Filamentous Thermosensitive Mutant Z: An Appealing Target for Emerging Pathogens and a Trek on Its Natural Inhibitors
Abstract
Simple Summary
Abstract
1. Introduction
- (I)
- Methicillin-resistant Staphylococcus aureus, popularly known as (MRSA), which accounts for ~12,000 [7] deaths per year and a health care cost of ~USD 5 billion per year [26]. Following MRSA, Vancomycin-resistant S. aureus (VRSA) is another challenge to tackle and has become a new problem in hospitals [27].
- (II)
- Pan drug-resistant and Multidrug-resistant, popularly known as PDR and MDR respectively, in Gram-negative bacterial strains of E. coli, K. pneumoniae, A. baumannii, P. aeruginosa, pose the threat of untreatable infections [28]. The avenues for finding novel antibiotics for these are limited due to their outer membrane, which prevents the entry of some antibiotics, and efflux pumps expel many of the remainders [29,30,31].
- (III)
- Multidrug-resistant and extensively drug-resistant strains of M. tuberculosis, also called MDR-TB and XDR-TB, pose a threat to the developing world specifically. The treatment requires a 2-year course of antibiotics and has serious side effects for patients. XDR-TB is more difficult to cure and often fatal [32,33].
2. Antibiotics and Bacterial Divisome Proteins as Emerging Therapeutic Targets
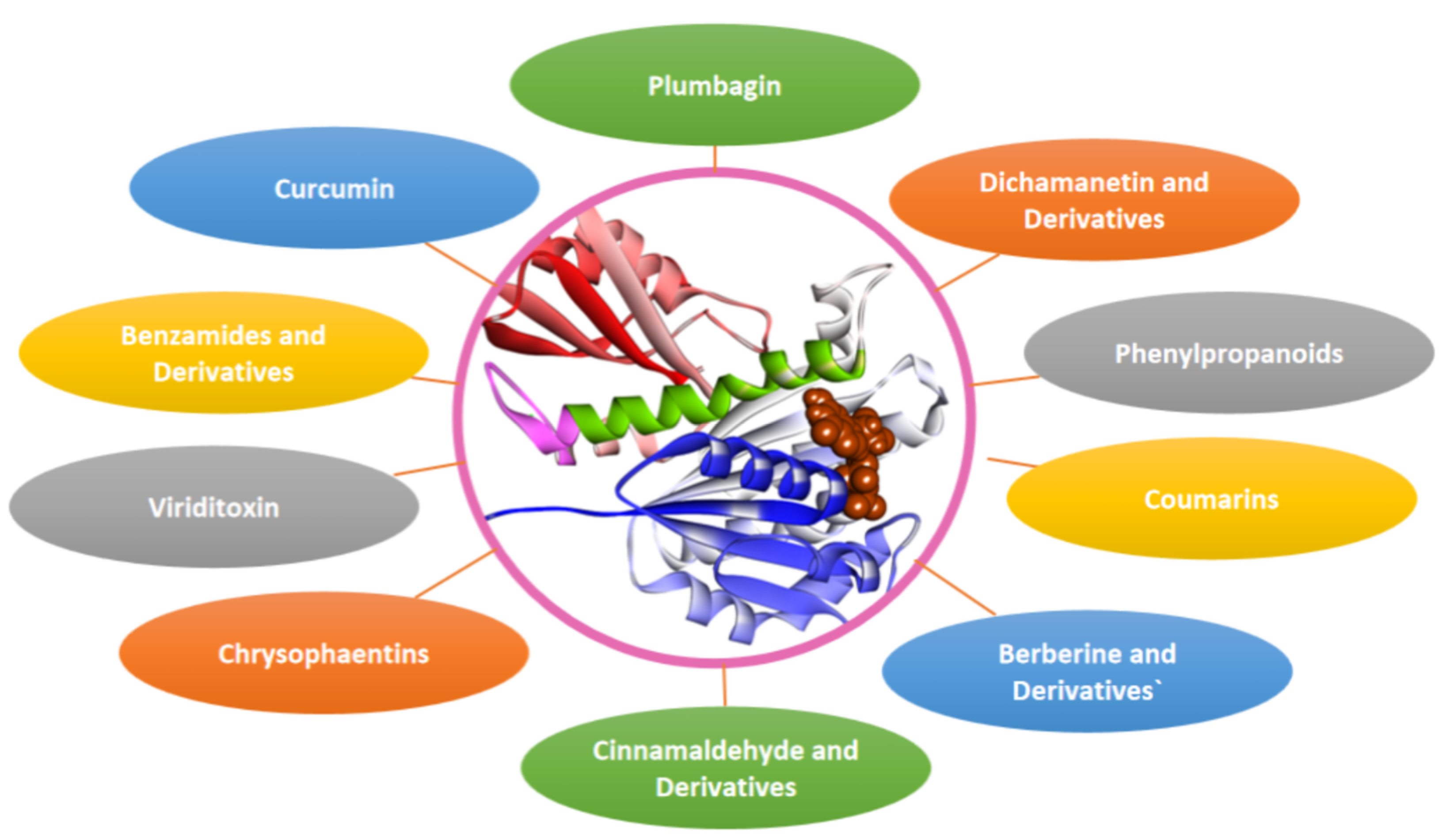
3. Fts-Z: An Appealing Antibacterial Target
Structural and Functional Aspect of Fts-Z
4. Fts-Z Inhibition: A Strategy to Combat AMR
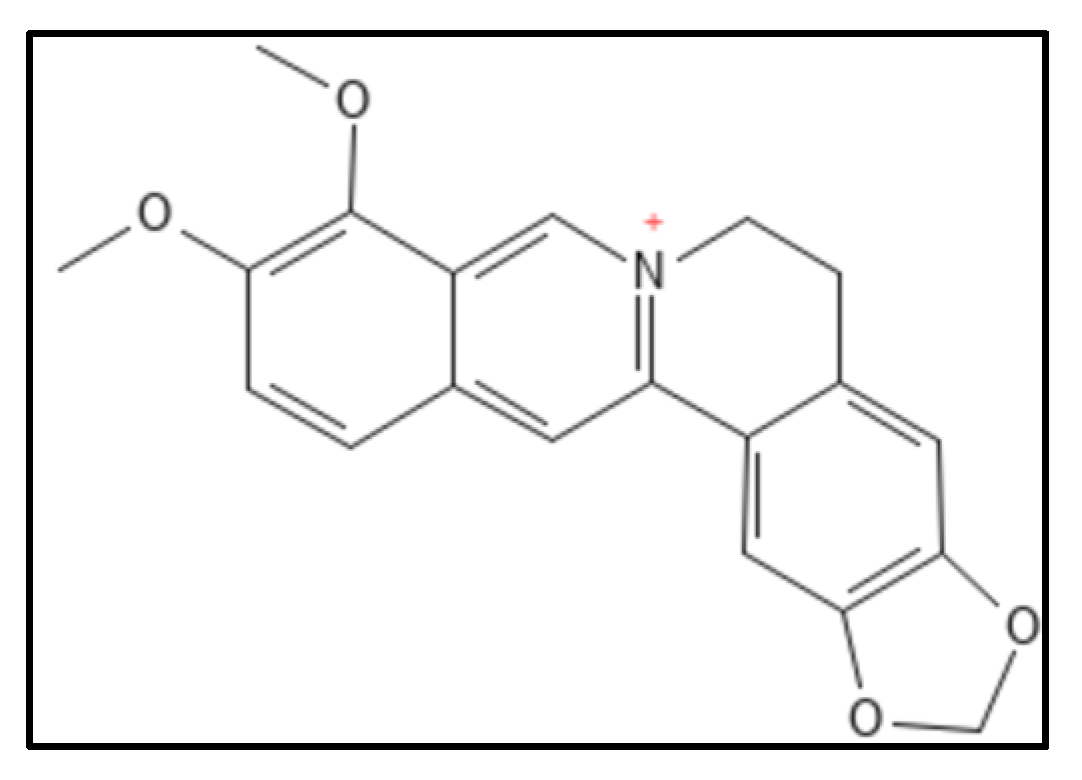
4.1. Berberine and Derivatives
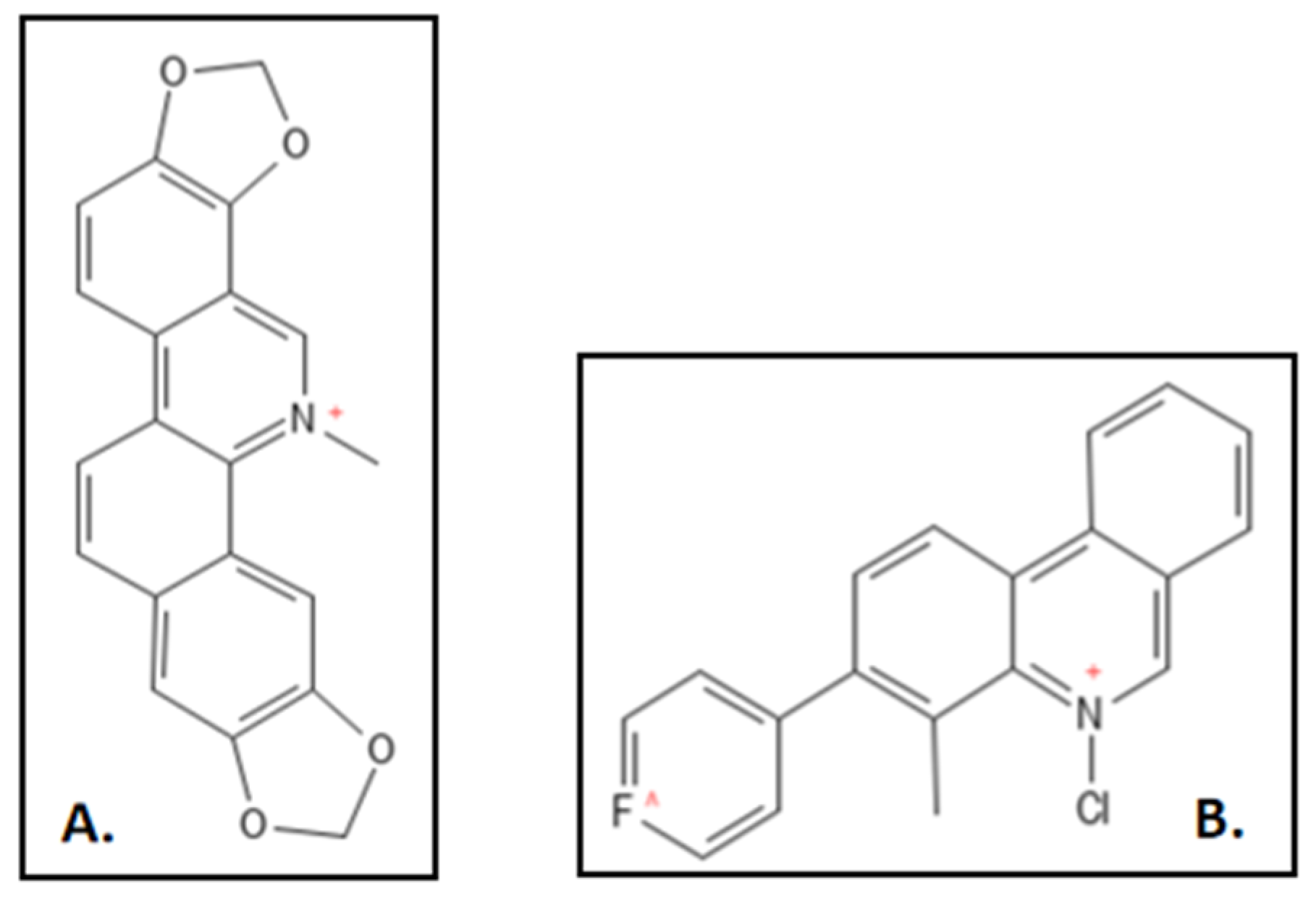
4.2. Sanguinarine
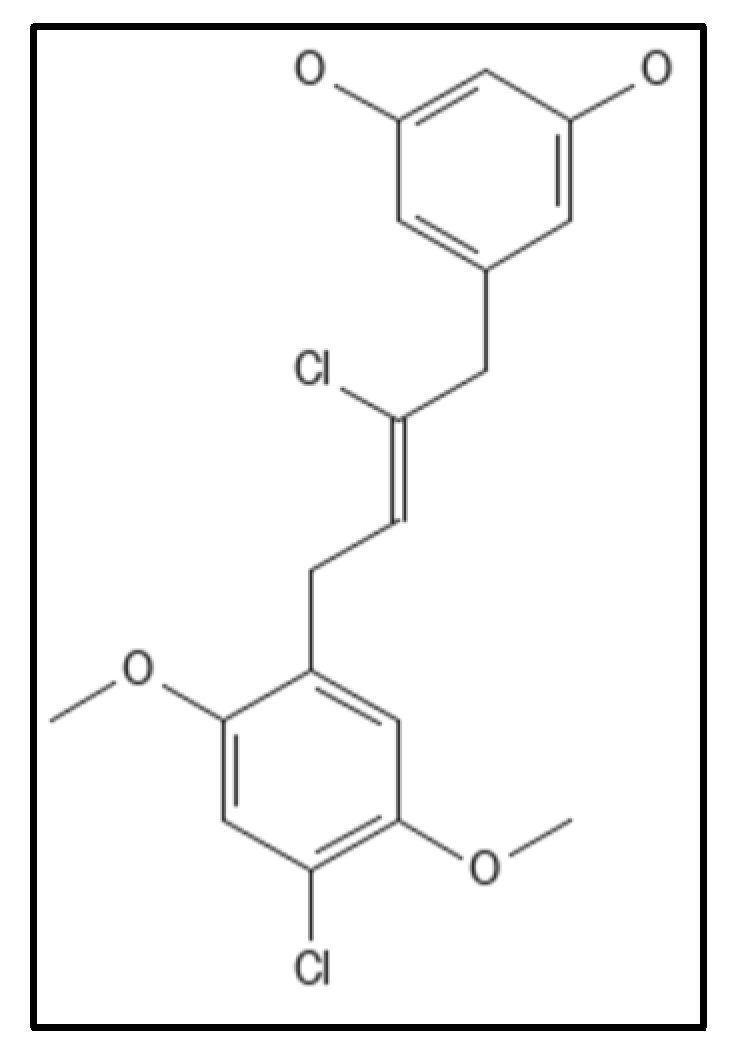
4.3. Cinnamaldehyde and Derivatives
4.4. Chrysophaentins
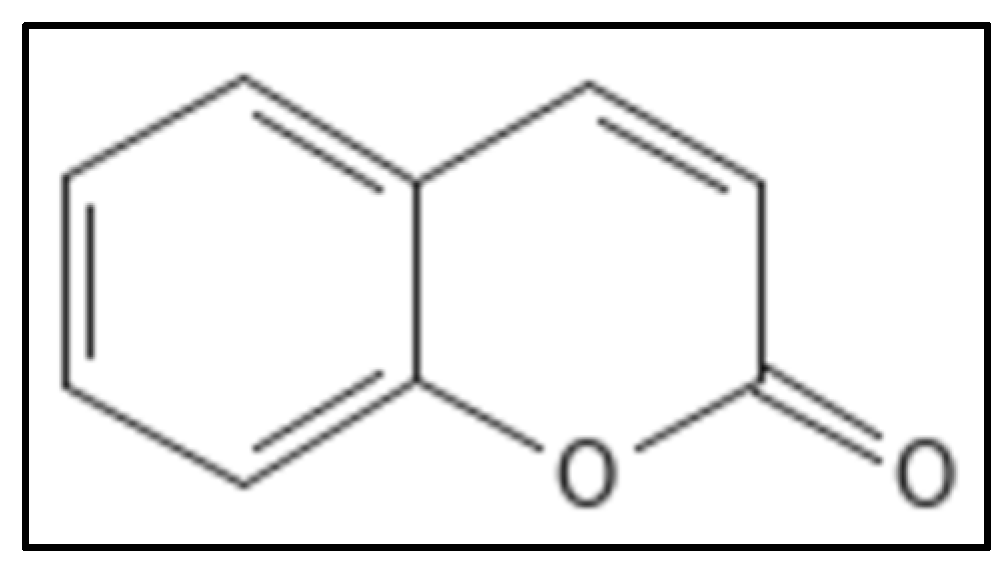
4.5. Coumarins
4.6. Curcumin
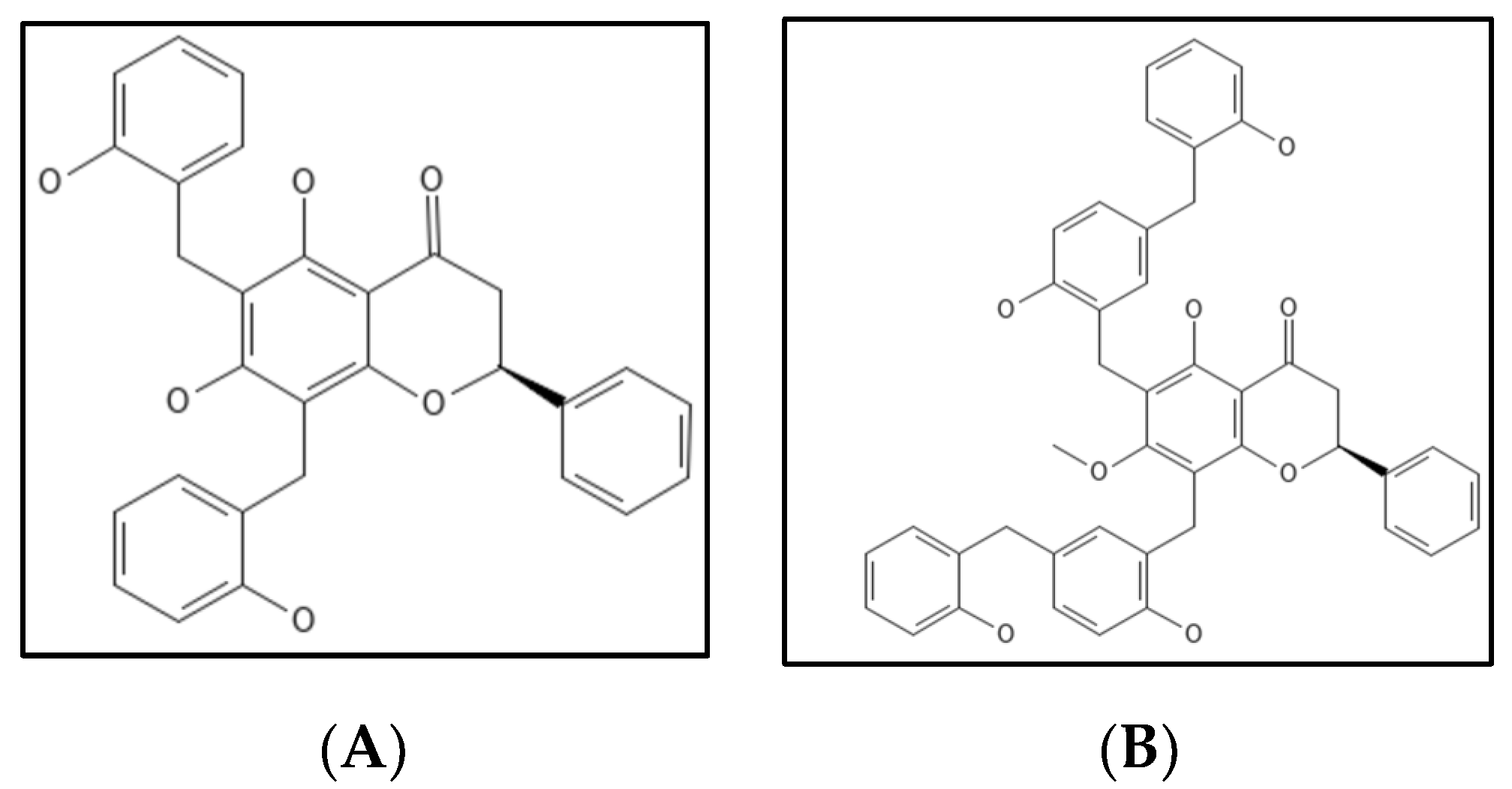
4.7. Dichamanetin and Derivatives
4.8. Doxorubicin
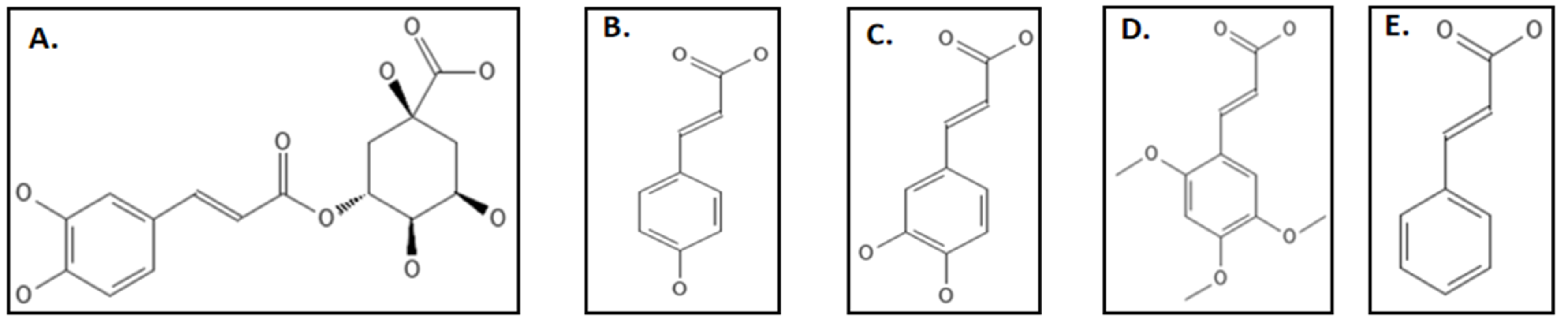
4.9. Phenylpropanoids
4.10. Plumbagin
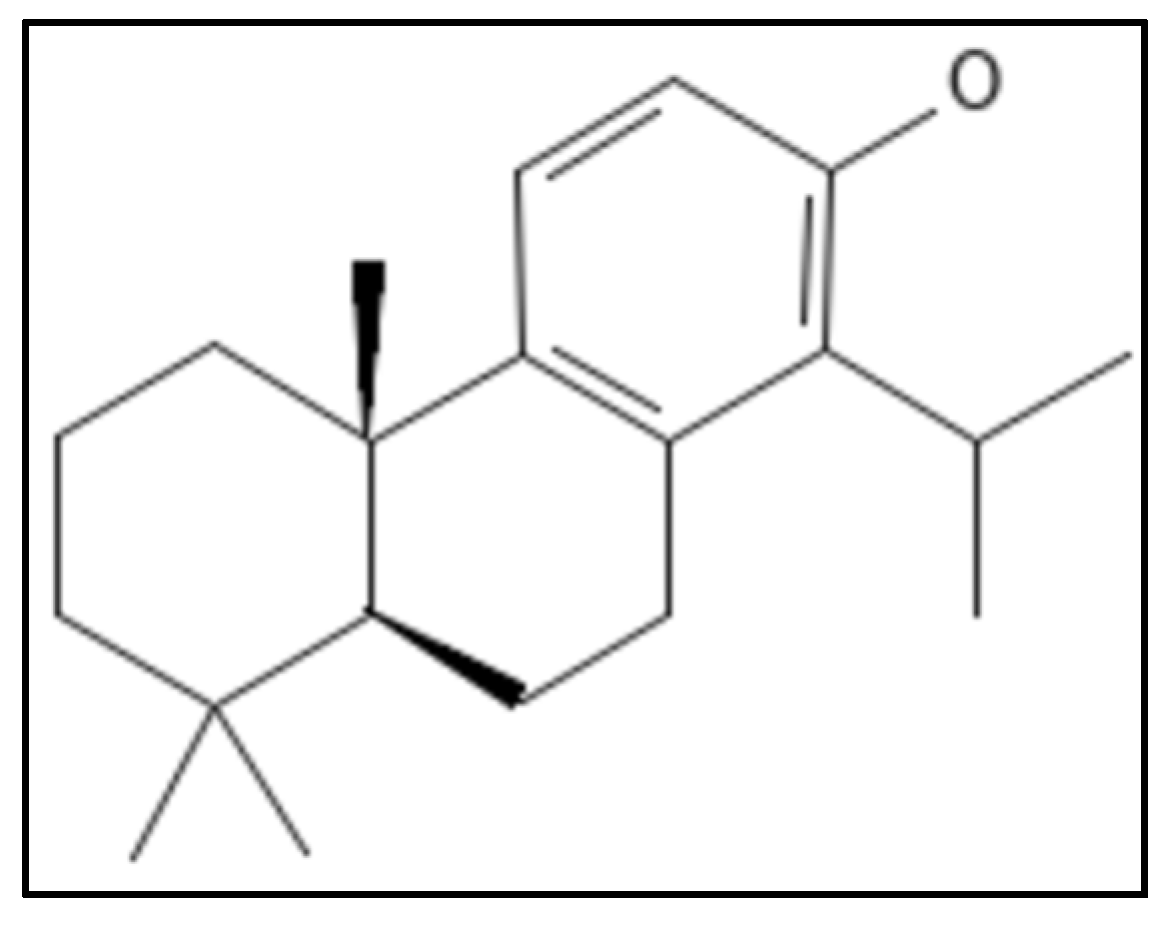
4.11. Totarol
4.12. Viriditoxin
4.13. Recent Reports
4.14. Natural Compounds over Synthetic Drugs: A Comparison
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Anti Microbial Resistance |
| ATCC | American Type Culture Collection |
| ESKAPE | Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species |
| GTP | Guanosine-5’-triphosphate |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| MGEs | Mobile Genetic Elements |
| NDM | New Delhi Metallo beta-lactamase-I |
| MDR | Multi Drug Resistance |
| PDR | Pan drug-resistant |
| VRSA | Vancomycin-resistant Staphylococcus aureus |
| WHO | World Health Organization |
| NP | Natural Products |
| VRE | Vancomycin-Resistant Enterococci |
References
- Arias, C.A.; Murray, B.E. Antibiotic-Resistant Bugs in the 21st Century—A Clinical Super-Challenge. N. Engl. J. Med. 2009, 360, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Nazaroff, W.W. Embracing microbes in exposure science. J. Expo. Sci. Environ. Epidemiol. 2018, 29, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Moyane, J.N.; Jideani, A.I.O.; Aiyegoro, O.A. Antibiotics usage in food-producing animals in South Africa and impact on human: Antibiotic resistance. Afr. J. Microbiol. Res. 2013, 7, 2990–2997. [Google Scholar]
- Ma, Y.X.; Wang, C.Y.; Li, Y.Y.; Li, J.; Wan, Q.Q.; Chen, J.H.; Tay, F.R.; Niu, L.N. Considerations and Caveats in Combating ESKAPE Pathogens against Nosocomial Infections. Adv. Sci. 2020, 7, 1901872. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Nanayakkara, A.K.; Boucher, H.W.; Fowler, V.G.; Jezek, A.; Outterson, K.; Greenberg, D.E. Antibiotic resistance in the patient with cancer: Escalating challenges and paths forward. CA Cancer J. Clin. 2021, 71, 488–504. [Google Scholar] [CrossRef]
- Wilsey, H.A.; Burgess, D.R.; Burgess, D.S. Focusing the Lens on the CAMERA Concepts: Early Combination β-Lactam and Vancomycin Therapy in Methicillin-Resistant Staphylococcus aureus Bacteremia. Antimicrob. Agents Chemother. 2020, 64, e00360-20. [Google Scholar] [CrossRef]
- Lima, L.M.; da Silva, B.N.M.; Barbosa, G.; Barreiro, E.J. β-lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 2020, 208, 112829. [Google Scholar] [CrossRef]
- Kim, S.W.; Bin Park, S.; Im, S.P.; Lee, J.S.; Jung, J.W.; Gong, T.W.; Lazarte, J.M.S.; Kim, J.; Seo, J.-S.; Kim, J.-H.; et al. Outer membrane vesicles from β-lactam-resistant Escherichia coli enable the survival of β-lactam-susceptible E. coli in the presence of β-lactam antibiotics. Sci. Rep. 2018, 8, 5402. [Google Scholar] [CrossRef]
- Yang, P.; Chen, Y.; Jiang, S.; Shen, P.; Lu, X.; Xiao, Y. Association between the rate of fluoroquinolones-resistant gram-negative bacteria and antibiotic consumption from China based on 145 tertiary hospitals data in 2014. BMC Infect. Dis. 2020, 20, 269. [Google Scholar] [CrossRef]
- Barrett, T.C.; Mok, W.W.K.; Murawski, A.M.; Brynildsen, M.P. Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic. Nat. Commun. 2019, 10, 1177. [Google Scholar] [CrossRef] [PubMed]
- Derbie, A.; Mekonnen, D.; Woldeamanuel, Y.; Abebe, T. Azithromycin resistant gonococci: A literature review. Antimicrob. Resist. Infect. Control 2020, 9, 138. [Google Scholar] [CrossRef] [PubMed]
