Simple Summary
Stock enhancement aggressively replenishes depleted wild finfish populations. However, stock enhancement of black sea bream in Taiwan with complex genetic sources, especially when successful, maintains genetic diversity but dramatically changes the genetic structure within and among wild populations.
Abstract
Stock enhancement, used for replenishing depleted wild finfish populations, is an aggressive approach. Stock enhancement projects in Taiwan involve black sea bream (Acanthopagrus schlegelii), a major commercial species. During 2004–2015, even management agencies conducted stock enhancement projects, leading to numerous private releases that have not been recorded. Stock enhancement by a private hatchery without accurate genetic records may lead to a genetic structure change in wild populations. Using allele frequencies at nine microsatellite loci, we studied the genetic effects of stock enhancement in 19 samples collected from populations in the hatcheries and the wild. In 458 individuals from nine hatchery samples, most populations showed weak but significant genetic differences and complex clusters in structure analysis, indicating dramatic stock change within and among hatcheries. The 10 wild populations (n = 773) also had a complex genetic composition and were genetically different among sampling sites and times. However, a simple and clear cluster in structure analysis was found for only one sampling site, which had no release history. Thus, stock enhancement with complex genetic sources helps maintain genetic diversity but dramatically changes the genetic structure within and among wild populations, especially when stock enhancement is successful.
1. Introduction
Advances in fishing technology have led to fish stocks, which are renewable fishery resources, being exhausted. Approximately half of all fish stocks have been deemed “fully exploited” or “overexploited” [1,2]. Pollution and habitat destruction due to human activities have drastically reduced the abundance and distribution of marine fish and invertebrate populations [2,3,4] and depleted fishery stocks in more than 100 species to date [5,6,7].
Aiming to improve fishery resources, Taiwan’s government promotes a massive stock enhancement program in its coastal waters every year [8,9]. Under this program, numerous artificial breeding fry are released, with a small number of fish labeled to enable survival rate evaluation [8,9,10]. Moreover, the government annually allocates a considerable amount of money to this program. However, the role of genetic factors in stock enhancement has been neglected [6,11,12]. Conventional markers (biological, physical, and chemical) cannot estimate the reproduction rate of released fish [5,6,13]. Stock enhancement programs should incorporate genetic information such as genetic stock structure and diversity. The official fishery organization (the Taiwan Fisheries Sustainable Development Association; TFSDA) in Taiwan procures fish fry for stock enhancement from one or several private hatcheries. According to several genetic studies, negative effects on natural populations were noted after successful stock enhancements [7,14]. These enhancements involved the release of hatchery populations or the escape of numerous relatively unfit [14,15,16,17] and not genetically diverse individuals, such as the Adriatic sturgeon [18], Korean starry flounder [19], and black sea bream from Japan [20]. The genetic effects of these hatchery fish in the wild have received considerable attention in developed countries. For example, Japan, the United States, and several countries in Europe have created official agencies for managing stock enhancement [7,21,22]. These agencies not only verify the reliability of external markers, but also evaluate the genetic composition of offspring and their reproductive rate through mark–recapture studies [5,7,22]. However, identifying fish sources is difficult when release records are complex and unclear, such as in Taiwan [8,9,10,11,12].
Black sea bream (Acanthopagrus schlegelii), an economically vital species for both fisheries and aquaculture, is widely distributed along West Pacific coasts from Japan and Korea to the East China Sea and Taiwan. In southern China, black sea bream males become sexually mature within 1 year, and 50% of them change sex by two years old. In this fish, reproduction occurs at 1–2 years of age in coastal waters and at river mouths [23]. The species is abundant off the west coast of Taiwan, where it is a popular sport fish. Notably, concern over the rapid decline in black sea bream stocks is growing because the related catch production declined from 718 tons in 2000 to 212 tons in 2015, according to the Fisheries Statistical Yearbook of Taiwan [24]. Black sea bream aquaculture began in the 1980s, and Taiwanese hatcheries are mainly located in the Kaohsiung and Ping-tung areas. Broodstocks are from main fishery areas, which are off the west coast of Taiwan (Chiayi–Yunlin–Changhua). Using reliable mass production techniques [8,9], massive hatchery juveniles are released into the wild to replenish the insufficient natural supply [8,9,10]. Black sea bream has thus become a dominant species for stock enhancement in Taiwan. During 2004–2015, more than 12 million hatchery black sea bream fry were released into the coastal water off Taiwan by TFSDA [8,9,10,11,12].
Although no genetic or hatchery information is available for stocks and fry in Taiwan during 2004–2015, investigating the genetic structure of hatchery and wild populations and assessing the effectiveness of stock enhancement are critical [8,9,10,11,12]. The analysis data allow study of the genetic diversity of fish fry and tracing of fish origins in the absence of hatchery information [11,12]. In this study, we used microsatellite DNA markers to distinguish cultured black sea bream populations from wild ones and to understand the genetic effects of stock enhancement on these wild populations.
