agReg-SNPdb-Plants: A Database of Regulatory SNPs for Agricultural Plant Species
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
- Selection of SNPs in the promoter and surrounding region: For each gene, we considered a promoter region of 7.5 kb upstream to 2.5 kb downstream from the transcription start site (TSS) and selected all SNPs located within that region. On the website, the user has the possibility to insert a user-defined promoter region with the default being −1 kb to +100 bp.
- Extraction of the SNP-flanking region: Using the reference genomes under study, we extracted 25 bp on each side of a SNP to obtain 51 bp long sequences with the SNP in the central position. During this step, we discarded sequences with a total length of less than 51 bp, sequences containing N’s, and sequences in which the nucleotide at position 26 differed from the reference allele of the SNP (as specified in the SNP catalog in GVF format [26]). The latter only occurred in the species tomato, Asian rice (Indica Group), and sorghum.
- Creation of search sequences: For each SNP, we created an additional copy of its 51 bp long sequence by replacing the reference allele with its alternate allele.
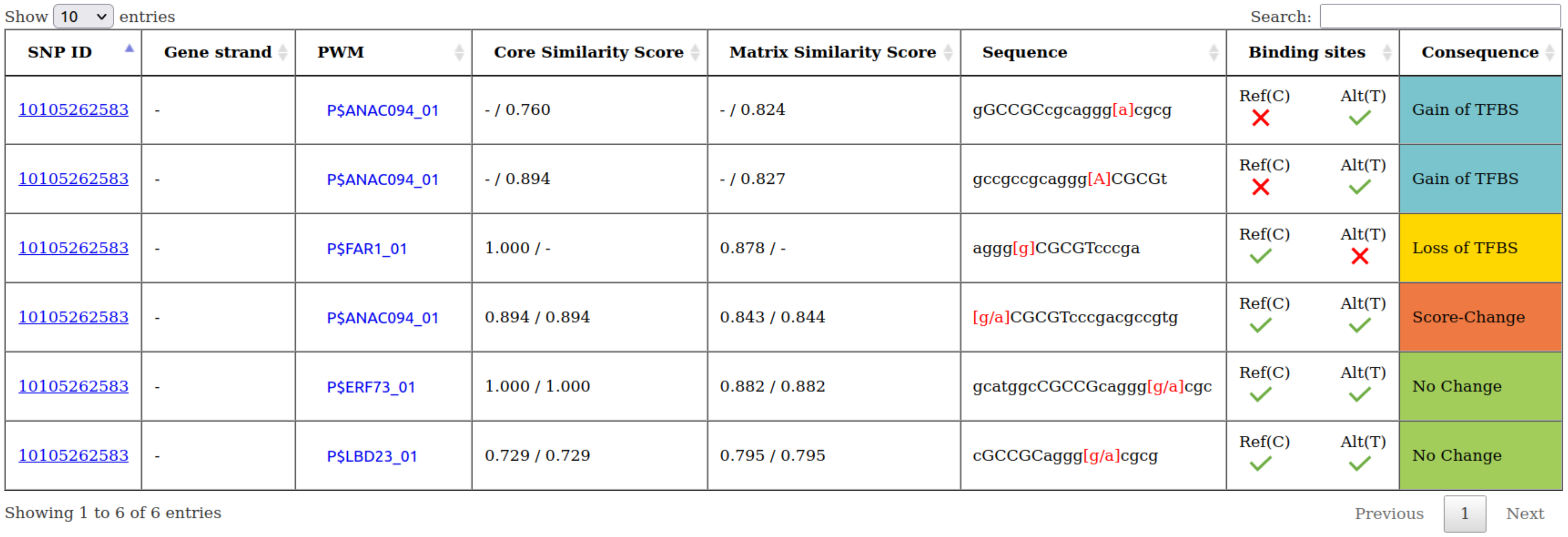
- Annotation of consequences: By comparing the two sets of predicted TFBSs, we assessed the consequences of each SNP on a specific TFBS. In particular, the effect of each SNP on a TFBS was assigned to one of the following consequences:
- Gain of TFBS: the TFBS exists only for the alternate allele of the SNP.
- Loss of TFBS: the TFBS exists only for the reference allele of the SNP.
- Score-Change: the TFBS exists for both alleles but with differing binding affinity as determined by the MATCH™ scores.
- No Change: the TFBS exists for both alleles with the same binding affinity.
