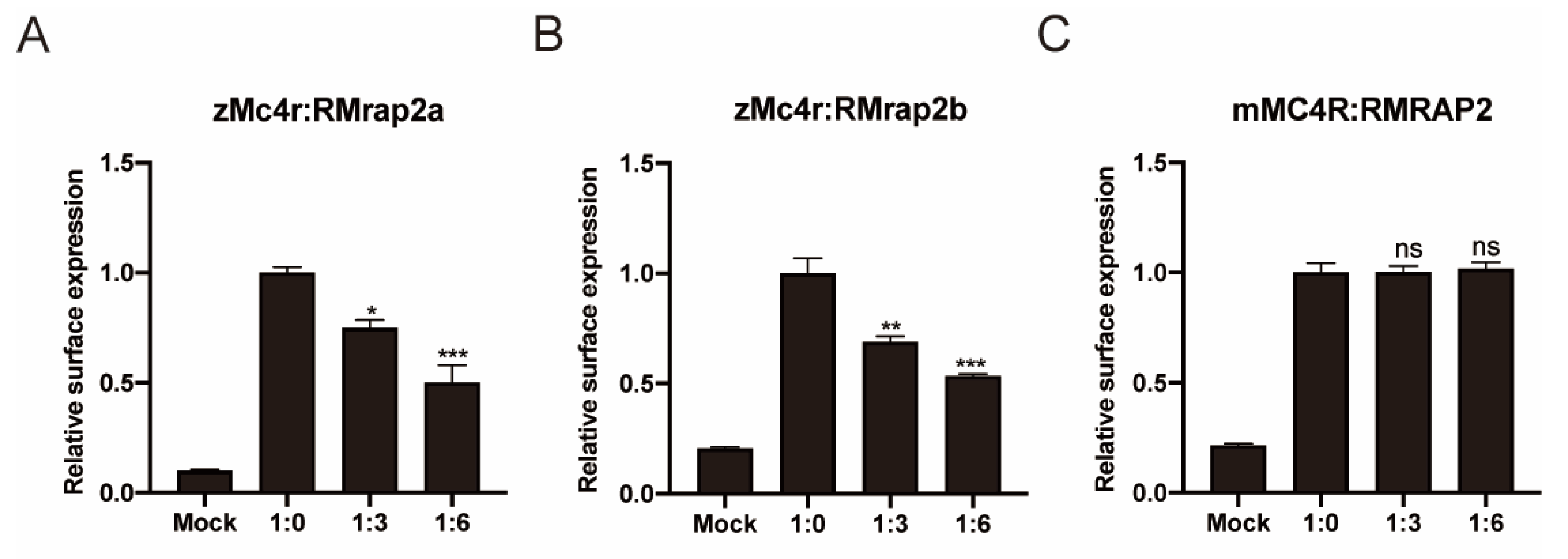
3.3. RMrap2a and RMrap2b Inhibit the Membrane Trafficking of zMc4r
3.4. RMrap2a/b Affects Pharmacological Activity of zMc4r
3.5. The Distinct Effects of Wild-Type and Reversed Mrap2a/b on Pharmacological Activity of zMc4r
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moscowitz, A.E.; Asif, H.; Lindenmaier, L.B.; Calzadilla, A.; Zhang, C.; Mirsaeidi, M. The Importance of Melanocortin Receptors and Their Agonists in Pulmonary Disease. Front. Med. 2019, 6, 145. [Google Scholar] [CrossRef]
- Xu, Y.; Guan, X.; Zhou, R.; Gong, R. Melanocortin 5 receptor signaling pathway in health and disease. Cell Mol. Life Sci. 2020, 77, 3831–3840. [Google Scholar] [CrossRef] [PubMed]
- Switonski, M.; Mankowska, M.; Salamon, S. Family of melanocortin receptor (MCR) genes in mammals-mutations, polymorphisms and phenotypic effects. J. Appl. Genet. 2013, 54, 461–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dores, R.M.; Londraville, R.L.; Prokop, J.; Davis, P.; Dewey, N.; Lesinski, N. Molecular evolution of GPCRs: Melanocortin/melanocortin receptors. J. Mol. Endocrinol. 2014, 52, T29–T42. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.X. Melanocortin receptors. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2411–2413. [Google Scholar] [CrossRef]
- Tao, Y.X. The melanocortin-4 receptor: Physiology, pharmacology, and pathophysiology. Endocr. Rev. 2010, 31, 506–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huszar, D.; Lynch, C.A.; Fairchild-Huntress, V.; Dunmore, J.H.; Fang, Q.; Berkemeier, L.R.; Lee, F. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 1997, 88, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Williams, M.D.; Nawaz, A.; Evans, M. Drug Therapy in Obesity: A Review of Current and Emerging Treatments. Diabetes Ther. 2020, 11, 1199–1216. [Google Scholar] [CrossRef] [Green Version]
- Greenfield, J.R.; Miller, J.W.; Keogh, J.M.; Henning, E.; Satterwhite, J.H.; Cameron, G.S.; Farooqi, I.S. Modulation of Blood Pressure by Central Melanocortinergic Pathways. N. Engl. J. Med. 2009, 360, 44–52. [Google Scholar] [CrossRef]
- Collet, T.H.; Dubern, B.; Mokrosinski, J.; Connors, H.; Keogh, J.M.; de Oliveira, E.M.; Van der Ploeg, L.H. Evaluation of a melanocortin-4 receptor (MC4R) agonist (Setmelanotide) in MC4R deficiency. Mol. Metab. 2017, 6, 1321–1329. [Google Scholar] [CrossRef]
- Li, L.; Liang, J.; Zhang, C.; Liu, T.; Zhang, C. Peripheral ctions and direct central-local communications of melanocortin 4 receptor signaling. J. Sport Health Sci. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Cone, R.D. Studies on the physiological functions of the melanocortin system. Endocr. Rev. 2006, 27, 736–749. [Google Scholar] [CrossRef]