2. Materials and Methods
2.1. Sample Preparation
In total, 1231 black sea bream specimens (458 from the hatcheries and 773 from the wild) were obtained from 2015 to 2017 (Table 1). Species were identified by following the method of Hsu et al. [25]. Fresh specimens—at least 20 individuals for each batch, including broodstock, juveniles, and subadults/adults—were sampled from three types of hatchery source: (1) a private hatchery for the TFSDA release project and without genetic information (KS_C1, KS_C2, and KS_C3); (2) an unknown hatchery for private (religious) release (PR_C1 and PR_C2); (3) aquaculture farms from offshore islands of Taiwan (KM_C and MT_C) and southern China (XM_C); and (4) an aquaculture farm from northern China (QD_C). Ten batches from eight field locations: (1) Miaoli County, site of the TFSDA release project during 2013–2015 (ML_W1, ML_W2, and ML_W3); (2) Yunlin County, Penghu County, Tainan City, and Taipei City, with a hatchery fish release history during 2004–2015 (YL_W, PH_W, TN_W, TP_W, and KM_W); (3) Chiayi County, with no hatchery fish release history during 2004–2015 (CY_W); and (4) Nagasaki Prefecture, Japan, used for comparison (JP_W; Table 1; Figure 1). The geographical locations of these populations, sampling locations with abbreviated population names, and sample size for each population are presented in Figure 1 and Figure 2 and Table 1. Small muscle tissue pieces (approximately 3–5 mm) were prepared from fresh (2% alcohol used for anesthesia) or frozen fish specimens, transported to our laboratory for molecular study, and preserved in 95% ethanol. The standard proteinase K/phenol method modified from an animal DNA extraction protocol was used. Moreover, DNA template quality was assessed through 0.8% agarose gel electrophoresis.

Table 1.
Summary of sample information. n = number of fish.
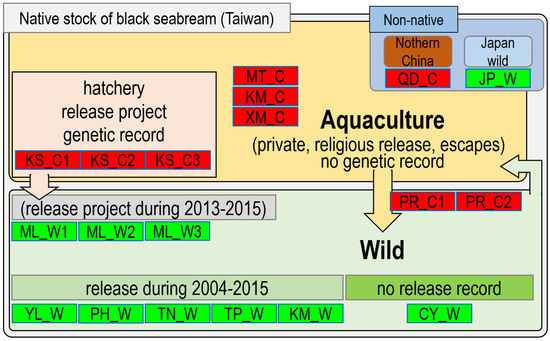
Figure 1.
Sampling, hatchery, and stock enhancement information for black sea bream.
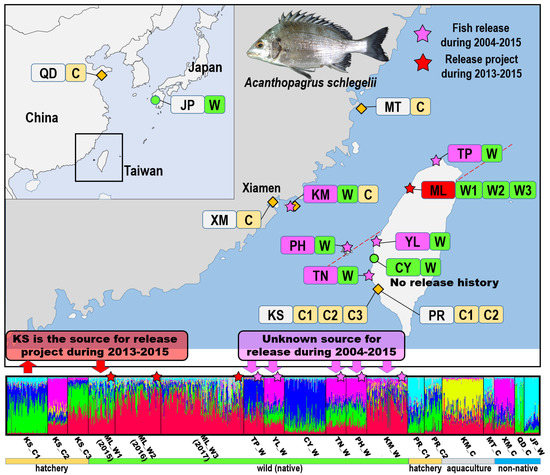
Figure 2.
Sampling locations and structure analysis of 19 black sea bream populations. KS: Kaohsiung City, private hatchery which provides juveniles for official release; PR: Kaohsiung City, from unknown hatchery for private release; TP: Taipei city; ML: Miaoli County; YL: Yunlin County; CY: Chiayi County; PH: Penghu County; TN: Tainan City; KM: Kinmen County; MT: Matsu islands; XM: Xiamen city; QD: Qingdao city; and JP: Nagasaki Prefecture. The estimated population structure based on the highest probability structure run at K = 8. Each individual is represented by a thin vertical line, which is partitioned into K colored segments that represent individuals’ estimated likelihood of membership in each of the K clusters.
2.2. SSR Markers
In accordance with Hsu et al. [26], nine microsatellite markers for black sea bream were selected and validated through multiplex polymerase chain reaction (PCR) to determine those ideal for genetic analysis (Table 2). PCR amplification was performed in 20 µL reaction volumes containing 5–10 ng of template DNA, 1× PCR buffer (10 mM Tris and 50 mM KCl, pH 9.0), 200 μM of each dNTP, 1.5 mM MgCl2, 0.5 U of Taq polymerase (Promega, Madison, WI, USA), and 4 pmol of each primer. Thereafter, PCR cycling was performed in an Autorisierter Thermocycler (Eppendorf, Hamburg, Germany) with initial denaturation at 95 °C for 2 min, which was followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at locus-specific temperatures for 30 s, extension at 72 °C for 30 s, and a final extension at 72 °C for 10 min. The fragments in PCR products were analyzed using an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA), with the output analyzed using GeneMapper software (version 4.0, Applied Biosystems, Foster City, CA, USA).

Table 2.
Nine selected microsatellite loci of black sea bream.