3. Results
3.1. Database
3.2. Web Interface
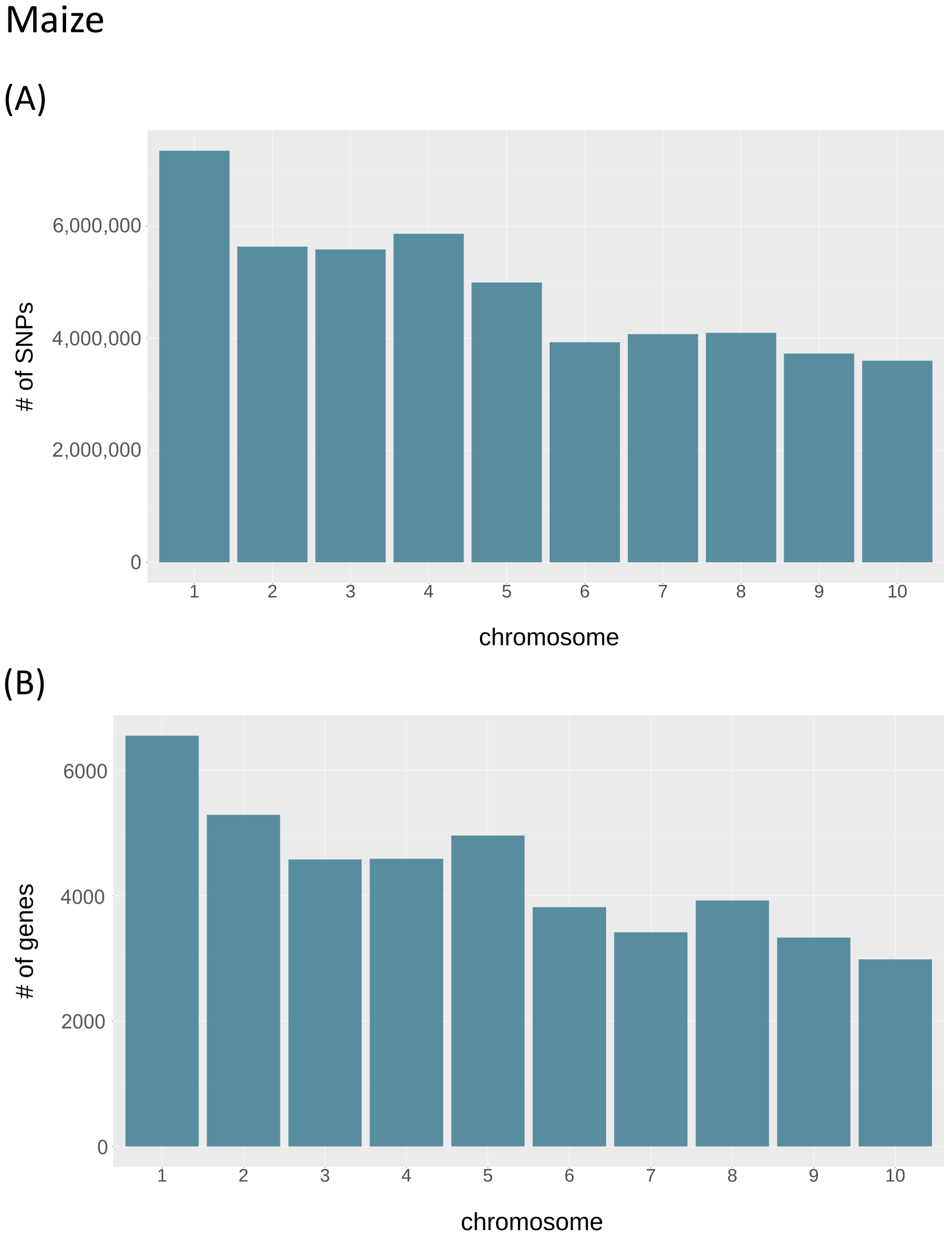
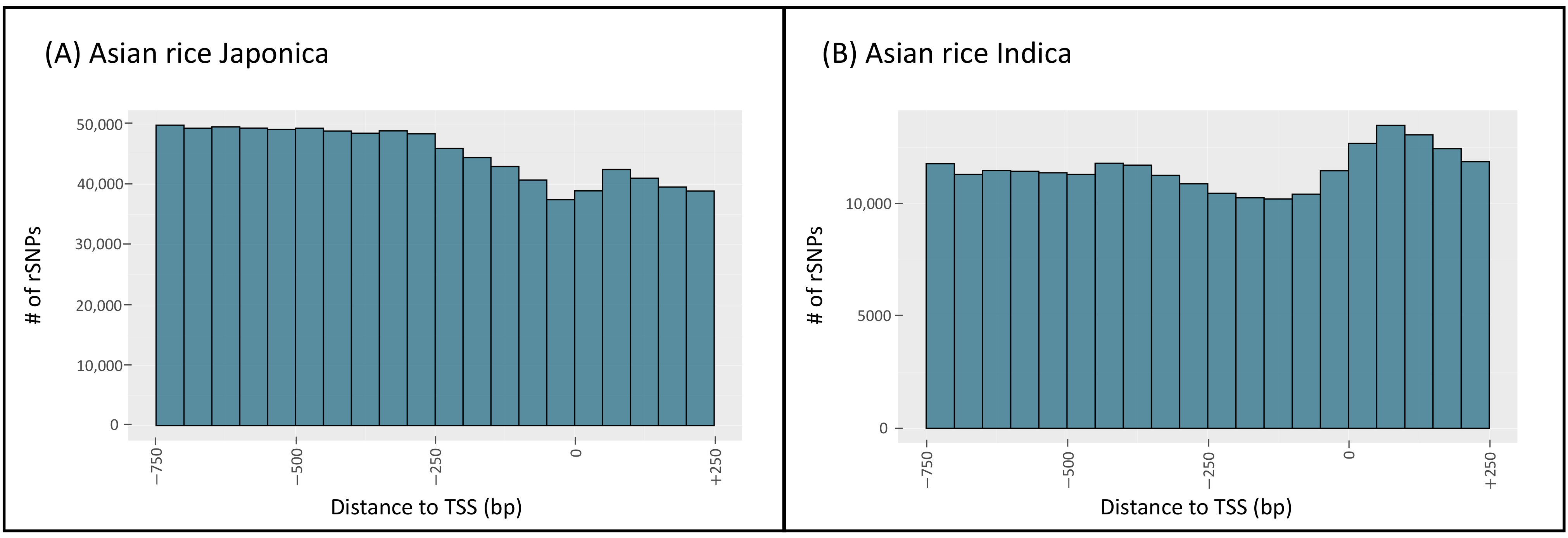
3.3. Statistical Overview of the Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SNP | single nucleotide polymorphism |
| rSNP | regulatory SNP |
| TF | transcription factor |
| TFBS | transcription factor binding site |
| TSS | transcription start site |
| bp | base pair |
| GWAS | genome-wide association study |
| eQTL | expression quantitative trait locus |
| PWM | position weight matrix |
| NGS | next generation sequencing |
| MAF | minor allele frequency |
References
- Begna, T. Global role of plant breeding in tackling climate change. Int. J. Agric. Sci. Food Technol. 2021, 7, 223–229. [Google Scholar]
- Ceccarelli, S.; Grando, S.; Maatougui, M.; Michael, M.; Slash, M.; Haghparast, R.; Rahmanian, M.; Taheri, A.; Al-Yassin, A.; Benbelkacem, A.; et al. Plant breeding and climate changes. J. Agric. Sci. 2010, 148, 627–637. [Google Scholar] [CrossRef]
- Klees, S.; Lange, T.M.; Bertram, H.; Rajavel, A.; Schlüter, J.S.; Lu, K.; Schmitt, A.O.; Gültas, M. In Silico Identification of the Complex Interplay between Regulatory SNPs, Transcription Factors, and Their Related Genes in Brassica napus L. Using Multi-Omics Data. Int. J. Mol. Sci. 2021, 22, 789. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yuan, Y.; Wang, H.; Yu, D.; Liu, Y.; Zhang, A.; Gowda, M.; Nair, S.K.; Hao, Z.; Lu, Y.; et al. Applications of genotyping-by-sequencing (GBS) in maize genetics and breeding. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.L.; Beesley, J.; French, J.D.; Dunning, A.M. Beyond GWASs: Illuminating the dark road from association to function. Am. J. Hum. Genet. 2013, 93, 779–797. [Google Scholar] [CrossRef]
- Klees, S.; Heinrich, F.; Schmitt, A.O.; Gültas, M. agReg-SNPdb: A Database of Regulatory SNPs for Agricultural Animal Species. Biology 2021, 10, 790. [Google Scholar] [CrossRef]
- Heinrich, F.; Wutke, M.; Das, P.P.; Kamp, M.; Gültas, M.; Link, W.; Schmitt, A.O. Identification of regulatory SNPs associated with vicine and convicine content of Vicia faba based on genotyping by sequencing data using deep learning. Genes 2020, 11, 614. [Google Scholar] [CrossRef]
