Proteomics Analysis in Japanese Medaka Oryzias latipes Exposed to Humic Acid Revealed Suppression of Innate Immunity and Coagulation Proteins
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Fish Maintenance
2.3. Experimental Procedure
2.4. Sampling
2.5. High-Performance Liquid Chromatography with Tandem Mass Spectrometry (HPLC–MS/MS) Analysis
2.6. Data Processing
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Melo, B.A.G.; Motta, F.L.; Santana, M.H.A. Humic acids: Structural properties and multiple functionalities for novel technological developments. Mater. Sci. Eng. C 2016, 62, 967–974. [Google Scholar] [CrossRef]
- Thurman, E.M. Amount of organic carbon in natural waters. In Organic Geochemistry of Natural Water; Thurman, E.M., Ed.; Springer: Dordrecht, The Netherlands, 1985; pp. 7–65. [Google Scholar] [CrossRef]
- Levshina, S. Geochemistry of organic matter in river waters of the Amur basin, Russia. Environ. Earth Sci. 2016, 75, 387. [Google Scholar] [CrossRef]
- Steinberg, C.E.; Kamara, S.; Prokhotskaya, V.Y.; Manusadžianas, L.; Karasyova, T.A.; Timofeyev, M.A.; Jie, Z.; Paul, A.; Meinelt, T.; Farjalla, V.F.; et al. Dissolved humic substances–Ecological driving forces from the individual to the ecosystem level. Freshw. Biol. 2006, 51, 1189–1210. [Google Scholar] [CrossRef]
- Steinberg, C.E.; Paul, A.; Pflugmacher, S.; Meinelt, T.; Klöcking, R.; Wiegand, C. Pure humic substances have the potential to act as xenobiotic chemicals—A review. Fresenius Environ. Bull. 2003, 12, 391–401. [Google Scholar]
- Steinberg, C.E.W.; Meinelt, T.; Timofeyev, M.A.; Bittner, M.; Menzel, R. Humic substances. Environ. Sci. Pollut. Res. 2008, 15, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieke, T.; Meinelt, T.; Hoseinifar, S.H.; Pan, B.; Straus, D.L.; Steinberg, C.E. Sustainable aquaculture requires environmental-friendly treatment strategies for fish diseases. Rev. Aquac. 2020, 12, 943–965. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, F.; Kumar, G.; Soliman, F.M.; Adly, M.A.; Soliman, H.A.; El-Matbouli, M.; Saleh, M. Proteomics for understanding pathogenesis, immune modulation and host pathogen interactions in aquaculture. Comp. Biochem. Physiol. D Genom. Proteom. 2019, 32, 100625. [Google Scholar] [CrossRef]
- Hoem, K.S.; Tveten, A.K. Current approaches in decoding the molecular mechanisms of long-term stress in adult farmed Atlantic salmon (Salmo salar). Rev. Aquac. 2020, 12, 1708–1720. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalisation and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [Green Version]
- UniProt. Proteomes-Oryzias Latipes (Japanese Rice Fish) (Japanese Killifish). Available online: https://www.uniprot.org/proteomes/UP000001038 (accessed on 25 April 2021).
- Yurchenko, V.; Morozov, A. Responses of hepatic biotransformation and antioxidant enzymes in Japanese medaka (Oryzias latipes) exposed to humic acid. Fish Physiol. Biochem. 2022, 48, 1–13. [Google Scholar] [CrossRef]
- Ke, R.; Luo, J.; Sun, L.; Wang, Z.; Spear, P.A. Predicting bioavailability and accumulation of organochlorine pesticides by Japanese medaka in the presence of humic acid and natural organic matter using passive sampling membranes. Environ. Sci. Technol. 2007, 41, 6698–6703. [Google Scholar] [CrossRef] [PubMed]
- Linnik, P.N.; Ivanechko, Y.S.; Linnik, R.P.; Zhezherya, V.A. Humic substances in surface waters of the Ukraine. Russ. J. Gen. Chem. 2013, 83, 2715–2730. [Google Scholar] [CrossRef]
- Test No. 203: Fish, Acute Toxicity Test. In OECD Guidelines for the Testing of Chemicals. Section 2; OECD Publishing: Paris, France, 2019. [CrossRef]
- Zolg, W. The proteomic search for diagnostic biomarkers: Lost in translation. Mol. Cell. Proteom. 2006, 5, 1720–1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diz, A.P.; Truebano, M.; Skibinski, D.O.F. The consequences of sample pooling in proteomics: An empirical study. Electrophoresis 2009, 30, 2967–2975. [Google Scholar] [CrossRef]
- Morro, B.; Doherty, M.K.; Balseiro, P.; Handeland, S.O.; MacKenzie, S.; Sveier, H.; Albalat, A. Plasma proteome profiling of freshwater and seawater life stages of rainbow trout (Oncorhynchus mykiss). PLoS ONE 2020, 15, e0227003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yurchenko, V.V.; Morozov, A.A.; Fedorov, R.A.; Bakina, L.G.; Zgoda, V.G.; Tikhonova, O.V. Dataset on the effects of environmentally relevant humic acid concentrations on the liver protein profile in Japanese medaka (Oryzias latipes). Data Brief 2022, 40, 107796. [Google Scholar] [CrossRef]
- Aguilan, J.T.; Kulej, K.; Sidoli, S. Guide for protein fold change and p-value calculation for non-experts in proteomics. Mol. Omics 2020, 16, 573–582. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Gierlinski, M.; Gastaldello, F.; Cole, C.; Barton, G.J. Proteus: An R package for downstream analysis of MaxQuant output. bioRxiv 2018, 1–10. [Google Scholar] [CrossRef] [Green Version]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2020, 49, D394–D403. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2020, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny 2.1. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 21 April 2022).