2.3. Population Genetic Analysis
The observed genetic diversity (Ho), expected genetic diversity (He), and FIS were calculated using Genalex 6.41 [32]. The chi-square Hardy–Weinberg equilibrium (HWE) test results, pairwise FST values, and associated p values were also determined using Genalex 6.41 [32]. To determine FST significance levels, multilocus genotypes were randomized between sample pairs (9999 permutations), and the significance after Bonferroni correction was calculated [33,34]. Genotyping errors and null alleles were estimated using Microchecker 2.2.3 [35]. To elucidate a population’s genetic structure on the basis of multilocus genotypes, an admixture model with correlated allele frequencies was developed using Structure version 2.3.4 [36,37]. Five independent runs were performed for the entire data set for K values (numbers of groups) ranging from 1 to 9. All runs were based on 1,000,000 iterations of burn-in, followed by an additional 5,000,000 iterations. The best estimation of the K value was obtained using Structure Harvester [38]. Summation and graphical representation of the structure analysis results were performed using Clumpak [39] and Structure Plot version 2.0 [40], respectively. The population structure estimation was based on the highest-probability structure obtained at K = 8. Hierarchical analysis of molecular variance (hierarchical AMOVA) was performed to partition the total genetic variance within and between regions, as described by Excoffier et al. [41]. Arlequin was used to perform AMOVA. The most-supported grouping can be automatically detected using SAMOVA (based on Arlequin 3.5) [42,43]. We calculated pairwise relatedness (unrelated, half-sibling, full-sibling, or parent–offspring) between each pair of individuals by using RELATED [44] and Wang’s [45] estimator. The expected relatedness values (r) were 0.5 between full-sibling pairs and parent–offspring pairs, 0.25 between half-sibling pairs, and 0 between unrelated individuals [44]. We prepared a kinship network by using a pairwise relatedness (r) of >0.4, which indicates related individuals [46]. Parentage assignment between the 94 broodstock (KS_C1) and 192 wild (recaptured) samples (ML_W3) was estimated using Cervus 3.0.7 [47]. For accuracy, parentage was assigned by employing a minimum of nine loci, 0.9; proportion of loci typed, 0.01; proportion of loci mistyped, 0.01; error rate in likelihood calculations, 0.01; and simulation of 100,000 offspring at an 80% confidence interval applying the LOD confidence determined.
3. Results
3.1. Genetic Diversity within Populations
Across the nine microsatellite markers, all the populations were successfully genotyped. No monomorphic loci were found among the 19 populations. In total, 170 alleles were detected from the samples (n = 1231). In all individuals, the marker AS324 exhibited the highest number of alleles per locus (31 alleles), whereas the marker CL011 exhibited the lowest number (12 alleles; Table 3). Fewer alleles were found in the hatchery populations (n = 388; 139 alleles) than in the wild populations (n = 739; 148 alleles); however, the allele frequency pattern was similar (Figure 3).

Table 3.
Number of alleles (Na) and effective alleles (Ne) at nine microsatellite loci in 19 black sea bream populations.
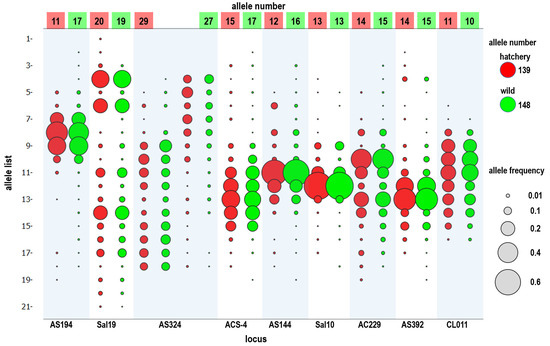
Figure 3.
Allele frequency of nine microsatellite loci in hatchery (n = 388) and wild (n = 739) black sea bream populations in Taiwan. Hatchery: KS_C1, KS_C2, KS_C3, PR_C1, PR_C2, KM_C, and MT_C; Wild: ML_W1, ML_W2, ML_W3, YL_W, PH_W, TN_W, TP_W, KM_W, and CY_W.
The marker AS324 in PH_W exhibited the highest number of alleles per locus and highest allele richness (21 alleles and 13.921, respectively), whereas the marker SaI10 in KM_C exhibited the lowest (4 alleles and 1.387, respectively; Table 4 and Table 5). The average heterozygosity (Ho) over all loci was between 0.565 and 0.725 among the 19 populations. CY_W exhibited the highest expected heterozygosity for all the loci, whereas XM_C exhibited the lowest (Table 4 and Table 5). The mean estimates of expected heterozygosity (He) over all loci were between 0.683 and 0.742 among the 19 populations. PR_C2 exhibited the highest expected heterozygosity for all the loci and XM_C and KS_C3 the lowest (Table 4 and Table 5). Of the 171 population–locus combinations, 67 displayed deviations from the HWE significant at the p < 0.001 level (Table 4 and Table 5), with no strong trends of deviation observed for specific loci (between 5 and 10). No possible genotyping errors were noted in any loci, and null alleles were found to be possible for only one locus (AC229) in KS_C1 (Table 4 and Table 5). The average FIS was between −0.051 and 0.164 among the 19 populations. The lowest FIS was found in TP_W and the highest in XM_C (Table 4 and Table 5).

Table 4.
Summary statistics for genetic variation at nine microsatellite loci in nine cultured black sea bream populations.

Table 5.
Summary statistics for genetic variation at nine microsatellite loci in 10 wild black sea bream populations.
3.2. Genetic Differentiation among Populations
Pairwise comparisons between sampling locations were performed. The pairwise FST values ranged from 0 (KS_C1–ML_W1) to 0.056 (XM_C–JP_W), and most of them were significant (p = 0), suggesting small but significant genetic differentiation (Table 6). Most comparisons among the 19 populations indicated low genetic differentiation (below 0.04), but the results of comparisons of other locations with QD_C and JP_W were high (QD_C–others: 0.012–0.052; JP_W–others: 0.018–0.056) (Table 6). Pairwise comparisons among Taiwan hatchery populations obtained values ranging from 0.005 to 0.037 (average 0.023); among Taiwan wild populations, 0.005 to 0.034 (average 0.017); and among Taiwan hatchery–wild populations, 0 to 0.039 (average 0.020; Table 6).