- Rojano, E.; Seoane, P.; Ranea, J.A.; Perkins, J.R. Regulatory variants: From detection to predicting impact. Brief. Bioinform. 2018, 20, 1639–1654. [Google Scholar] [CrossRef]
- Degtyareva, A.O.; Antontseva, E.V.; Merkulova, T.I. Regulatory SNPs: Altered Transcription Factor Binding Sites Implicated in Complex Traits and Diseases. Int. J. Mol. Sci. 2021, 22, 6454. [Google Scholar] [CrossRef]
- Nishizaki, S.S.; Ng, N.; Dong, S.; Porter, R.S.; Morterud, C.; Williams, C.; Asman, C.; Switzenberg, J.A.; Boyle, A.P. Predicting the effects of SNPs on transcription factor binding affinity. Bioinformatics 2020, 36, 364–372. [Google Scholar] [CrossRef]
- Martin, V.; Zhao, J.; Afek, A.; Mielko, Z.; Gordân, R. QBiC-Pred: Quantitative predictions of transcription factor binding changes due to sequence variants. Nucleic Acids Res. 2019, 47, W127–W135. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Hudson, R.; Harrison, C.; Craven, M.; Keleş, S. atSNP Search: A web resource for statistically evaluating influence of human genetic variation on transcription factor binding. Bioinformatics 2018, 35, 2657–2659. [Google Scholar] [CrossRef] [PubMed]
- Amlie-Wolf, A.; Tang, M.; Mlynarski, E.E.; Kuksa, P.P.; Valladares, O.; Katanic, Z.; Tsuang, D.; Brown, C.D.; Schellenberg, G.D.; Wang, L.-S. INFERNO: Inferring the molecular mechanisms of noncoding genetic variants. Nucleic Acids Res. 2018, 46, 8740–8753. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Wang, J. rSNPBase 3.0: An updated database of SNP-related regulatory elements, element-gene pairs and SNP-based gene regulatory networks. Nucleic Acids Res. 2017, 46, D1111–D1116. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Ambrosini, G.; Bucher, P. SNP2TFBS–A database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res. 2016, 45, D139–D144. [Google Scholar] [CrossRef]
- Coetzee, S.G.; Coetzee, G.A.; Hazelett, D.J. motifbreakR: An R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics 2015, 31, 3847–3849. [Google Scholar] [CrossRef]
- Guo, Y.; Conti, D.V.; Wang, K. Enlight: Web-based integration of GWAS results with biological annotations. Bioinformatics 2014, 31, 275–276. [Google Scholar] [CrossRef][Green Version]