- Farha, M.A.; Macnair, C.R.; Carfrae, L.A.; El Zahed, S.S.; Ellis, M.J.; Tran, H.-K.R.; McArthur, A.G.; Brown, E.D. Overcoming Acquired and Native Macrolide Resistance with Bicarbonate. ACS Infect. Dis. 2020, 6, 2709–2718. [Google Scholar] [CrossRef]
- Wang, J.; Galgoci, A.; Kodali, S.; Herath, K.B.; Jayasuriya, H.; Dorso, K.; Vicente, F.; González, A.; Cully, D.; Bramhill, D.; et al. Discovery of a Small Molecule that Inhibits Cell Division by Blocking FtsZ, a Novel Therapeutic Target of Antibiotics. J. Biol. Chem. 2003, 278, 44424–44428. [Google Scholar] [CrossRef]
- Gasparrini, A.J.; Markley, J.L.; Kumar, H.; Wang, B.; Fang, L.; Irum, S.; Symister, C.T.; Wallace, M.; Burnham, C.A.D.; Andleeb, S.; et al. Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance. Commun. Biol. 2020, 3, 241. [Google Scholar] [CrossRef] [PubMed]
- Jahantigh, M.; Samadi, K.; Dizaji, R.E.; Salari, S. Antimicrobial resistance and prevalence of tetracycline resistance genes in Escherichia coli isolated from lesions of colibacillosis in broiler chickens in Sistan, Iran. BMC Vet. Res. 2020, 16, 267. [Google Scholar] [CrossRef]
- Kresken, M.; Klare, I.; Wichelhaus, T.A.; Wohlfarth, E.; Layer-Nicolaou, F.; Neumann, B.; Werner, G. Glycopeptide resistance in Enterococcus spp. and coagulase-negative staphylococci from hospitalised patients in Germany: Occurrence, characteristics and dalbavancin susceptibility. J. Glob. Antimicrob. Resist. 2022, 28, 102–107. [Google Scholar] [CrossRef]
- Arthur, M.; Quintiliani, R. Regulation of VanA- and VanB-Type Glycopeptide Resistance in Enterococci. Antimicrob. Agents Chemother. 2001, 45, 375–381. [Google Scholar] [CrossRef]
- Hou, S.Y.; Wu, D.; Feng, X.H. Polymyxin monotherapy versus polymyxin-based combination therapy against carbapenem-resistant Klebsiella pneumoniae: A systematic review and meta-analysis. J. Glob. Antimicrob. Resist. 2020, 23, 197–202. [Google Scholar] [CrossRef]
- Christina, S.; Jorgensen, J.; Rybak, M.J. Meropenem and Vaborbactam: Stepping up the Battle against Carbapenem-resistant Enterobacteriaceae. Pharmacotherapy 2018, 38, 444–461. [Google Scholar] [CrossRef]
- Clancy, C.J.; Nguyen, M.H. Estimating the Size of the U.S. Market for New Antibiotics with Activity against Carbapenem-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2019, 63, e01733-19. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Courvalin, P.; Dantas, G.; Davies, J.; Eisenstein, B.; Huovinen, P.; Jacoby, G.A.; Kishony, R.; Kreiswirth, B.N.; Kutter, E.; et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011, 9, 894–896. [Google Scholar] [CrossRef] [PubMed]
- IACG Interagency Coordination Group on Antimicrobial Resistance. No Time to Wait: Securing the Future from Drug-Resistant Infections Report to the Secretary-General of the United Nations; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903. [Google Scholar] [CrossRef] [PubMed]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- Tiberi, S.; Zumla, A.; Migliori, G.B. Multi-Drug/Extensively Drug Resistant Tuberculosis-Epidemiology, Clinical Features, Management and Treatment. Infect. Dis. Clin. 2019, 33, 1063–1085. [Google Scholar] [CrossRef]
- Lee, B.Y.; Singh, A.; David, M.Z.; Bartsch, S.M.; Slayton, R.B.; Huang, S.S.; Zimmer, S.M.; Potter, M.A.; Macal, C.M.; Lauderdale, D.S.; et al. The economic burden of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). Clin. Microbiol. Infect. 2013, 19, 528–536. [Google Scholar] [CrossRef]
- Zhen, X.; Lundborg, C.S.; Zhang, M.; Sun, X.; Li, Y.; Hu, X.; Gu, S.; Gu, Y.; Wei, J.; Dong, H. Clinical and economic impact of methicillin-resistant Staphylococcus aureus: A multicentre study in China. Sci. Rep. 2020, 10, 3900. [Google Scholar] [CrossRef]
- Adrizain, R.; Suryaningrat, F.; Alam, A.; Setiabudi, D. Incidence of multidrug-resistant, extensively drug-resistant and pan-drug-resistant bacteria in children hospitalized at Dr. Hasan Sadikin general hospital Bandung Indonesia. In Proceedings of the IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2018; Volume 125, p. 12077. [Google Scholar]
- Oliva, A.; Giacobbe, D.R.; Di Luca, M.; Miller, N.S. New insights into infections due to multidrug resistant gram negative bacteria: The interplay between lab and clinic. BioMed Res. Int. 2018, 2018, 8905874. [Google Scholar] [CrossRef]
- Gill, J.; Arora, S.; Khanna, S.; Kumar, K.V.S. Prevalence of multidrug-resistant, extensively drug-resistant, and pandrug-resistant Pseudomonas aeruginosa from a tertiary level Intensive Care Unit. J. Glob. Infect. Dis. 2016, 8, 155–159. [Google Scholar] [CrossRef]
- Cillóniz, C.; Dominedò, C.; Torres, A. Multidrug Resistant Gram-Negative Bacteria in Community-Acquired Pneumonia. In Annual Update in Intensive Care and Emergency Medicine; Springer: Cham, Switzerland, 2019; pp. 459–475. [Google Scholar]
- World Health Organization. WHO: Global Tuberculosis Report; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Singh, V.; Chibale, K. Strategies to Combat Multi-Drug Resistance in Tuberculosis. Acc. Chem. Res. 2021, 54, 2361–2376. [Google Scholar] [CrossRef]
- Jubeh, B.; Breijyeh, Z.; Karaman, R. Antibacterial Prodrugs to Overcome Bacterial Resistance. Molecules 2020, 25, 1543. [Google Scholar] [CrossRef] [PubMed]