Table 6.
Pairwise FST values (below the diagonal) and associated p values (above the diagonal) among 19 black sea bream populations collected from Taiwanese hatcheries and the wild.
3.3. Genetic Structure of Populations
An initial AMOVA indicated low differentiation (FST = 0.022) with 2.17% genetic variation distributed among the 19 populations (Table 7). However, the overall FST differentiation was significant among the populations, based on 999 permutations (p = 0 < 0.001; Table 7). Even if JP_W and QD_C were removed, the overall FST was still significant among the remaining populations (p = 0 < 0.001; Table 7). Lower genetic differentiation and higher population connectivity were noted across the wild sampling sites (average FST = 0.015; gene flow (Nm) = 15.929), whereas higher genetic differentiation and lower population connectivity were noted across the hatchery sampling sites (average FST = 0.022; Nm = 10.895; Table 7).

Table 7.
Analysis of molecular variance (AMOVA) among different black sea bream groups.
A hierarchical AMOVA (SAMOVA) with the highest ΦCT was performed, and all groupings were significantly supported by permutations (0.05 ≥ p ≥ 0.01; Table 8). In the 19 populations analyzed, JP_W, QD_C, and others were separated (K = 3, ΦCT = 0.021, p = 0.009, variance 2.09). In the 17 populations (i.e., those other than JP_W and QD_C), the PH_W–TN_W and KS_C2–YL_W pairs were grouped (K = 15, ΦCT = 0.015, p = 0.001, variance 1.47). In the nine wild populations, when K = 6, the following was determined to be the optimal grouping by using the SAMOVA program [(ML_W2; ML_W3; KM_W); (PH_W; TN_W); (TP_W); (CY_W); and (ML_W1); (YL_W)]; these groups exhibited the highest intergroup variance (1.20%; Table 8). In the eight hatchery populations, when K = 7, [(KS_C); (PR_C1; PR_C2); (MT_C); (KS_C2); (XM_C); (KS_C3); and (KM_C)] was determined to have the highest ΦCT and variance (0.017 and 1.65%, respectively; Table 8).

Table 8.
Hierarchical AMOVA of 19 black sea bream populations collected from hatcheries and the wild, with analysis performed using SAMOVA.
In the STRUCTURE analysis, the best estimation of the K value (number of groups) was eight, and this corresponded to a stable representation (data not shown; Figure 2). No distinct clade pattern was noted across all populations, but the patterns for CY_W (blue), QD_C (green), and JP_W (light blue) were more clear. Despite KS (KS_C1, KS_C2, and KS_C3) and ML (ML_W1, ML_W2, and ML_W3) being from the same sampling location with three consecutive years (2015–2017), no distinct clade pattern was observed. There is a clear genetic structure change among KS and ML populations (Figure 2).
In the kinship network analysis based on pairwise relatedness within the 19 populations, some individuals displayed close kinship in the hatchery (KM_C, KS_C1, and KS_C3) and wild populations (CY_W, ML_W2, and ML_W3; Figure 4). However, the kinship network had a relaxed structure in seven wild populations and a concentrated structure in nine hatchery populations, indicating more inbreeding in the hatcheries (Figure 5). Parentage analysis showed that 12–49 individuals (3%–13%; 95%–80% confidence) in ML_W3 were possibly related to individuals in KS_C1 (at least a single parent; Table 9).
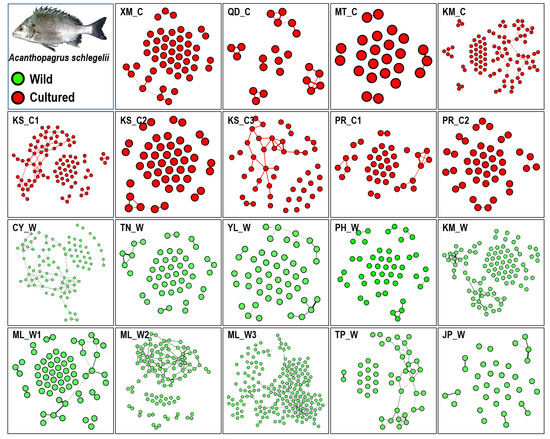
Figure 4.
Kinship network based on pairwise relatedness within 19 populations. Wild populations: ML_W1, ML_W2, ML_W3, YL_W, PH_W, TN_W, TP_W, KM_W, CY_W, and JP_W; Cultured populations: KS_C1, KS_C2, KS_C3, PR_C1, PR_C2, KM_C, MT_C, XM_C, and QD_C. Pairwise relatedness® > 0.4.
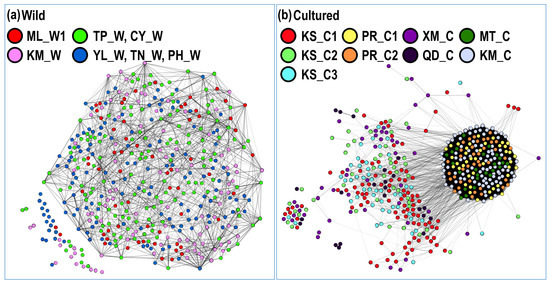
Figure 5.