- UniProtKB. Available online: https://www.uniprot.org/uniprot/ (accessed on 21 April 2022).
- Andreeva, A.M.; Serebryakova, M.V.; Lamash, N.E. Oligomeric protein complexes of apolipoproteins stabilize the internal fluid environment of organism in redfins of the Tribolodon genus [Pisces; Cypriniformes, Cyprinidae]. Comp. Biochem. Physiol. D Genom. Proteom. 2017, 22, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.A.; Arnold, G.J.; Nynca, J.; Fröhlich, T.; Otte, K.; Ciereszko, A. Characterization of carp seminal plasma proteome in relation to blood plasma. J. Proteom. 2014, 98, 218–232. [Google Scholar] [CrossRef] [PubMed]
- Leroy, B.; Toubeau, G.; Falmagne, P.; Wattiez, R. Identification and characterization of new protein chemoattractants in the frog skin secretome. Mol. Cell. Proteom. 2006, 5, 2114–2123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, N.L.; Anderson, N.G. The human plasma proteome: History, character, and diagnostic prospects. Mol. Cell. Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef] [Green Version]
- PANTHER Classification System. Available online: http://www.pantherdb.org/ (accessed on 25 January 2022).
- InterPro. Classification of Protein Families. Available online: https://www.ebi.ac.uk/interpro/ (accessed on 25 January 2022).
- Linder, M.C. Ceruloplasmin and other copper binding components of blood plasma and their functions: An update. Metallomics 2016, 8, 887–905. [Google Scholar] [CrossRef]
- Hseu, Y.-C.; Kumar, K.J.S.; Chen, C.-S.; Cho, H.-J.; Lin, S.-W.; Shen, P.-C.; Lin, C.-W.; Lu, F.-J.; Yang, H.-L. Humic acid in drinking well water induces inflammation through reactive oxygen species generation and activation of nuclear factor-κB/activator protein-1 signaling pathways: A possible role in atherosclerosis. Toxicol. Appl. Pharmacol. 2014, 274, 249–262. [Google Scholar] [CrossRef]
- Alak, G.; Atamanalp, M.; Topal, A.; Arslan, H.; Oruç, E.; Altun, S. Histopathological and biochemical effects of humic acid against cadmium toxicity in brown trout gills and muscles. Turk. J. Fish. Aquat. Sci. 2013, 13, 315–320. [Google Scholar] [CrossRef]
- Deng, J.; Lin, B.; Zhang, X.; Guo, L.; Chen, L.; Li, G.; Wang, Q.; Yu, C.; Mi, H. Effects of dietary sodium humate on growth, antioxidant capacity, non-specific immune response, and resistance to Aeromonas hydrophila in genetic improvement of farmed tilapia (GIFT, Oreochromis niloticus). Aquaculture 2020, 520, 734788. [Google Scholar] [CrossRef]
- Engelking, L.R. Textbook of Veterinary Physiological Chemistry, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar] [CrossRef]
- Liu, W.; Rodgers, G.P. Olfactomedin 4 expression and functions in innate immunity, inflammation, and cancer. Cancer Metastasis Rev. 2016, 35, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Lim, W.; Song, G. Differential expression of vitelline membrane outer layer protein 1: Hormonal regulation of expression in the oviduct and in ovarian carcinomas from laying hens. Mol. Cell. Endocrinol. 2015, 399, 250–258. [Google Scholar] [CrossRef]
- Goodfellow, B.J.; Freire, F.; Carvalho, A.L.; Aveiro, S.S.; Charbonnier, P.; Moulis, J.M.; Delgado, L.; Ferreira, G.C.; Rodrigues, J.E.; Poussin-Courmontagne, P.; et al. The SOUL family of heme-binding proteins: Structure and function 15 years later. Coord. Chem. Rev. 2021, 448, 214189. [Google Scholar] [CrossRef]
- Farhana, A.; Lappin, S.L. Biochemistry, Lactate Dehydrogenase. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK557536/ (accessed on 21 April 2022).