- Yang, L.K.; Tao, Y.X. Biased signaling at neural melanocortin receptors in regulation of energy homeostasis. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2486–2495. [Google Scholar] [CrossRef]
- Lotta, L.A.; Mokrosiński, J.; de Oliveira, E.M.; Li, C.; Sharp, S.J.; Luan, J.A.; Farooqi, I.S. Human Gain-of-Function MC4R Variants Show Signaling Bias and Protect against Obesity. Cell 2019, 177, 597–607.e9. [Google Scholar] [CrossRef] [Green Version]
- Cooray, S.N.; Almiro Do Vale, I.; Leung, K.Y.; Webb, T.R.; Chapple, J.P.; Egertová, M.; Clark, A.J. The melanocortin 2 receptor accessory protein exists as a homodimer and is essential for the function of the melanocortin 2 receptor in the mouse y1 cell line. Endocrinology 2008, 149, 1935–1941. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Li, L.; Zheng, J.; Wang, M.; Jiang, B.; Zhai, Y.; Zhang, C. Pharmacological modulation of the cAMP signaling of two isoforms of melanocortin-3 receptor by melanocortin receptor accessory proteins in the tetrapod Xenopus laevis. Endocr. Connect. 2021, 10, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, Y.; Zhu, M.; Xu, B.; Guo, W.; Lyu, Y.; Zhang, C. Pharmacological modulation of melanocortin-4 receptor by melanocortin receptor accessory protein 2 in Nile tilapia. Gen. Comp. Endocrinol. 2019, 282, 113219. [Google Scholar] [CrossRef] [PubMed]
- Tao, M.; Ji, R.L.; Huang, L.; Fan, S.Y.; Liu, T.; Liu, S.J.; Tao, Y.X. Regulation of Melanocortin-4 Receptor Pharmacology by Two Isoforms of Melanocortin Receptor Accessory Protein 2 in Topmouth Culter (Culter alburnus). Front. Endocrinol. 2020, 11, 538. [Google Scholar] [CrossRef]
- Sebag, J.A.; Hinkle, P.M. Opposite effects of the melanocortin-2 (MC2) receptor accessory protein MRAP on MC2 and MC5 receptor dimerization and trafficking. J. Biol. Chem. 2009, 284, 22641–22648. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Deng, Y.; Zhang, Z.; Cao, B.; Li, J.; Sun, C.; Wang, Y. Melanocortin Receptor 4 (MC4R) Signaling System in Nile Tilapia. Int. J. Mol. Sci. 2020, 21, 7036. [Google Scholar] [CrossRef]
- Rouault, A.A.J.; Lee, A.A.; Sebag, J.A. Regions of MRAP2 required for the inhibition of orexin and prokineticin receptor signaling. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2322–2329. [Google Scholar] [CrossRef] [PubMed]
- Berruien, N.N.A.; Smith, C.L. Emerging roles of melanocortin receptor accessory proteins (MRAP and MRAP2) in physiology and pathophysiology. Gene 2020, 757, 144949. [Google Scholar] [CrossRef] [PubMed]