Kinship network based on pairwise relatedness: (a) seven wild populations and (b) nine cultured populations. Wild populations: ML_W1, YL_W, PH_W, TN_W, TP_W, KM_W, and CY_W; Cultured populations: KS_C1, KS_C2, KS_C3, PR_C1, PR_C2, KM_C, and MT_C. Pairwise relatedne®(r) > 0.4.

Table 9.
Parentage analysis of black sea bream collected from hatcheries (KS_C1) and the wild (ML_W3), with analysis performed using Cervus.
4. Discussion
4.1. Genetic Difference among/between Hatchery and Wild Populations
Black sea bream, A. schlegelii, is a crucial aquaculture species in East Asia, from Taiwan, China, and Korea to Japan. Black sea bream aquaculture began in the 1980s, and Taiwanese hatcheries are located in areas near Kaohsiung. Broodstocks are obtained from the main fishery areas, which are off the Penghu Islands and the west coast of Taiwan (Yunlin–Chiayi–Tainan–Kaohsiung). Black sea bream has been cultured for more than 30 years in Taiwan. This is the first study analyzing the genetic diversity of cultured and wild black sea bream populations in Taiwan coastal waters. Nine hatchery populations were collected, including one in northern China (as an outgroup population), QD_C (Qingdao City); one in southern China, XM_C (Xiamen city): two from Taiwan’s offshore islands, KM_C (Kinmen) and MT_C (Matsu); and five from southern Taiwan, PR_C1, PR_C2, KS_C1, KS_C2, and KS_C3 (Kaohsiung City). According to the allele number (Na), observed heterozygosity (Ho), and expected heterozygosity (He) of nine microsatellite loci, the genetic diversity of the cultured populations was slightly lower than that of the wild populations, except for some such as KS_C1 (Table 4 and Table 5). The allele frequency pattern was similar between the wild and hatchery populations (Figure 3). Genetic differences among the hatchery populations were generally larger than those among the wild populations; however, high gene flow still existed among the hatchery populations (Nm = 10.895; Table 6 and Table 7). This indicates maintenance of a high degree of genetic diversity among cultured black sea bream, and this can avoid inbreeding effects. The hatcheries used to produce juveniles for the release project during 2013–2015 (KS_C1, KS_C2, and KS_C3) imported new stock from an unknown source. Although significant changes were observed in the genetic structure, fish larvae from the unknown hatchery (PR_C1, PC_C2, and KS_C1) exhibited small genetic differences. Therefore, hatchery information is not always reliable, especially because fish larvae may come from several hatcheries simultaneously, or broodstock may change after a natural disaster. As expected, smaller genetic differences were observed among hatcheries in Taiwan than among hatcheries in Taiwan’s offshore islands, southern China, and northern China. Generally, inbreeding effects easily arise in hatchery populations due to their small effective population size, which clearly means that hatchery larvae released into the wild could reduce the genetic diversity of wild populations. For example, in one of the world’s largest marine stock early programs involving the red sea bream (Pagrus major) in Kagoshima Bay, Japan, the released hatchery fish clearly reduced the genetic diversity of the wild population [48]. Due to the application of a special type of stock enhancement in Taiwan (fish from private farms not from official institutes), more hatcheries (stock) could contribute to the genetic diversity, thus preventing inbreeding.
When hatchery fish are cultured, high gene flow among hatchery populations and between wild populations cannot usually be maintained. As broodstocks are not changed each year, no random mating occurs. After several generations, hatchery populations tend to show different genetic structures to wild populations [12]. Our study of the silver sea bream (Rhabdosargus sarba) presented two distinct clusters (hatchery and wild population clusters) [12]. As silver sea bream and black sea bream are both Sparidae fish and have a similar culture-related history in Taiwan, they are generally considered to have the same genetic structure. Unexpectedly, unlike silver sea bream, wild and hatchery black sea bream cannot be clearly separated into two clusters. The genetic structure of black sea bream did not have the same pattern among and between hatchery and wild populations. This may have had several causes; one is that the hatcheries of silver sea bream are few, located only in southern Taiwan, and maintain high communication (gene flow among silver sea bream hatcheries is high at 32.677). However, there are more hatcheries of black sea bream and they are located in different areas (i.e., southern Taiwan, offshore islands, and even China). Moreover, low but effective communication is maintained among and between populations in black sea bream hatcheries (Nm = 10.895). Second, black sea bream is more abundant and widely distributed in Taiwan’s coastal waters than silver sea bream. The stock population was initially established independently and from different areas and may have helped to maintain genetic diversity.
4.2. Dramatic Change in Genetic Structure after Fish Release
We collected 10 wild populations from the following: Japan (as an outgroup population)—JP_W (Nagasaki City); Taiwan’s offshore islands—KM_W (Kinmen) and PH_W (Penghu); northern Taiwan—ML_W1, ML_W2, ML_W3 (Miaoli City), and TP_W (Taipei City); and southern Taiwan—YL_W (Yunlin City), CY_W (Chiayi City), and TN_W (Tainan City). The fish from Taiwan’s offshore islands, northern Taiwan, and southern Taiwan were expected to have a clear genetic structure such as one population (e.g., silver sea bream) [12] or two populations (northern Taiwan vs. Penghu–southern Taiwan; e.g., rabbitfish, Siganus fuscescens) [49]. However, no clear pattern of genetic structure was observed among wild populations and the three wild populations (namely ML_W2, PH_W, and CY_W) that showed higher pairwise FST. Notably, the hatchery populations KS_C1, PR_C1, and PR_C2 exhibited a lower pairwise FST with other wild populations than with ML_W2, PH_W, and CY_W. This indicated that the hatchery populations KS_C1, PR_C1, and PR_C2 had high gene flow with the wild populations, and changes in their genetic structure were mainly caused by fish release.