- Silber, N.; Pan, S.; Schäkermann, S.; Mayer, C.; Brötz-Oesterhelt, H.; Sass, P. Cell division protein FTSZ is unfolded for N-terminal degradation by antibiotic-activated CLPP. mBio 2020, 11, e01006-20. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Gonzalez, R.A.; Leung, C.Y.; Chan, B.K.; Turner, P.E.; Weitz, J.S. Quantitative Models of Phage-Antibiotic Combination Therapy. mSystems 2020, 5, e00756-19. [Google Scholar] [CrossRef]
- Zhu, M.; Tse, M.W.; Weller, J.; Chen, J.; Blainey, P.C. The future of antibiotics begins with discovering new combinations. Ann. N. Y. Acad. Sci. 2021, 1496, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-S.; Choi, T.-R.; Bhatia, S.K.; Lee, S.M.; Park, S.L.; Lee, H.S.; Kim, Y.-G.; Kim, J.-S.; Kim, W.; Yang, Y.-H. Combination Therapy Using Low-Concentration Oxacillin with Palmitic Acid and Span85 to Control Clinical Methicillin-Resistant Staphylococcus aureus. Antibiotics 2020, 9, 682. [Google Scholar] [CrossRef]
- Thwaites, M.; Hall, D.; Stoneburner, A.; Shinabarger, D.; Serio, A.W.; Krause, K.M.; Marra, A.; Pillar, C. Activity of plazomicin in combination with other antibiotics against multidrug-resistant Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2018, 92, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Coates, A.R.M.; Hu, Y.; Holt, J.; Yeh, P. Antibiotic combination therapy against resistant bacterial infections: Synergy, rejuvenation and resistance reduction. Expert Rev. Anti-Infect. Ther. 2020, 18, 5–15. [Google Scholar] [CrossRef]
- Schmid, A.; Wolfensberger, A.; Nemeth, J.; Schreiber, P.W.; Sax, H.; Kuster, S.P. Monotherapy versus combination therapy for multidrug-resistant Gram-negative infections: Systematic Review and Meta-Analysis. Sci. Rep. 2019, 9, 15290. [Google Scholar] [CrossRef]
- Belete, T.M. Novel targets to develop new antibacterial agents and novel alternatives to antibacterial agents. Hum. Microbiome J. 2019, 11, 100052. [Google Scholar] [CrossRef]
- Mendes, A. Tackling antibiotic resistance. Br. J. Community Nurs. 2019, 24, 612–613. [Google Scholar] [CrossRef]
- Bbosa, G.S.; Mwebaza, N.; Odda, J.; Kyegombe, D.B.; Ntale, M. Antibiotics/antibacterial drug use, their marketing and promotion during the post-antibiotic golden age and their role in emergence of bacterial resistance. Health 2014, 06, 410–425. [Google Scholar] [CrossRef]
- Morrison, J.J.; Conti, J.; Camberg, J.L. Assembly and architecture of Escherichia coli divisome proteins FtsA and FtsZ. J. Biol. Chem. 2022, 298, 101663. [Google Scholar] [CrossRef] [PubMed]
- Boes, A.; Olatunji, S.; Breukink, E.; Terrak, M. Regulation of the peptidoglycan polymerase activity of PBP1b by antagonist actions of the core divisome proteins FtsBLQ and FtsN. mBio 2019, 10, e01912-18. [Google Scholar] [CrossRef] [PubMed]
- Seidel, L. Investigating the Structure of the Escherichia coli Divisome Protein FtsK via Covariance Analysis. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2020. [Google Scholar]
- Glas, M.; Eiso, A.B.; Hollander, J.; Siegal, G.; Luirink, J.; de Esch, I. Interrogating the essential bacterial cell division protein FtsQ with fragments using target immobilized NMR screening (TINS). Int. J. Mol. Sci. 2019, 20, 3684. [Google Scholar] [CrossRef] [PubMed]
- Marmont, L.S.; Bernhardt, T.G. A conserved subcomplex within the bacterial cytokinetic ring activates cell wall synthesis by the FtsW-FtsI synthase. Proc. Natl. Acad. Sci. USA 2020, 117, 23879–23885. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Lamothe, R.; Sherratt, D.J. The bacterial cell cycle, chromosome inheritance and cell growth. Nat. Rev. Microbiol. 2019, 17, 467–478. [Google Scholar] [CrossRef]
- Geissler, B.; Margolin, W. Evidence for functional overlap among multiple bacterial cell division proteins: Compensating for the loss of FtsK. Mol. Microbiol. 2005, 58, 596–612. [Google Scholar] [CrossRef]
- Lock, R.L.; Harry, E.J. Cell-division inhibitors: New insights for future antibiotics. Nat. Rev. Drug Discov. 2008, 7, 324–338. [Google Scholar] [CrossRef]
- Chaudhary, R.; Mishra, S.; Kota, S.; Misra, H. Molecular interactions and their predictive roles in cell pole determination in bacteria. Crit. Rev. Microbiol. 2021, 47, 141–161. [Google Scholar] [CrossRef]
- Goehring, N.W.; Beckwith, J. Diverse paths to midcell: Assembly of the bacterial cell division machinery. Curr. Biol. 2005, 15, R514–R526. [Google Scholar] [CrossRef]
- Choi, Y.; Kim, J.; Yoon, H.J.; Jin, K.S.; Ryu, S.; Lee, H.H. Structural Insights into the FtsQ/FtsB/FtsL Complex, a Key Component of the Divisome. Sci. Rep. 2018, 8, 18061. [Google Scholar] [CrossRef] [PubMed]
- Vicente, M.; Rico, A.I.; Martínez-Arteaga, R.; Mingorance, J. Septum enlightenment: Assembly of bacterial division proteins. J. Bacteriol. 2006, 188, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Duggirala, S.; Nankar, R.; Rajendran, S. Phytochemicals as Inhibitors of Bacterial Cell Division Protein FtsZ: Coumarins Are Promising Candidates. Appl. Biochem. Biotechnol. 2014, 174, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Yao, Z.; Goehring, N.W.; Kishony, R.; Kahne, J.B. Rapid β-lactam-induced lysis requires successful assembly of the cell division machinery. Proc. Natl. Acad. Sci. USA 2009, 106, 21872–21877. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, S.; Sahu, S.K. FtsZ inhibitors as a new genera of antibacterial agents. Bioorg. Chem. 2019, 91, 103169. [Google Scholar] [CrossRef]