- Rouault, A.A.; Rosselli-Murai, L.K.; Hernandez, C.C.; Gimenez, L.E.; Tall, G.G.; Sebag, J.A. The GPCR accessory protein MRAP2 regulates both biased signaling and constitutive activity of the ghrelin receptor GHSR1a. Sci. Signal. 2020, 13, eaax4569. [Google Scholar] [CrossRef] [Green Version]
- Chaly, A.L.; Srisai, D.; Gardner, E.E.; Sebag, J.A. The Melanocortin Receptor Accessory Protein 2 promotes food intake through inhibition of the Prokineticin Receptor-1. Elife 2016, 5, e12397. [Google Scholar] [CrossRef] [Green Version]
- Srisai, D.; Yin, T.C.; Lee, A.A.; Rouault, A.A.; Pearson, N.A.; Grobe, J.L.; Sebag, J.A. MRAP2 regulates ghrelin receptor signaling and hunger sensing. Nat. Commun. 2017, 8, 713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Li, X.; Zhou, Y.; Cui, L.; Li, J.; Wu, C.; Wang, Y. The interaction of MC3R and MC4R with MRAP2, ACTH, α-MSH and AgRP in chickens. J. Endocrinol. 2017, 234, 155–174. [Google Scholar] [CrossRef]
- Wen, Z.Y.; Liu, T.; Qin, C.J.; Zou, Y.C.; Wang, J.; Li, R.; Tao, Y.X. MRAP2 Interaction with Melanocortin-4 Receptor in SnakeHead (Channa argus). Biomolecules 2021, 11, 481. [Google Scholar] [CrossRef] [PubMed]
- Asai, M.; Ramachandrappa, S.; Joachim, M.; Shen, Y.; Zhang, R.; Nuthalapati, N.; Majzoub, J.A. Loss of Function of the Melanocortin 2 Receptor Accessory Protein 2 Is Associated with Mammalian Obesity. Science 2013, 341, 275–278. [Google Scholar] [PubMed] [Green Version]
- Tai, X.; Xue, S.; Zhang, C.; Liu, Y.; Chen, J.; Han, Y.; Zhang, C. Pharmacological evaluation of MRAP proteins on Xenopus neural melanocortin signaling. J. Cell Physiol. 2021, 236, 6344–6361. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, Y.; Zheng, J.; Kuang, Z.; Zhang, C.; Li, N.; Zhang, C. Pharmacological modulation of dual melanocortin-4 receptor signaling by melanocortin receptor accessory proteins in the Xenopus laevis. J. Cell Physiol. 2021, 236, 5980–5993. [Google Scholar] [CrossRef]
- Sebag, J.A.; Zhang, C.; Hinkle, P.M.; Bradshaw, A.M.; Cone, R.D. Developmental Control of the Melanocortin-4 Receptor by MRAP2 Proteins in Zebrafish. Science 2013, 341, 278–281. [Google Scholar] [CrossRef] [Green Version]
- Josep Agulleiro, M.; Cortés, R.; Fernández-Durán, B.; Navarro, S.; Guillot, R.; Meimaridou, E.; Cerdá-Reverter, J.M. Melanocortin 4 receptor becomes an ACTH receptor by coexpression of melanocortin receptor accessory protein 2. Mol. Endocrinol. 2013, 27, 1934–1945. [Google Scholar] [CrossRef]
- Sebag, J.A.; Hinkle, P.M. Regulation of G Protein–Coupled Receptor Signaling: Specific Dominant-Negative Effects of Melanocortin 2 Receptor Accessory Protein 2. Sci. Signal. 2010, 3, ra28. [Google Scholar] [CrossRef] [Green Version]