Stock enhancement has been found to induce genetic structure changes in fishes such as brown trout and red sea bream [48,50]. When the genetic differences between hatchery and wild populations are considerable, the genetic structure changes greatly with a greater number of releases and longer release duration. By contrast, when the genetic difference (FST) between KS_C1 and ML_W1 is 0, the effect on the genetic structure should be minor. However, according to the present STRUCTURE analysis, KS_C1 was different from ML_W1, and stock enhancement led to evident genetic changes over three consecutive years (ML_W1, ML_W2, and ML_W3; Figure 2). In addition, the genetic structure of sea bream from the sampling sites was inconsistent with their geographical distribution along the western coast of Taiwan. CY_W is geographically close to YL_W, TN_W, and PH_W, but the genetic structures of fish from these populations were found to be different (Figure 2). For YL_W, TN_W, and PH_W, several records of the official release from 2004 to 2015 were available. CY_W, YL_W, and TN_W were probably affected by stock enhancement and therefore exhibited no clear clusters with relative complexity in STRUCTURE analysis. For sampling sites ML_W, TP_W, and KM_W, which belonged to the northern region, several official releases were noted during 2004–2015. Long geographical distance and stock enhancement may have led to the lack of consistent and clear clusters in the STRUCTURE analysis. In addition to the large number of juveniles released over the past decade, escapes were another potential source of continued gene flow. Earth pond farming aquaculture is mainly performed in the coastal waters of southern Taiwan, and escapes happen after typhoons. Escaped farmed fish may affect natural populations and the broodstock (Holmer). One- to two-year-old silver sea bream can mature and undergo protandrous (male-to-female) sex changes later [23]. Escaped farmed fish are relatively big at 1–2 years of age and have a higher survival rate than juveniles used for stock enhancement. However, those fish were all traced to hatchery-reared stock (Figure 1). In general, fish were released over the entire western coast and outer islands of Taiwan from 2004 to 2015, resulting in complex genetic structures of wild populations that are inconsistent with their geographic distribution. Among them, no release history was found for only CY_W, which presented a simple cluster exhibiting few or no effects of stock enhancement (Figure 2).
4.3. Stock Enhancement of Black Sea Bream in Taiwan
Through a literature review, Araki and Schmid [51] summarized 50 years of data about the effects of hatcheries on fish and stock enhancement. They reported a clear reduction in genetic variation in hatchery populations. However, this result is completely different from the result of stock enhancement of black sea bream in Taiwan. In this study, we investigated 19 hatchery and wild populations and found that the frequency distribution of microsatellites in the hatchery and wild populations was similar, and the allele number remained at a high level (Figure 3). Additionally, we used pairwise relatedness to prepare a kinship network for evaluating the genetic relationship among each population, all hatchery populations, and all wild populations. We considered individuals with a relatedness (r) of >0.4 as related, almost excluding unrelated individuals. Weng et al. [52] used 11 microsatellites for parentage analysis in giant grouper (Epinephelus lanceolatus), and their relatedness value (r) of >0.25 accurately excluded unrelated individuals. In each population, we could not find obvious inbreeding groups, and further guaranteed stock enhancement should not reduce the diversity of the wild population (Figure 4). However, the kinship network indicated that the relationships among hatchery individuals were closer than those among wild individuals (Figure 5). Pairwise relatedness must be introduced to monitor stock enhancement programs and avoid unexpected inbreeding in hatcheries and large-scale programs.
Stock enhancement of black sea bream in Hiroshima Bay, Japan, is a successful example [53]. Gonzalez et al. [54] estimated that hatchery black sea bream contributed 12.5% and 13.5% to the wild population, and even as high as 58.9% in Jeong et al. [30]. In case of stock enhancement in Daya Bay, China, the contribution rate was low (approximately 1.18%), as assessed by Wang et al. [55]. Thus, stock enhancement varies widely depending on the location and method used for assessing it. In this study, the contribution rate of stock enhancement in ML_W was between 3% and 13% (Table 9). Private release (religious release) in Taiwan, another major contributor of stock, is estimated to have contributed approximately one-third of the total number of released fish. On evaluating two batches of religious release, Lee et al. [56] found that hatchery fish contributed 61% to the wild population. Regardless of the contribution rate, this study found that frequent large-scale release (official and private) in Taiwan has significantly changed the genetic structure of wild populations. Chiayi (CY_W), the main oyster production area, is a location in which no release has been recorded. Considering the influence of the oyster industry, no official stock enhancement is performed in this area, which enables investigation of the genetic structure of an area with no or few stock enhancements in Taiwan. As oyster farms are also a crucial reproductive base for black sea bream [57], the stock enhancement of black sea bream in Hiroshima Bay may have negatively affected oysters and other fishes [51,58]. Thus, not only the survival and contribution rates of released fish, but also the impacts of the release on the environment and ecology should be determined [7,59].