- Anca, I.-A.; Lumini, E.; Ghignone, S.; Salvioli, A.; Bianciotto, V.; Bonfante, P. The ftsZ Gene of the Endocellular Bacterium “Candidatus Glomeribacter gigasporarum” Is Preferentially Expressed during the Symbiotic Phases of Its Host Mycorrhizal Fungus. Mol. Plant Microbe Interact. MPMI 2009, 22, 302–310. [Google Scholar] [CrossRef]
- Tegos, G.; Mylonakis, E. Antimicrobial Drug Discovery: Emerging Strategies; CAB International: Wallingford, UK, 2012; ISBN 9781845939434. [Google Scholar]
- Haranahalli, K.; Tong, S.; Ojima, I. Recent advances in the discovery and development of antibacterial agents targeting the cell-division protein FtsZ. Bioorg. Med. Chem. 2016, 24, 6354–6369. [Google Scholar] [CrossRef]
- Maier, S.K.; Scherer, S.; Loessner, M.J. Long-chain polyphosphate causes cell lysis and inhibits Bacillus cereus septum formation, which is dependent on divalent cations. Appl. Environ. Microbiol. 1999, 65, 3942–3949. [Google Scholar] [CrossRef]
- Prajwala, B.; Kanthesh, M.B.; Gopenath, T.S. Studies on Inhibitory Potentials of Tinospora Cordifolia Plant Extract against Pathogenic Bacteria; The Greens Trust: Turuvekere, India, 2021; ISBN 81-951096-2-3. [Google Scholar]
- Rahman, M.U.; Wang, P.; Wang, N.; Chen, Y. A key bacterial cytoskeletal cell division protein FtsZ as a novel therapeutic antibacterial drug target. Bosn. J. Basic Med. Sci. 2020, 20, 310–318. [Google Scholar]
- Margolin, W.; Krupka, M. Unite to divide: Oligomerization of tubulin and actin homologs regulates initiation of bacterial cell division. F1000Research 2018, 7, 235. [Google Scholar] [CrossRef]
- Gandhi, M. Bacterial Cell Division Machinery: An Insight for Development of New Antibacterial Agent Targeting Cell Division Machinery for Development of Novel Inhibitors View Project COVID-19 View Project Shashikant Ray; Springer: Singapore, 2019. [Google Scholar] [CrossRef]
- Liao, Y.; Ithurbide, S.; Löwe, J.; Duggin, I.G. Two FtsZ proteins orchestrate archaeal cell division through distinct functions in ring assembly and constriction. bioRxiv 2020. [Google Scholar] [CrossRef]
- Volpi, S.; Cancelli, U.; Neri, M.; Corradini, R. Multifunctional delivery systems for peptide nucleic acids. Pharmaceuticals 2021, 14, 14. [Google Scholar] [CrossRef] [PubMed]
- Booth, S.; Lewis, R.J. Structural basis for the coordination of cell division with the synthesis of the bacterial cell envelope. Protein Sci. 2019, 28, 2042–2054. [Google Scholar] [CrossRef] [PubMed]
- Redfearn, J.C. A Comprehensive Model of the Structure and Function of the FtsZ Ring of Escherichia coli; Kent State University: Kent, OH, USA, 2016; pp. 1–200. [Google Scholar]
- Xiao, J.; Goley, E.D. Redefining the roles of the FtsZ-ring in bacterial cytokinesis. Curr. Opin. Microbiol. 2016, 34, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Harry, E.J. Bacterial cell division: Regulating Z-ring formation. Mol. Microbiol. 2001, 40, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Domadia, P.N.; Bhunia, A.; Sivaraman, J.; Swarup, S.; Dasgupta, D. Berberine targets assembly of Escherichia coli cell division protein FtsZ. Biochemistry 2008, 47, 3225–3234. [Google Scholar] [CrossRef]
- Domadia, P.; Swarup, S.; Bhunia, A.; Sivaraman, J.; Dasgupta, D. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem. Pharmacol. 2007, 74, 831–840. [Google Scholar] [CrossRef]
- Huang, Q.; Kirikae, F.; Kirikae, T.; Pepe, A.; Amin, A.; Respicio, L.; Slayden, R.A.; Tonge, P.J.; Ojima, I. Targeting FtsZ for antituberculosis drug discovery: Noncytotoxic taxanes as novel antituberculosis agents. J. Med. Chem. 2006, 49, 463–466. [Google Scholar] [CrossRef][Green Version]
- St. George, M.; Ayoub, A.T.; Banerjee, A.; Churchill, C.D.M.; Winter, P.; Klobukowski, M.; Cass, C.E.; Ludueña, R.F.; Tuszynski, J.A.; Damaraju, S. Designing and Testing of Novel Taxanes to Probe the Highly Complex Mechanisms by Which Taxanes Bind to Microtubules and Cause Cytotoxicity to Cancer Cells. PLoS ONE 2015, 10, e0129168. [Google Scholar] [CrossRef]
- Araújo-Bazán, L.; Ruiz-Avila, L.B.; Andreu, D.; Huecas, S.; Andreu, J.M. Cytological profile of antibacterial FtsZ inhibitors and synthetic peptide MciZ. Front. Microbiol. 2016, 7, 1558. [Google Scholar] [CrossRef]
- Rai, D.; Singh, J.K.; Roy, N.; Panda, D. Curcumin inhibits FtsZ assembly: An attractive mechanism for its antibacterial activity. Biochem. J. 2008, 410, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Erickson, H.P.; Anderson, D.E.; Osawa, M. FtsZ in Bacterial Cytokinesis: Cytoskeleton and Force Generator All in One. Microbiol. Mol. Biol. Rev. 2010, 74, 504–528. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Bian, L.; Ma, X.; Meng, Y.; Chen, C.S.; Ur Rahman, M.; Zhang, T.; Li, Z.; Wang, P.; Chen, Y. Assembly properties of the bacterial tubulin homolog FtsZ from the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 2019, 294, 16309–16319. [Google Scholar] [CrossRef] [PubMed]
- Mingorance, J.; Rivas, G.; Vélez, M.; Gómez-Puertas, P.; Vicente, M. Strong FtsZ is with the force: Mechanisms to constrict bacteria. Trends Microbiol. 2010, 18, 348–356. [Google Scholar] [CrossRef]
- Casiraghi, A.; Suigo, L.; Valoti, E.; Straniero, V. Targeting bacterial cell division: A binding site-centered approach to the most promising inhibitors of the essential protein FtsZ. Antibiotics 2020, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Dai, K.; Lutkenhaus, J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc. Natl. Acad. Sci. USA 1993, 90, 1053–1057. [Google Scholar] [CrossRef]