- Santana-Garcia, W.; Rocha-Acevedo, M.; Ramirez-Navarro, L.; Mbouamboua, Y.; Thieffry, D.; Thomas-Chollier, M.; Contreras-Moreira, B.; van Helden, J.; Medina-Rivera, A. RSAT variation-tools: An accessible and flexible framework to predict the impact of regulatory variants on transcription factor binding. Comput. Struct. Biotechnol. J. 2019, 17, 1415–1428. [Google Scholar] [CrossRef]
- Zuo, C.; Shin, S.; Keleş, S. atSNP: Transcription factor binding affinity testing for regulatory SNP detection. Bioinformatics 2015, 31, 3353–3355. [Google Scholar] [CrossRef]
- Pagès, H. BSgenome: Infrastructure for Biostrings-based genome data packages and support for efficient SNP representation. R Package 2016, 1, 10-18129. [Google Scholar]
- Jacquemin, J.; Bhatia, D.; Singh, K.; Wing, R.A. The International Oryza Map Alignment Project: Development of a genus-wide comparative genomics platform to help solve the 9 billion-people question. Curr. Opin. Plant Biol. 2013, 16, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Brondani, C.; Rangel, P.; Brondani, R.; Ferreira, M. QTL mapping and introgression of yield-related traits from Oryza glumaepatula to cultivated rice (Oryza Sativa) using microsatellite markers. Theor. Appl. Genet. 2002, 104, 1192–1203. [Google Scholar] [CrossRef] [PubMed]
- Bolser, D.M.; Staines, D.M.; Perry, E.; Kersey, P.J. Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomic data. In Plant Genomics Databases; Humana Press: New York, NY, USA, 2017; pp. 1–31. [Google Scholar]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Rajavel, A.; Klees, S.; Schlüter, J.S.; Bertram, H.; Lu, K.; Schmitt, A.O.; Gültas, M. Unravelling the Complex Interplay of Transcription Factors Orchestrating Seed Oil Content in Brassica napus L. Int. J. Mol. Sci. 2021, 22, 1033. [Google Scholar] [CrossRef] [PubMed]
- Reese, M.G.; Moore, B.; Batchelor, C.; Salas, F.; Cunningham, F.; Marth, G.T.; Stein, L.; Flicek, P.; Yandell, M.; Eilbeck, K. A standard variation file format for human genome sequences. Genome Biol. 2010, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Genome Variation Format 1.10. Available online: https://github.com/The-Sequence-Ontology/Specifications/blob/master/gvf.md (accessed on 24 March 2022).