5. Conclusions
In Taiwan, official stock enhancement and private religious release of black sea bream are conducted frequently and on a large scale. Such diverse and unpredictable fish larvae prevent the decline of overall diversity. Although determining the short-term effect of stock enhancement in Taiwan is difficult, the contribution of stock enhancement to wild populations is evidenced by changes in the genetic structure and the inconsistency of such structure.
Author Contributions
Conceptualization, T.-H.H., H.-T.L. and C.-W.H.; methodology, T.-H.H.; software, T.-H.H.; validation, T.-H.H., H.-T.L., H.-J.L., C.-H.L., H.-Y.G. and C.-W.H.; formal analysis, T.-H.H., H.-T.L. and C.-W.H.; investigation, T.-H.H. and H.-T.L.; resources, T.-H.H., H.-T.L., H.-J.L., C.-H.L., H.-Y.G. and C.-W.H.; data curation, T.-H.H.; writing—original draft preparation, T.-H.H. writing—review and editing, T.-H.H., H.-T.L. and C.-W.H.; visualization, T.-H.H.; supervision, C.-W.H.; project administration, T.-H.H., C.-H.L. and C.-W.H.; and funding acquisition, T.-H.H., C.-H.L. and C.-W.H.; All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Center of Excellence for the Oceans (National Taiwan Ocean University), which were financially supported by the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education, ROC (Taiwan). This work was also supported by the Fisheries Agency, Council of Agriculture, Executive Yuan, ROC (Taiwan) and the Taiwan Ocean Conservation and Fisheries Sustainability Foundation (109toffrest001, 110toffrest001, 111toffrest001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful to Jou-Yun Chen for technical support.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Cressey, D. Aquaculture: Future Fish. Nature 2009, 458, 398–400. [Google Scholar] [CrossRef] [PubMed]
- Sumaila, U.R.; Tai, T.C. End Overfishing and Increase the Resilience of the Ocean to Climate Change. Front. Mar. Sci. 2020, 7, 523. [Google Scholar] [CrossRef]
- Schiermeier, Q. How Many More Fish in the Sea? Nature 2002, 419, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Pauly, D. Beyond Duplicity and Ignorance in Global Fisheries. Sci. Mar. 2009, 73, 215–224. [Google Scholar] [CrossRef]
- Bell, J.D.; Leber, K.M.; Blankenship, H.L.; Loneragan, N.R.; Masuda, R. A New Era for Restocking, Stock Enhancement and Sea Ranching of Coastal Fisheries Resources. Rev. Fish. Sci. 2008, 16, 1–9. [Google Scholar] [CrossRef]
- Liao, I.C.; Su, M.S.; Leano, E.M. Status of Research in Stock Enhancement and Sea Ranching. Rev. Fish Biol. Fish. 2003, 13, 151–163. [Google Scholar] [CrossRef]
- Kitada, S. Lessons from Japan Marine Stock Enhancement and Sea Ranching Programmes over 100 Years. Rev. Aquacult. 2020, 12, 1944–1969. [Google Scholar] [CrossRef]
- Liao, I.C. Status, Problems and Prospects of Stock Enhancement in Taiwan. Hydrobiologia 1997, 352, 167–180. [Google Scholar] [CrossRef]
- Adan, R.I.Y. Stock Enhancement in Japan and Taiwan. SEAFDEC Asian Aquac. 2001, 23, 20–21, 40. [Google Scholar]
- Chang, W.-C.; Lee, Y.-C.; Shih, C.-H.; Chu, T.-J.; Chang, P.-H. Population Size and Stocking Contribution Rates for Marked and Recaptured Black Porgy Acanthopagrus Schlegelli, in Northwestern Taiwan, 2005–2008. Fish. Res. 2011, 109, 252–256. [Google Scholar] [CrossRef]
- Hsu, T.-H.; Huang, C.-W.; Lin, C.-H.; Lee, H.-T.; Pan, C.-Y. Tracing the Origin of Fish without Hatchery Information: Genetic Management of Stock Enhancement for Mangrove Red Snapper (Lutjanus Argentimaculatus) in Taiwan. Fish. Aquat. Sci. 2020, 23, 13. [Google Scholar] [CrossRef]
- Hsu, T.-H.; Huang, C.-W.; Lee, H.-T.; Kuo, Y.-H.; Liu, K.-M.; Lin, C.-H.; Gong, H.-Y. Population Genetic Analysis for Stock Enhancement of Silver Sea Bream (Rhabdosargus Sarba) in Taiwan. Fishes 2020, 5, 19. [Google Scholar] [CrossRef]
- Ward, R.D. The importance of identifying spatial population structure in restocking and stock enhancement programmes. Fish. Res. 2006, 80, 9–18. [Google Scholar] [CrossRef]
- Satake, A.; Araki, H. Stocking of captive-bred fish can cause long-term population decline and gene pool replacement: Predictions from a population dynamics model incorporating density-dependent mortality. Theor. Ecol. 2012, 5, 283–296. [Google Scholar] [CrossRef]
- Baskett, M.L.; Burgess, S.C.; Waples, R.S. Assessing strategies to minimize unintended fitness consequences of aquaculture on wild populations. Evol. Appl. 2013, 6, 1090–1108. [Google Scholar] [CrossRef]