- Murata, K.; Kinoshita, M. Targeted deletion of liver-expressed Choriogenin L results in the production of soft eggs and infertility in medaka. Oryzias latipes. Zool. Lett. 2022, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Magkrioti, C.; Galaris, A.; Kanellopoulou, P.; Stylianaki, E.-A.; Kaffe, E.; Aidinis, V. Autotaxin and chronic inflammatory diseases. J. Autoimmun. 2019, 104, 102327. [Google Scholar] [CrossRef]
- Smith, N.C.; Rise, M.L.; Christian, S.L. A comparison of the innate and adaptive immune systems in cartilaginous fish, ray-finned fish, and lobe-finned fish. Front. Immunol. 2019, 10, 2292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carland, T.M.; Gerwick, L. The C1q domain containing proteins: Where do they come from and what do they do. Dev. Comp. Immunol. 2010, 34, 785–790. [Google Scholar] [CrossRef]
- Boshra, H.; Li, J.; Sunyer, J.O. Recent advances on the complement system of teleost fish. Fish Shellfish Immunol. 2006, 20, 239–262. [Google Scholar] [CrossRef]
- Barnum, S.R.; Schein, T.N. The complement system. In The Complement Factsbook, 2nd ed.; Barnum, S.R., Schein, T.N., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 7–20. [Google Scholar] [CrossRef]
- Schröder-Braunstein, J.; Kirschfink, M. Complement deficiencies and dysregulation: Pathophysiological consequences, modern analysis, and clinical management. Mol. Immunol. 2019, 114, 299–311. [Google Scholar] [CrossRef]
- Magnadóttir, B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006, 20, 137–151. [Google Scholar] [CrossRef]
- Gialeli, C.; Gungor, B.; Blom, A.M. Novel potential inhibitors of complement system and their roles in complement regulation and beyond. Mol. Immunol. 2018, 102, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Louvado, A.; Cleary, D.F.; Pereira, L.F.; Coelho, F.J.; Pousão-Ferreira, P.; Ozório, R.O.; Gomes, N.C. Humic substances modulate fish bacterial communities in a marine recirculating aquaculture system. Aquaculture 2021, 544, 737121. [Google Scholar] [CrossRef]
- Elton, C.S. The Ecology of Invasions by Animals and Plants, 2nd ed.; Springer: Cham, Switzerland; Berlin/Heidelberg Germany, 2020. [Google Scholar]
- Mohammed, H.H.; Arias, C.R. Potassium permanganate elicits a shift of the external fish microbiome and increases host susceptibility to columnaris disease. Vet. Res. 2015, 46, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavares-Dias, M.; Oliveira, S.R. A review of the blood coagulation system of fish. Rev. Bras. Biociência 2009, 7, 205–224. Available online: http://www.ufrgs.br/seerbio/ojs/index.php/rbb/article/view/1144 (accessed on 13 March 2022).