- Mukherjee, A.; Lutkenhaus, J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998, 17, 462–469. [Google Scholar] [CrossRef]
- Battaje, R.R.; Panda, D. Lessons from bacterial homolog of tubulin, FtsZ for microtubule dynamics. Endocr. Relat. Cancer 2017, 24, T1–T21. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Biswas, K.; Bandyopadhyay, U.; Banerjee, R.K. Turmeric and curcumin: Biological actions and medicinal applications. Curr. Sci. 2004, 87, 44–53. [Google Scholar]
- Shreaz, S.; Wani, W.A.; Behbehani, J.M.; Raja, V.; Irshad, M.; Karched, M.; Ali, I.; Siddiqi, W.A.; Hun, L.T. Cinnamaldehyde and its derivatives, a novel class of antifungal agents. Fitoterapia 2016, 112, 116–131. [Google Scholar] [CrossRef]
- Kumar, K.; Awasthi, D.; Berger, W.T.; Tonge, P.J.; Slayden, R.A.; Ojima, I. Discovery of anti-TB agents that target the cell-division protein FtsZ. Future Med. Chem. 2010, 2, 1305–1323. [Google Scholar] [CrossRef] [PubMed]
- Dhaked, H.P.S.; Bhattacharya, A.; Yadav, S.; Dantu, S.C.; Kumar, A.; Panda, D. Mutation of Arg191 in FtsZ Impairs Cytokinetic Abscission of Bacillus subtilis Cells. Biochemistry 2016, 55, 5754–5763. [Google Scholar] [CrossRef] [PubMed]
- Sankhwar, R.; Yadav, S.; Kumar, A.; Gupta, R.K. Application of nano-curcumin as a natural antimicrobial agent against gram-positive pathogens. J. Appl. Nat. Sci. 2021, 13, 110–126. [Google Scholar] [CrossRef]
- Keffer, J.L.; Bifulco, G.; Lloyd, J.R.; Bewley, C.A.; Huecas, S.; Hammill, J.T.; Wipf, P.; Andreu, J.M.; Bewley, C.A. Chrysophaentins are competitive inhibitors of FtsZ and inhibit Z-ring formation in live bacteria. J. Am. Chem. Soc. 2013, 132, 5673–5678. [Google Scholar] [CrossRef]
- Li, X.; Sheng, J.; Huang, G.; Ma, R.; Yin, F.; Song, D.; Zhao, C.; Ma, S. Design, synthesis and antibacterial activity of cinnamaldehyde derivatives as inhibitors of the bacterial cell division protein FtsZ. Eur. J. Med. Chem. 2015, 97, 32–41. [Google Scholar] [CrossRef]
- Korkina, L.; Kostyuk, V.; De Luca, C.; Pastore, S. Plant Phenylpropanoids as Emerging Anti-Inflammatory Agents. Mini Rev. Med. Chem. 2011, 11, 823–835. [Google Scholar] [CrossRef]
- Sun, N.; Chan, F.Y.; Lu, Y.J.; Neves, M.A.C.; Lui, H.K.; Wang, Y.; Chow, K.Y.; Chan, K.F.; Yan, S.C.; Leung, Y.C.; et al. Rational design of berberine-based FtsZ inhibitors with broad-spectrum antibacterial activity. PLoS ONE 2014, 9, e97514. [Google Scholar] [CrossRef]
- Cathie, I.A.B. Neonatal jaundice. Med. World 1947, 67, 104–106. [Google Scholar] [CrossRef]
- Beuria, T.K.; Santra, M.K.; Panda, D. Sanguinarine blocks cytokinesis in bacteria by inhibiting FtsZ assembly a bundling. Biochemistry 2005, 44, 16584–16593. [Google Scholar] [CrossRef]
- Pilch, D.S.; Lavoie, E.J.; Kaul, M.; Parhi, A. Benzo [C] Phenanthridines as Antimicrobial Agents. U.S. Patent 8,741,917, 2010. [Google Scholar]
- Musa, M.A.; Badisa, V.L.D.; Latinwo, L.M.; Cooperwood, J.; Sinclair, A.; Abdullah, A. Cytotoxic activity of new acetoxycoumarin derivatives in cancer cell lines. Anticancer Res. 2011, 31, 2017–2022. [Google Scholar]
- Matos, M.J.; Vazquez-Rodriguez, S.; Santana, L.; Uriarte, E.; Fuentes-Edfuf, C.; Santos, Y.; Muñoz-Crego, A. Synthesis and Structure-Activity Relationships of Novel Amino/Nitro Substituted 3-Arylcoumarins as Antibacterial Agents. Molecules 2013, 18, 1394. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ma, R.; Bi, F.; Zhang, F.; Hu, C.; Venter, H.; Semple, S.J.; Ma, S. Novel 5-methyl-2-phenylphenanthridium derivatives as FtsZ-targeting antibacterial agents from structural simplification of natural product sanguinarine. Bioorg. Med. Chem. Lett. 2018, 28, 1825–1831. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Ashaq, M. Safety and Toxicity of Botanical Medicines: A critical Appraisal. Int. J. All Res. Educ. Sci. Methods 2021, 9, 2455–6211. [Google Scholar]
- Mitra, K.; Chadha, A.; Doble, M. Pharmacophore based approach to screen and evaluate novel Mycobacterium cell division inhibitors targeting FtsZ-A modelling and experimental study. Eur. J. Pharm. Sci. 2019, 135, 103–112. [Google Scholar] [CrossRef]
- Riveiro, M.; De Kimpe, N.; Moglioni, A.; Vazquez, R.; Monczor, F.; Shayo, C.; Davio, C. Coumarins: Old compounds with novel promising therapeutic perspectives. Curr. Med. Chem. 2010, 17, 1325–1338. [Google Scholar] [CrossRef]
- Sridevi, D.; Sudhakar, K.U.; Ananthathatmula, R.; Nankar, R.P.; Doble, M. Mutation at G103 of MtbFtsZ altered their sensitivity to coumarins. Front. Microbiol. 2017, 8, 578. [Google Scholar] [CrossRef]
- Alnami, A.; Norton, R.S.; Pena, H.P.; Haider, S.; Kozielski, F. Conformational Flexibility of A Highly Conserved Helix Controls Cryptic Pocket Formation in FtsZ. J. Mol. Biol. 2021, 433, 167061. [Google Scholar] [CrossRef]
- Kaur, S.; Modi, N.H.; Panda, D.; Roy, N. Probing the binding site of curcumin in Escherichia coli and Bacillus subtilis FtsZ-A structural insight to unveil antibacterial activity of curcumin. Eur. J. Med. Chem. 2010, 45, 4209–4214. [Google Scholar] [CrossRef]
- Urgaonkar, S.; La Pierre, H.S.; Meir, I.; Lund, H.; RayChaudhuri, D.; Shaw, J.T. Synthesis of antimicrobial natural products targeting FtsZ: (±)-dichamanetin and (±)-2‴-hydroxy-5″- benzylisouvarinol-B. Org. Lett. 2005, 7, 5609–5612. [Google Scholar] [CrossRef]