- Generic Feature Format Version 3. Available online: https://github.com/The-Sequence-Ontology/Specifications/blob/master/gff3.md (accessed on 24 March 2022).
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Kel, A.E.; Gößling, E.; Cheremushkin, E.; Kel-Margoulis, O.V.; Wingender, E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003, 31, 3576–3579. [Google Scholar] [CrossRef]
- Wingender, E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Briefi. Bioinform. 2008, 9, 326–332. [Google Scholar] [CrossRef]
- Triska, M.; Solovyev, V.; Baranova, A.; Kel, A.; Tatarinova, T.V. Nucleotide patterns aiding in prediction of eukaryotic promoters. PLoS ONE 2017, 12, e0187243. [Google Scholar] [CrossRef]
- Jaiswal, V.; Gahlaut, V.; Mathur, S.; Agarwal, P.; Khandelwal, M.K.; Khurana, J.P.; Tyagi, A.K.; Balyan, H.S.; Gupta, P.K. Identification of novel SNP in promoter sequence of TaGW2-6A associated with grain weight and other agronomic traits in wheat (Triticum aestivum L.). PLoS ONE 2015, 10, e0129400. [Google Scholar] [CrossRef]
- Shi, L.; Weng, J.; Liu, C.; Song, X.; Miao, H.; Hao, Z.; Xie, C.; Li, M.; Zhang, D.; Bai, L.; et al. Identification of promoter motifs regulating ZmeIF4E expression level involved in maize rough dwarf disease resistance in maize (Zea mays L.). Mol. Genet. Genom. 2013, 288, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Konishi, S.; Izawa, T.; Lin, S.Y.; Ebana, K.; Fukuta, Y.; Sasaki, T.; Yano, M. An SNP caused loss of seed shattering during rice domestication. Science 2006, 312, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Ryan, N.M.; Morris, S.W.; Porteous, D.J.; Taylor, M.S.; Evans, K.L. SuRFing the genomics wave: An R package for prioritising SNPs by functionality. Genome Med. 2014, 6, 79. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Liu, Z.; Lou, S.; Bedford, J.; Mu, X.J.; Yip, K.Y.; Khurana, E.; Gerstein, M. FunSeq2: A framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 2014, 15, 480. [Google Scholar] [CrossRef]
- Riva, A. Large-scale computational identification of regulatory SNPs with rSNP-MAPPER. Proc. Bmc Genom. Biomed Cent. 2012, 13, S7. [Google Scholar] [CrossRef]
- Kwon, A.T.; Arenillas, D.J.; Hunt, R.W.; Wasserman, W.W. oPOSSUM-3: Advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 Genes Genomes Genet. 2012, 2, 987–1002. [Google Scholar] [CrossRef]
- Coetzee, S.G.; Rhie, S.K.; Berman, B.P.; Coetzee, G.A.; Noushmehr, H. FunciSNP: An R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res. 2012, 40, e139. [Google Scholar] [CrossRef]
- Ho Sui, S.J.; Mortimer, J.R.; Arenillas, D.J.; Brumm, J.; Walsh, C.J.; Kennedy, B.P.; Wasserman, W.W. oPOSSUM: Identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005, 33, 3154–3164. [Google Scholar] [CrossRef]
- Stepanova, M.; Tiazhelova, T.; Skoblov, M.; Baranova, A. A comparative analysis of relative occurrence of transcription factor binding sites in vertebrate genomes and gene promoter areas. Bioinformatics 2005, 21, 1789–1796. [Google Scholar] [CrossRef]