- Eigenbrot, C. Structure, function, and activation of coagulation factor VII. Curr. Protein Pept. Sci. 2002, 3, 287–299. [Google Scholar] [CrossRef]
- Palta, S.; Saroa, R.; Palta, A. Overview of the coagulation system. Indian J. Anaesth. 2014, 58, 515–523. [Google Scholar] [CrossRef]
- Doolittle, R.F. Coagulation in vertebrates with a focus on evolution and inflammation. J. Innate Immun. 2011, 3, 9–16. [Google Scholar] [CrossRef]
- Harvison, P.J. Thrombin as a target. In xPharm: The Comprehensive Pharmacology Reference; Enna, S.J., Bylund, D.B., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–6. [Google Scholar] [CrossRef]
- Amara, U.; Flierl, M.A.; Rittirsch, D.; Klos, A.; Chen, H.; Acker, B.; Brückner, U.B.; Nilsson, B.; Gebhard, F.; Lambris, J.D.; et al. Molecular intercommunication between the complement and coagulation systems. J. Immunol. 2010, 185, 5628–5636. [Google Scholar] [CrossRef] [Green Version]
- Esmon, C.T.; Xu, J.; Lupu, F. Innate immunity and coagulation. J. Thromb. Haemost. 2011, 9, 182–188. [Google Scholar] [CrossRef] [Green Version]
- Jerez-Cepa, I.; Ruiz-Jarabo, I. Physiology: An important tool to assess the welfare of aquatic animals. Biology 2021, 10, 61. [Google Scholar] [CrossRef]
- Okuda, S.; Watanabe, Y.; Moriya, Y.; Kawano, S.; Yamamoto, T.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Araki, N.; Yoshizawa, A.C.; et al. jPOSTrepo: An international standard data repository for proteomes. Nucleic Acids Res. 2017, 45, D1107–D1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
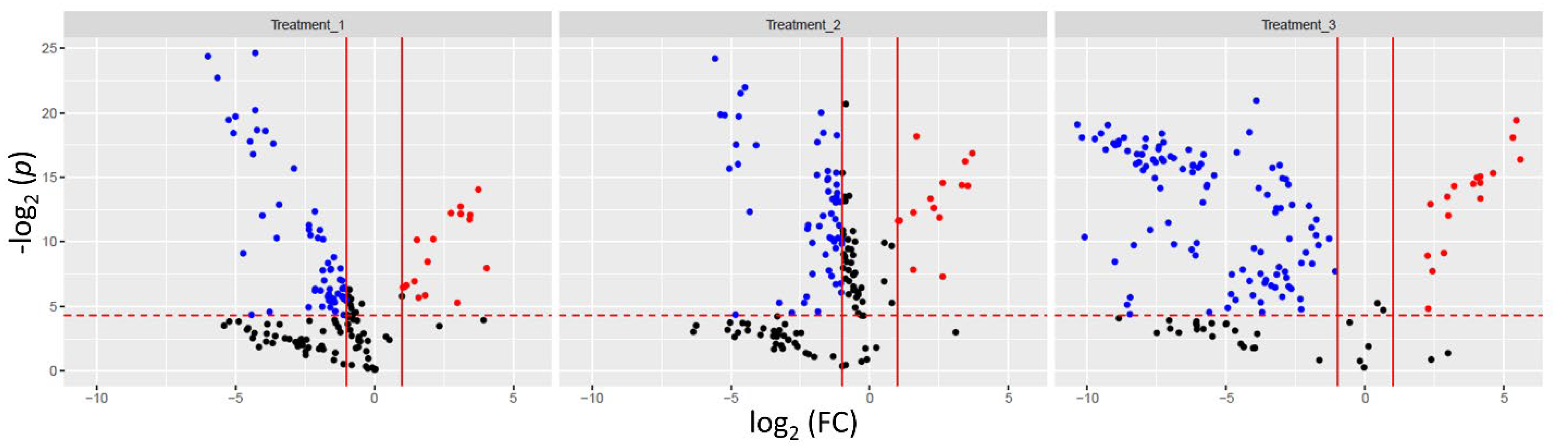