- Panda, P.; Taviti, A.C.; Satpati, S.; Kar, M.M.; Dixit, A.; Beuria, T.K. Doxorubicin inhibits E. coli division by interacting at a novel site in FtsZ. Biochem. J. 2015, 471, 335–346. [Google Scholar] [CrossRef]
- Hemaiswarya, S.; Doble, M. Synergistic interaction of phenylpropanoids with antibiotics against bacteria. J. Med. Microbiol. 2010, 59, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sheng, J.; Song, D.; Guo, L.; Ma, S. Phenylacrylamides as Novel FtsZ-targeted Potential Antimicrobials. Lett. Drug Des. Discov. 2014, 12, 234–240. [Google Scholar] [CrossRef]
- Sun, J.; Li, M.H.; Wang, X.Y.; Zhang, Y.; Yuan, R.J.; Liu, H.Y.; Zhu, H.L. Vanillin derivatives as the selective small molecule inhibitors of FtsZ. Med. Chem. Res. 2014, 23, 2985–2994. [Google Scholar] [CrossRef]
- Sun, J.; Yin, Y.; Sheng, G.H.; Yang, Z.B.; Zhu, H.L. Synthesis, molecular modeling and structural characterization of vanillin derivatives as antimicrobial agents. J. Mol. Struct. 2013, 1039, 214–218. [Google Scholar] [CrossRef]
- Mathew, R.; Kruthiventi, A.K.; Prasad, J.V.; Kumar, S.P.; Srinu, G.; Chatterji, D. Inhibition of mycobacterial growth by plumbagin derivatives. Chem. Biol. Drug Des. 2010, 76, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, D.T.; Andrei, C.C.; Saridakis, H.O.; De Jesus Faria, T.; Vinhato, E.; Carvalho, K.E.; Daniel, J.F.S.; Machado, S.L.; Saridakis, D.P.; Braz-Filho, R. Antimicrobial activity and chemical investigation of Brazilian Drosera. Mem. Inst. Oswaldo Cruz 2004, 99, 753–755. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Jindal, B.; Singh, P.; Datta, A.; Panda, D. Plumbagin inhibits cytokinesis in Bacillus subtilis by inhibiting FtsZ assembly—A mechanistic study of its antibacterial activity. FEBS J. 2013, 280, 4585–4599. [Google Scholar] [CrossRef]
- De Paiva, S.R.; Figueiredo, M.R.; Aragão, T.V.; Coelho Kaplan, M.A. Antimicrobial Activity in Vitro of Plumbagin Isolated from Plumbago Species. Mem. Inst. Oswaldo Cruz 2003, 98, 959–961. [Google Scholar] [CrossRef]
- Acharya, B.R.; Bhattacharyya, B.; Chakrabarti, G. The natural naphthoquinone plumbagin exhibits antiproliferative activity and disrupts the microtubule network through tubulin binding. Biochemistry 2008, 47, 7838–7845. [Google Scholar] [CrossRef]
- Jaiswal, R.; Beuria, T.K.; Mohan, R.; Mahajan, S.K.; Panda, D. Totarol inhibits bacterial cytokinesis by perturbing the assembly dynamics of FtsZ. Biochemistry 2007, 46, 4211–4220. [Google Scholar] [CrossRef]
- Kim, M.B.; Shaw, J.T. Synthesis of antimicrobial natural products targeting FtsZ: (+)-totarol and related totarane diterpenes. Org. Lett. 2010, 12, 3324–3327. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Zhao, C.; Li, D.; Cao, J.; Ju, Z.; Kim, E.L.; Jung, Y.S.; Jung, J.H. Viriditoxin stabilizes microtubule polymers in SK-OV-3 cells and exhibits antimitotic and antimetastatic potential. Mar. Drugs 2020, 18, 445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.Y.; Wu, Y.Y.; Zhang, M.Y.; Cheng, J.; Dube, B.; Yu, H.J.; Zhang, Y.X. New antimicrobial compounds produced by Seltsamia galinsogisoli sp. nov., isolated from Galinsoga parviflora as potential inhibitors of FtsZ. Sci. Rep. 2019, 9, 8319. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ye, H.; Gao, X.; Feng, Y.; Shao, W.; Qi, P.; Wu, Z.; Liu, L.; Wang, P.; Yang, S. The discovery of natural 4’-demethylepipodophyllotoxin from renewable Dysosma versipellis species as a novel bacterial cell division inhibitor for controlling intractable diseases in rice. Ind. Crops Prod. 2021, 174, 114182. [Google Scholar] [CrossRef]
- Mahanty, S.; Rathinasamy, K. The natural anthraquinone dye purpurin exerts antibacterial activity by perturbing the FtsZ assembly. Bioorg. Med. Chem. 2021, 50, 116463. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cui, Y.; Lu, L.; Gong, Y.; Han, W.; Piao, G. Natural indole-containing alkaloids and their antibacterial activities. Arch. Pharm. 2020, 353, 2000120. [Google Scholar] [CrossRef]
- Margalit, D.N.; Romberg, L.; Mets, R.B.; Hebert, A.M.; Mitchison, T.J.; Kirschner, M.W.; RayChaudhuri, D. Targeting cell division: Small-molecule inhibitors of FtsZ GTPase perturb cytokinetic ring assembly and induce bacterial lethality. Proc. Natl. Acad. Sci. USA 2004, 101, 11821–11826. [Google Scholar] [CrossRef]
- Gurnani, M.; Rath, P.; Chauhan, A.; Ranjan, A.; Ghosh, A.; Lal, R.; Mukerjee, N.; Aljarba, N.H.; Alkahtani, S.; Rajput, V.D.; et al. Inhibition of Filamentous Thermosensitive Mutant-Z Protein in Bacillus subtilis by Cyanobacterial Bioactive Compounds. Molecules 2022, 27, 1907. [Google Scholar] [CrossRef]
- Possoz, C.; Newmark, J.; Sorto, N.; Sherratt, D.J.; Tolmasky, M.E. Sublethal concentrations of the aminoglycoside amikacin interfere with cell division without affecting chromosome dynamics. Antimicrob. Agents Chemother. 2007, 51, 252–256. [Google Scholar] [CrossRef]
- Czaplewski, L.G.; Collins, I.; Boyd, E.A.; Brown, D.; East, S.P.; Gardiner, M.; Fletcher, R.; Haydon, D.J.; Henstock, V.; Ingram, P.; et al. Antibacterial alkoxybenzamide inhibitors of the essential bacterial cell division protein FtsZ. Bioorg. Med. Chem. Lett. 2009, 19, 524–527. [Google Scholar] [CrossRef]