- Chen, V.; Bruno, A.E.; Britt, L.L.; Hernandez, C.C.; Gimenez, L.E.; Peisley, A.; Millhauser, G.L. Membrane orientation and oligomerization of the melanocortin receptor accessory protein 2. J. Biol. Chem. 2020, 295, 16370–16379. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, M.; Chen, Y.; Zhang, C. Pharmacological modulation of two melanocortin-5 receptors by MRAP2 proteins in zebrafish. J. Mol. Endocrinol. 2018, 62, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Zhai, Y.; Lu, L.; Zhang, C.; Li, N.; Xue, S.; Zhang, C. Elucidation of the dimeric interplay of dual MRAP2 proteins in the zebrafish. J. Cell Physiol. 2021, 236, 6472–6480. [Google Scholar] [CrossRef]
- Wang, M.; Pi, L.; Lei, X.; Li, L.; Xu, J.; Kuang, Z.; Zhang, C. Functional Characterization of the Internal Symmetry of MRAP2 Antiparallel Homodimer. Front. Endocrinol. 2021, 12, 750797. [Google Scholar] [CrossRef]
- Liang, J.; Li, L.; Jin, X.; Xu, B.; Pi, L.; Liu, S.; Zhang, C. Pharmacological effect of human melanocortin-2 receptor accessory protein 2 variants on hypothalamic melanocortin receptors. Endocrine 2018, 61, 94–104. [Google Scholar] [CrossRef]
- Yu, J.; Gimenez, L.E.; Hernandez, C.C.; Wu, Y.; Wein, A.H.; Han, G.W.; Stevens, R.C. Determination of the melanocortin-4 receptor structure identifies Ca2+ as a cofactor for ligand binding. Science 2020, 368, 428–433. [Google Scholar] [CrossRef]
- Zhang, Y.; Weber, J.K.; Zhou, R. Folding and Stabilization of Native-Sequence-Reversed Proteins. Sci. Rep. 2016, 6, 25138. [Google Scholar] [CrossRef] [Green Version]




| EC50 | |||||
|---|---|---|---|---|---|
| Figure 5 | 1:0 | 1:3 | 1:6 | ||
| A | zMc4r:RMrap2a (α-MSH) | 1.51 × 10−9 [±1.06 × 10−9] | 4.01 × 10−10 ns [±2.49 10−10] | 6.62 × 10−11 *** [±9.93 10−11] | |
| B | zMc4r:RMrap2b (α-MSH) | 1.23 × 10−9 [±0.55 × 10−9] | 9.42 10−10 * [±11.80 10−10] | 1.28 10−9 *** [±1.56 10−9] | |
| C | mMC4R:RMRAP2 (α-MSH) | 5.79 10−9 [±2.83 10−9] | 5.47 10−9 ns [±2.06 10−9] | 7.46 10−9 ns [±5.43 10−9] | |
| D | zMc4r:RMrap2a (SHU9119) | 1.04 10−8 [±0.67 10−8] | 6.97 10−9 *** [±5.21 10−9] | 8.59 10−9 *** [±10.16 10−9] | |
| E | zMc4r:RMrap2b (SHU9119) | 1.11 10−8 [±0.75 10−8] | 1.80 10−8 ns [±1.74 10−8] | 7.68 10−9 ** [±6.31 10−9] | |
| F | mMC4R:RMRAP2 (SHU9119) | 8.17 10−11 [±6.02 10−11] | 8.37 10−11 ns [±11.60 10−11] | 4.20 10−11 ns [±11.56 10−11] | |
| Figure 6 | 1:0:0 | 1:2:4 | 1:3:3 | 1:4:2 | |
| A | zMc4r:Mrap2a:RMrap2a (α-MSH) | 2.60 10−9 [±1.74 10−9] | 5.26 10−10 ns [±3.01 10−10] | 8.17 10−10 ns [±2.61 10−10] | 1.07 10−9 ns [±0.37 10−9] |
| B | zMc4r:Mrap2b:RMrap2b (α-MSH) | 9.44 10−10 [±6.12 10−10] | 3.60 10−10 * [±4.00 10−10] | 3.02 10−10 * [±3.73 10−10] | 2.50 10−10 * [±3.02 10−9] |