- Lange, T.M.; Heinrich, F.; Enders, M.; Wolf, M.; Schmitt, A.O. In silico quality assessment of SNPs—A case study on the Axiom® Wheat genotyping arrays. Curr. Plant Biol. 2020, 21, 100140. [Google Scholar] [CrossRef]
- Treangen, T.J.; Salzberg, S.L. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat. Rev. Genet. 2012, 13, 36–46. [Google Scholar] [CrossRef] [PubMed]
| Plant | Assembly Version | Download Date (DD/MM/YYYY) |
|---|---|---|
| Helianthus annuus (sunflower) | HanXRQr1.0 | 11/08/2021 |
| Hordeum vulgare (barley) | MorexV3_pseudomolecules_assembly | 12/22/2021 |
| Oryza glaberrima (African rice) | Oryza_glaberrima_V1 | 11/08/2021 |
| Oryza glumipatula (wild rice) | Oryza_glumaepatula_v1.5 | 11/08/2021 |
| Oryza sativa Indica (Asian rice Indica) | ASM465v1 | 12/22/2021 |
| Oryza sativa Japonica (Asian rice Japonica) | IRGSP-1.0 | 11/08/2021 |
| Solanum lycopersicum (tomato) | SL3.0 | 12/22/2021 |
| Sorghum bicolor (sorghum) | Sorghum_bicolor_NCBIv3 | 12/22/2021 |
| Triticum aestivum (bread wheat) | IWGSC | 11/08/2021 |
| Triticum turgidum (durum wheat) | Svevo.v1 | 11/08/2021 |
| Vitis vinifera (grape) | 12X | 11/08/2021 |
| Zea mays (maize) | Zm-B73-REFERENCE-NAM-5.0 | 11/08/2021 |
| Plant | snp_info | gene_info | snp_region | TFBS_results |
|---|---|---|---|---|
| African rice | 7,567,669 | 33,164 | 7,341,550 | 8,336,778 |
| Asian rice Indica | 4,340,785 | 37,878 | 4,589,915 | 4,441,820 |
| Asian rice Japonica | 25,135,669 | 37,960 | 20,155,983 | 20,940,720 |
| Barley | 12,771,762 | 35,106 | 2,545,069 | 2,736,205 |
| Bread wheat | 18,093,867 | 107,889 | 13,334,911 | 19,733,723 |
| Durum wheat | 1,815,904 | 66,559 | 1,121,107 | 1,734,495 |
| Grape | 400,940 | 29,971 | 334,500 | 290,793 |
| Maize | 48,830,598 | 44,289 | 15,439,220 | 13,101,269 |
| Rapeseed | 670,028 | 406,325 | 5,110,349 | 506,859 |
| Sorghum | 8,081,051 | 34,023 | 6,414,543 | 3,118,613 |
| Sunflower | 11,834 | 52,191 | 2335 | 1498 |
| Tomato | 60,973,560 | 33,869 | 28,709,218 | 10,347,415 |
| Wild rice | 4,865,161 | 35,735 | 4,752,796 | 5,154,313 |
| Total | 193,558,828 | 954,959 | 109,851,496 | 90,444,501 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klees, S.; Heinrich, F.; Schmitt, A.O.; Gültas, M. agReg-SNPdb-Plants: A Database of Regulatory SNPs for Agricultural Plant Species. Biology 2022, 11, 684. https://doi.org/10.3390/biology11050684
Klees S, Heinrich F, Schmitt AO, Gültas M. agReg-SNPdb-Plants: A Database of Regulatory SNPs for Agricultural Plant Species. Biology. 2022; 11(5):684. https://doi.org/10.3390/biology11050684
Chicago/Turabian StyleKlees, Selina, Felix Heinrich, Armin Otto Schmitt, and Mehmet Gültas. 2022. "agReg-SNPdb-Plants: A Database of Regulatory SNPs for Agricultural Plant Species" Biology 11, no. 5: 684. https://doi.org/10.3390/biology11050684
APA StyleKlees, S., Heinrich, F., Schmitt, A. O., & Gültas, M. (2022). agReg-SNPdb-Plants: A Database of Regulatory SNPs for Agricultural Plant Species. Biology, 11(5), 684. https://doi.org/10.3390/biology11050684