- Läppchen, T.; Pinas, V.A.; Hartog, A.F.; Koomen, G.J.; Schaffner-Barbero, C.; Andreu, J.M.; Trambaiolo, D.; Löwe, J.; Juhem, A.; Popov, A.V.; et al. Probing FtsZ and Tubulin with C8-Substituted GTP Analogs Reveals Differences in Their Nucleotide Binding Sites. Chem. Biol. 2008, 15, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Kaul, M.; Zhang, Y.; Parhi, A.K.; Lavoie, E.J.; Tuske, S.; Arnold, E.; Kerrigan, J.E.; Pilch, D.S. Enterococcal and streptococcal resistance to PC190723 and related compounds: Molecular insights from a FtsZ mutational analysis. Biochimie 2013, 95, 1880–1887. [Google Scholar] [CrossRef] [PubMed]
- White, E.L.; Suling, W.J.; Ross, L.J.; Seitz, L.E.; Reynolds, R.C. 2-Alkoxycarbonylaminopyridines: Inhibitors of Mycobacterium tuberculosis FtsZ. J. Antimicrob. Chemother. 2002, 50, 111–114. [Google Scholar] [CrossRef][Green Version]
- Reynolds, R.C.; Srivastava, S.; Ross, L.J.; Suling, W.J.; White, E.L. A new 2-carbamoyl pteridine that inhibits mycobacterial FtsZ. Bioorg. Med. Chem. Lett. 2004, 14, 3161–3164. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Robinson, C.A.; Howe, A.G.; Mazor, T.; Wood, P.A.; Urgaonkar, S.; Hebert, A.M.; RayChaudhuri, D.; Shaw, J.T. N-Benzyl-3-sulfonamidopyrrolidines as novel inhibitors of cell division in E. coli. Bioorg. Med. Chem. Lett. 2007, 17, 6651–6655. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khadkikar, P.; Goud, N.S.; Mohammed, A.; Ramamoorthy, G.; Ananthathatmula, R.; Doble, M.; Rizvi, A.; Banerjee, S.; Ravi, A.; Alvala, M. An efficient and facile green synthesis of bisindole methanes as potential Mtb FtsZ inhibitors. Chem. Biol. Drug Des. 2018, 92, 1933–1939. [Google Scholar] [CrossRef]
- Sun, N.; Ban, L.; Li, M.; Fang, Z.; Li, X.; Yao, W.; Pan, J.; Lu, Y.; Liu, Z.; Wong, W.L. Probing the benzofuroquinolinium derivative as a potent antibacterial agent through the inhibition of FtsZ activity. J. Pharmacol. Sci. 2018, 138, 83–85. [Google Scholar] [CrossRef]
- Feher, M.; Schmidt, J.M. Property distributions: Differences between drugs, natural products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci. 2003, 43, 218–227. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Barnes, E.C.; Kumar, R.; Davis, R.A. The use of isolated natural products as scaffolds for the generation of chemically diverse screening libraries for drug discovery. Nat. Prod. Rep. 2016, 33, 372–381. [Google Scholar] [CrossRef]
- Petre, A.A.; Nenciu, F.; Vanghele, N.A.; Stanciu, M.M.; Mihalache, D.B.; Grigore, I.A.; Vladuţoiu, L. Study on the effects and changes of soil degradation under the influence of antibiotics. E3S Web Conf. 2020, 180, 03018. [Google Scholar] [CrossRef]
- Moreno-Bondi, M.C. Antibiotics in food and environmental samples. Anal. Bioanal. Chem. 2009, 395, 875–876. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lima, T.; Domingues, S.; Silva, G.J. Da Manure as a potential hotspot for antibiotic resistance dissemination by horizontal gene transfer events. Vet. Sci. 2020, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Kanaan, M.H.G. Prevalence, Resistance to Antimicrobials, and Antibiotypes of Arcobacter Species Recovered from Retail Meat in Wasit Marketplaces in Iraq. Int. J. One Health 2021, 7, 142–150. [Google Scholar] [CrossRef]
- De Cesare, A.; Oliveri, C.; Lucchi, A.; Savini, F.; Manfreda, G.; Sala, C. Pilot Study on Poultry Meat from Antibiotic Free and Conventional Farms: Can Metagenomics Detect Any Difference? Foods 2022, 11, 249. [Google Scholar] [CrossRef]
- Conde-Cid, M.; Núñez-Delgado, A.; Fernández-Sanjurjo, M.J.; Álvarez-Rodríguez, E.; Fernández-Calviño, D.; Arias-Estévez, M. Tetracycline and sulfonamide antibiotics in soils: Presence, fate and environmental risks. Processes 2020, 8, 1479. [Google Scholar] [CrossRef]
- Carvalho, I.T.; Santos, L. Antibiotics in the aquatic environments: A review of the European scenario. Environ. Int. 2016, 94, 736–757. [Google Scholar] [CrossRef]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; de Schaetzen, M.A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial resistance in the food chain: A review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef]
- Klees, S.; Effelsberg, N.; Stührenberg, B.; Mellmann, A.; Schwarz, S.; Köck, R. Prevalence and epidemiology of multidrug-resistant pathogens in the food chain and the urban environment in northwestern germany. Antibiotics 2020, 9, 708. [Google Scholar] [CrossRef]
- Proietti, P.C.; Stefanetti, V.; Musa, L.; Zicavo, A.; Dionisi, A.M.; Bellucci, S.; Mensa, A.L.; Menchetti, L.; Branciari, R.; Ortenzi, R.; et al. Genetic profiles and antimicrobial resistance patterns of salmonella infantis strains isolated in italy in the food chain of broiler meat production. Antibiotics 2020, 9, 814. [Google Scholar] [CrossRef]
- Pan, M.; Wong, C.K.C.; Chu, L.M. Distribution of antibiotics in wastewater-irrigated soils and their accumulation in vegetable crops in the Pearl River Delta, Southern China. J. Agric. Food Chem. 2014, 62, 11062–11069. [Google Scholar] [CrossRef] [PubMed]
- Reybroeck, W. Residues of Antibiotics and Sulphonamides in Honey on. Apiacta 2003, 38, 23–30. [Google Scholar]
- Shih, Y.-L.; Rothfield, L. The Bacterial Cytoskeleton. Microbiol. Mol. Biol. Rev. 2006, 70, 729. [Google Scholar] [CrossRef] [PubMed]
- McCormick, J.R. Cell division is dispensable but not irrelevant in Streptomyces. Curr. Opin. Microbiol. 2009, 12, 689–698. [Google Scholar] [CrossRef]