| C | mMC4R:MRAP2:RMRAP2 (α-MSH) | 6.48 10−9 [±4.15 10−9] | 1.62 10−8 ns [±1.91 10−8] | 2.81 10−8 ** [±3.43 10−8] | 2.41 10−8 ns [±12.62 10−8] |
| D | zMc4r:Mrap2a:RMrap2a (SHU9119) | 1.23 10−8 [±1.42 10−8] | 1.30 10−8 ns [±0.55 10−8] | 9.03 10−9 * [±3.97 10−9] | 7.53 10−9 * [±3.16 10−9] |
| E | zMc4r:Mrap2b:RMrap2b (SHU9119) | 8.78 10−9 [±11.20 10−9] | 2.87 10−9 *** [±4.11 10−9] | 3.85 10−9 *** [±3.40 10−9] | 3.99 10−9 *** [±2.01 10−9] |
| F | mMC4R:MRAP2:RMRAP2 (SHU9119) | 2.49 10−9 [±3.05 10−9] | 1.02 10−9 ns [±0.39 10−9] | 5.47 10−10 ns [±2.29 10−10] | 3.83 10−10 ns [±1.37 10−10] |
| Figure 5 | 1:0 | 1:3 | 1:6 | |||
|---|---|---|---|---|---|---|
| A | zMc4r:RMrap2a | constitutive activity | 100% | 109.1% | 129.7% | |
| maximal activity | 134.1% | 130.0% | 137.1% | |||
| B | zMc4r:RMrap2b | constitutive activity | 100% | 128.3% | 126.7% | |
| maximal activity | 132.0% | 111.4% | 122.1% | |||
| C | mMC4R:RMRAP2 | constitutive activity | 100% | 114.2% | 123.1% | |
| maximal activity | 245.2% | 199.4% | 175.1% | |||
| Figure 6 | 1:0:0 | 1:2:4 | 1:3:3 | 1:4:2 | ||
| A | zMc4r:Mrap2a:RMrap2a | constitutive activity | 100% | 119.7% | 96.7% | 72.7% |
| maximal activity | 160.3% | 230.5% | 280.3% | 400% | ||
| B | zMc4r:Mrap2b:RMrap2b | constitutive activity | 100% | 126.7% | 119.7% | 112.3% |
| maximal activity | 142.6% | 133.9% | 137.5% | 147.8% | ||
| C | mMC4R:MRAP2:RMRAP2 | constitutive activity | 100% | 142.0% | 167.0% | 161.3% |
| maximal activity | 180.6% | 142.8% | 134.8% | 124.7% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Wang, M.; Fu, Y.; Zhang, C.; Kuang, Z.; Bian, S.; Wan, R.; Qu, S.; Zhang, C. Reversion of MRAP2 Protein Sequence Generates a Functional Novel Pharmacological Modulator for MC4R Signaling. Biology 2022, 11, 874. https://doi.org/10.3390/biology11060874
Xu J, Wang M, Fu Y, Zhang C, Kuang Z, Bian S, Wan R, Qu S, Zhang C. Reversion of MRAP2 Protein Sequence Generates a Functional Novel Pharmacological Modulator for MC4R Signaling. Biology. 2022; 11(6):874. https://doi.org/10.3390/biology11060874
Chicago/Turabian StyleXu, Jing, Meng Wang, Yanbin Fu, Cong Zhang, Zhe Kuang, Shan Bian, Rui Wan, Shen Qu, and Chao Zhang. 2022. "Reversion of MRAP2 Protein Sequence Generates a Functional Novel Pharmacological Modulator for MC4R Signaling" Biology 11, no. 6: 874. https://doi.org/10.3390/biology11060874
APA StyleXu, J., Wang, M., Fu, Y., Zhang, C., Kuang, Z., Bian, S., Wan, R., Qu, S., & Zhang, C. (2022). Reversion of MRAP2 Protein Sequence Generates a Functional Novel Pharmacological Modulator for MC4R Signaling. Biology, 11(6), 874. https://doi.org/10.3390/biology11060874