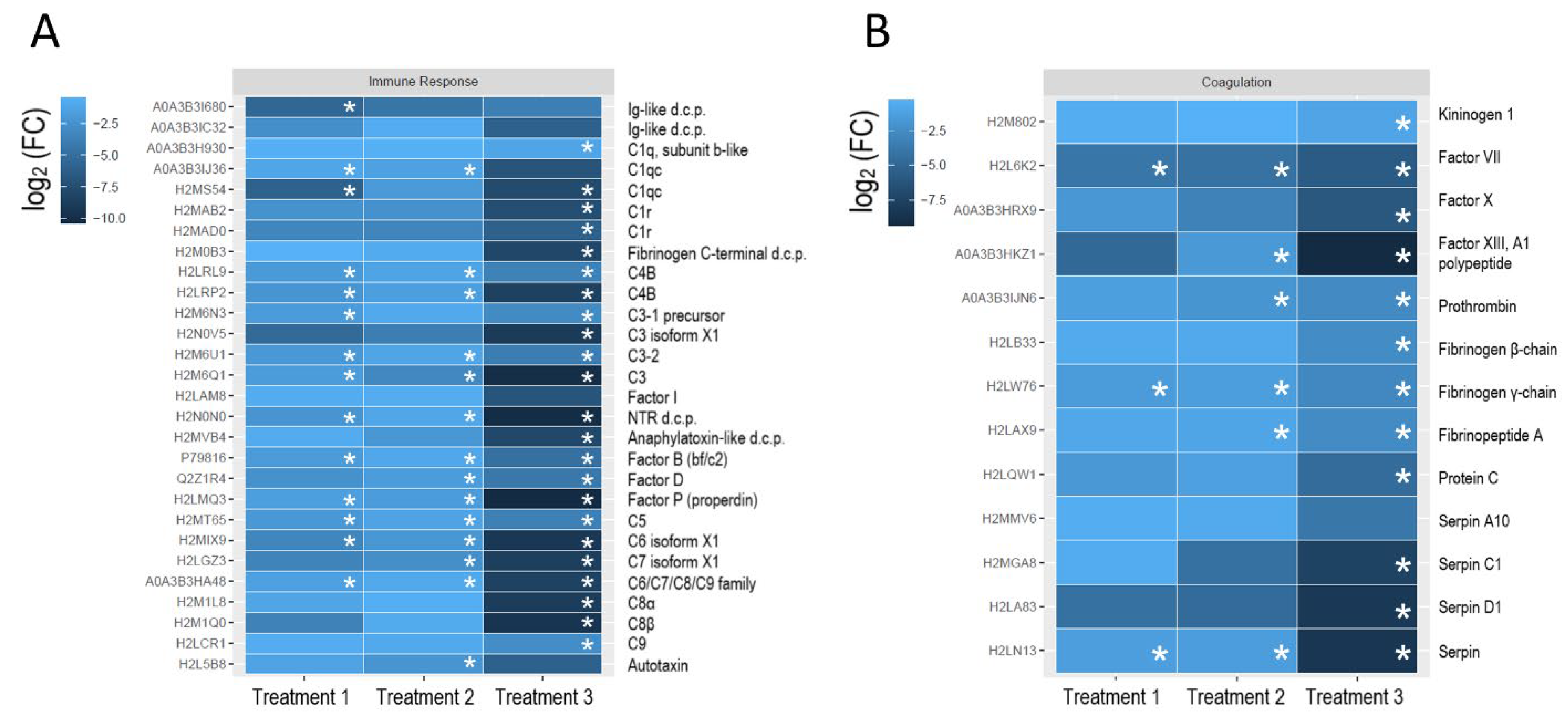
| Treatment | HA, mg/L, Nominal | Organic Carbon, mg C/L, 0 h | Organic Carbon, mg C/L, 78 h | pH (±SD) | O2, mg/L, (±SD) | W, g, (±SD) |
|---|---|---|---|---|---|---|
| Control | 0 | <LOQ | <LOQ | 8.14 ± 0.16 | 6.6 ± 0.3 | 0.49 ± 0.08 |
| 1 | 5 | <LOQ | <LOQ | 7.98 ± 0.43 | 6.6 ± 0.3 | 0.50 ± 0.07 |
| 2 | 40 | 4.5 | 6.6 | 8.10 ± 0.24 | 6.6 ± 0.2 | 0.50 ± 0.11 |
| 3 | 80 | 9.4 | 9.4 | 8.16 ± 0.22 | 6.7 ± 0.2 | 0.50 ± 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yurchenko, V.V.; Morozov, A.A.; Kiriukhin, B.A. Proteomics Analysis in Japanese Medaka Oryzias latipes Exposed to Humic Acid Revealed Suppression of Innate Immunity and Coagulation Proteins. Biology 2022, 11, 683. https://doi.org/10.3390/biology11050683
Yurchenko VV, Morozov AA, Kiriukhin BA. Proteomics Analysis in Japanese Medaka Oryzias latipes Exposed to Humic Acid Revealed Suppression of Innate Immunity and Coagulation Proteins. Biology. 2022; 11(5):683. https://doi.org/10.3390/biology11050683
Chicago/Turabian StyleYurchenko, Victoria V., Alexey A. Morozov, and Bogdan A. Kiriukhin. 2022. "Proteomics Analysis in Japanese Medaka Oryzias latipes Exposed to Humic Acid Revealed Suppression of Innate Immunity and Coagulation Proteins" Biology 11, no. 5: 683. https://doi.org/10.3390/biology11050683
APA StyleYurchenko, V. V., Morozov, A. A., & Kiriukhin, B. A. (2022). Proteomics Analysis in Japanese Medaka Oryzias latipes Exposed to Humic Acid Revealed Suppression of Innate Immunity and Coagulation Proteins. Biology, 11(5), 683. https://doi.org/10.3390/biology11050683