- Jukič, M.; Gobec, S.; Sova, M. Reaching toward underexplored targets in antibacterial drug design. Drug Dev. Res. 2019, 80, 6–10. [Google Scholar] [CrossRef]
- Silber, N.; Matos De Opitz, C.L.; Mayer, C.; Sass, P. Cell division protein FtsZ: From structure and mechanism to antibiotic target. Future Microbiol. 2020, 15, 801–831. [Google Scholar] [CrossRef]
- Ferrer-González, E.; Huh, H.; Al-Tameemi, H.M.; Boyd, J.M.; Lee, S.H.; Pilch, D.S. Impact of ftsz inhibition on the localization of the penicillin binding proteins in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 2021, 203, 204–225. [Google Scholar] [CrossRef]
- Carro, L. Recent progress in the development of small-molecule FtsZ inhibitors as chemical tools for the development of novel antibiotics. Antibiotics 2019, 8, 217. [Google Scholar] [CrossRef]









| Protein | Function | Interacting Protein |
|---|---|---|
| Fts-Z | Cytoskeleton protein, Self-polymerizing GTPase which forms Z-ring/proto ring and recruitment of other proteins | Fts-A, Zip-A, Fts-K |
| Fts-A | ATP binding protein which anchors and stabilizes Fts-Z filaments | Fts-N, Fts-Z |
| Zip-A | Provides membrane anchorage for Fts-Z by bunding the filaments | Fts-Z |
| Fts-K | DNA segregation at C terminal, stabilizing component and linker between early and late stages of division. | Fts-A, Fts-I, Fts-L, Fts-Q, Fts-Z |
| Fts-Q | Periplasmic functional domain, Peptidoglycan synthesis, Complex formation with Fts-B and Fts-L | Fts-B, Fts-I, Fts-L, Fts-N, Fts-W |
| Fts-L | Complex formation with Fts-B and Fts-Q, Peptidoglycan synthesis | Fts-B, Fts-I, Fts-K, Fts-Q |
| Fts-B | Complex formation with Fts-L and Fts-Q, Peptidoglycan synthesis | Fts-L, Fts-Q |
| Fts-W | Translocation of lipid precursors across membrane, Peptidoglycan synthesis | Fts-I, Fts-L, Fts-N, Fts-Q, Fts-Z |
| Fts-I | Transpeptidase, Cross links peptidoglycan strains | Fts-N, Fts-Q, Fts-W |
| Fts-N | Triggers septation during peptidoglycan synthesis, Periplasmic functional domain | Fts-A, Fts-I, Fts-Q, Fts-W |
| Compound | Target Organism | Action Mechanism | Assay Used | MIC/IC50 | Ref. |
|---|---|---|---|---|---|
| Zantrin | Broad range | Z1, Z2, Z4 destabilize Fts-Z assembly; Z5 hyper stabilize Fts-Z assembly. | Real time enzyme coupled fluorescent GTPase assay. | E. coli = 4–25 µM M. tuberculosis = 30–70 µM | [129] |
| Amikacin | E. coli | Z-ring perturbation | Cell elongation | 4 µg mL−1 | [130] |
| A-189 | E. coli, S. aureus | GTPase inhibition and Z-ring assembly inhibition | Anucleate cell blue assay. | 16 µg mL−1 | [131] |
| GTP analogue | E. coli, S. aureus | GTPase inhibition | Spectrophotometric coupled enzymatic assay | MeOGTP-IC50 = 15 μM | [132] |
| PC190723 | B. subtilis, S. aureus (MRSA) | Binds H7 loop affecting GTPase activity causing Z-ring mis localization | Whole cell-based assay leading to filamentous phenotype | 0.5 µg mL−1 | [133] |
| SRI-3072 | M. tuberculosis | Inhibition of Fts-Z polymerization and GTPase activity | Antimicrobial assay | 19 µM | [134] |
| Taxanes | M. tuberculosis | Stabilizes Fts-Z against depolymerization | Real-time PCR based assay, Cell filamentation | 1.25−2.5 μM | [76] |
| 2-carbamoyl pteridine | M. tuberculosis | GTPase activity inhibition and Fts-Z polymerization | GTPase activity and Fts-Z polymerization through in vitro technique | 2 µg mL−1 | [135] |
| 534F6 derivatives | E. coli | Cell division inhibition | Microorganisms lacking MDR pumps were used for screening | >80 μM | [136] |
| Bis indole methane (2,2′-bisindole) | M. tuberculosis, M. segmentis, H37Rv strain | Cell division inhibition | GTPase activity, Cell proliferation, Antimycobacterial property | 62.5 µg mL−1 | [137] |
| 5-Methyl-11-((3-(3-(4-methylpyridine))-propyl benzo[d] thiazol-2(3H)-ylidene) methyl) benzofuro[3,2-b] quinolin-5-ium iodide) | B. subtilis, S. aureus, E. coli | GTPase activity, Polymerization of Fts-Z | Cell based antibiotic screening assay, Biochemical assay, Light scattering assay | 0.5, 2, 4 µg mL−1 respectively | [138] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurnani, M.; Chauhan, A.; Ranjan, A.; Tuli, H.S.; Alkhanani, M.F.; Haque, S.; Dhama, K.; Lal, R.; Jindal, T. Filamentous Thermosensitive Mutant Z: An Appealing Target for Emerging Pathogens and a Trek on Its Natural Inhibitors. Biology 2022, 11, 624. https://doi.org/10.3390/biology11050624
Gurnani M, Chauhan A, Ranjan A, Tuli HS, Alkhanani MF, Haque S, Dhama K, Lal R, Jindal T. Filamentous Thermosensitive Mutant Z: An Appealing Target for Emerging Pathogens and a Trek on Its Natural Inhibitors. Biology. 2022; 11(5):624. https://doi.org/10.3390/biology11050624
Chicago/Turabian StyleGurnani, Manisha, Abhishek Chauhan, Anuj Ranjan, Hardeep Singh Tuli, Mustfa F. Alkhanani, Shafiul Haque, Kuldeep Dhama, Rup Lal, and Tanu Jindal. 2022. "Filamentous Thermosensitive Mutant Z: An Appealing Target for Emerging Pathogens and a Trek on Its Natural Inhibitors" Biology 11, no. 5: 624. https://doi.org/10.3390/biology11050624
APA StyleGurnani, M., Chauhan, A., Ranjan, A., Tuli, H. S., Alkhanani, M. F., Haque, S., Dhama, K., Lal, R., & Jindal, T. (2022). Filamentous Thermosensitive Mutant Z: An Appealing Target for Emerging Pathogens and a Trek on Its Natural Inhibitors. Biology, 11(5), 624. https://doi.org/10.3390/biology11050624