- Milot, E.; Perrier, C.; Papillon, L.; Dodson, J.J.; Bernatchez, L. Reduced fitness of Atlantic salmon released in the wild after one generation of captive breeding. Evol. Appl. 2013, 6, 472–485. [Google Scholar] [CrossRef]
- Naish, K.A.; Seamons, T.R.; Dauer, M.B.; Hauser, L.; Quinn, T.P. Relationship between effective population size, inbreeding and adult fitness-related traits in a steelhead (Oncorhynchus mykiss) population released in the wild. Mol. Ecol. 2013, 22, 1295–1309. [Google Scholar] [CrossRef]
- Boscari, E.; Congiu, L. The need for genetic support in restocking activities and ex situ conservation programmes: The case of the Adriatic sturgeon (Acipenser naccarii Bonaparte, 1836) in the Ticino River Park. J. Appl. Ichthyol. 2014, 30, 1416–1422. [Google Scholar] [CrossRef]
- An, H.S.; Nam, M.M.; Myeong, J.I.; An, C.M. Genetic diversity and differentiation of the Korean starry flounder (Platichthys stellatus) between and within cultured stocks and wild populations inferred from microsatellite DNA analysis. Mol. Biol. Rep. 2014, 41, 7281–7292. [Google Scholar] [CrossRef]
- Blanco Gonzalez, E.; Umino, T. Fine-scale genetic structure derived from stocking black sea bream, Acanthopagrus schlegelii (Bleeker, 1854), in Hiroshima Bay, Japan. J. Appl. Ichthyol. 2009, 25, 407–410. [Google Scholar] [CrossRef]
- Le Vay, L.; Carvalho, G.R.; Quinitio, E.T.; Lebata, J.H.; Ut, V.N.; Fushimi, H. Quality of hatchery-reared juveniles for marine fisheries stock enhancement. Aquaculture 2007, 268, 169–180. [Google Scholar] [CrossRef]
- Lorenzen, K.; Leber, K.M.; Blankenship, H.L. Responsible approach to marine stock enhancement: An update. Rev. Fish. Sci. 2010, 18, 189–210. [Google Scholar] [CrossRef]
- Law, C.S.W.; Sadovy de Mitcheson, Y. Age and growth of black seabream Acanthopagrus schlegelii (Sparidae) in Hong Kong and adjacent waters of the northern South China Sea. J. Fish Biol. 2018, 93, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Acanthopagrus schlegelii in Fisheries Statistics Yearbook of Taiwan Area. Available online: https://fishdb.sinica.edu.tw/chi/yearrpt2.php?id=26 (accessed on 10 January 2022).
- Hsu, T.-H.; Madrid, A.G.G.; Burridge, C.P.; Cheng, H.-Y.; Gwo, J.-C. Resolution of the Acanthopagrus black seabream complex based on mitochondrial and amplified fragment-length polymorphism analyses. J. Fish Biol. 2011, 79, 1182–1192. [Google Scholar] [CrossRef]
- Hsu, T.-H.; Lee, H.-T.; Chen, J.-Y.; Gong, H.-Y.; Huang, C.-W. Development of Microsatellite Multiplex PCR Assays for the Black Seabream (Acanthopagrus Schlegelii). J. Fish. Soc. Taiwan 2018, 45, 65–75. [Google Scholar] [CrossRef]
- Kim, W.J.; Kim, K.K.; Kim, Y.K.; Shin, E.H.; Kim, S.G. Isolation and characterization of 20 polymorphic microsatellite loci in the black seabream, Acanthopagrus schlegeli. Mol. Ecol. Resour. 2010, 10, 404–408. [Google Scholar] [CrossRef]
- Reid, K.; Hoareau, T.B.; Bloomer, P. High-throughput microsatellite marker development in two sparid species and verification of their transferability in the family Sparidae. Mol. Ecol. Resour. 2012, 12, 740–752. [Google Scholar] [CrossRef]
- Liu, Y.-G.; Liu, L.-X.; Wu, Z.-X.; Lin, H.; Li, B.-F.; Sun, X.-Q. Isolation and characterization of polymorphic microsatellite loci in black sea bream (Acanthopagrus schlegeli) by cross-species amplification with six species of the Sparidae family. Aquat. Living Resour. 2007, 20, 257–262. [Google Scholar] [CrossRef]
- Jeong, D.-S.; Gonzalez, E.B.; Morishima, K.; Arai, K.; Umino, T. Parentage assignment of stocked black sea bream Acanthopagrus schlegelii in Hiroshima Bay using microsatellite DNA markers. Fish. Sci. 2007, 73, 823–830. [Google Scholar] [CrossRef][Green Version]
- Yang, Y.; Cao, X.; Feng, R.; Chen, S.; Zhang, Z.; Ren, W.; Xu, S. Isolation and characterization of thirteen polymorphic microsatellite loci from black porgy (Acanthopagrus schlegeli). J. Genet. 2014, 93, e97–e99. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.R. Analysing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Borrell, Y.J.; Pinera, J.A.; Sanchez Prado, J.A.; Blanco, G. Mitochondrial DNA and microsatellite genetic differentiation in the European anchovy Engraulis encrasicolus L. ICES J. Mar. Sci. 2012, 69, 1357–1371. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Ramasamy, R.K.; Ramasamy, S.; Bindroo, B.B.; Naik, V.G. STRUCTURE PLOT: A program for drawing elegant STRUCTURE bar plots in user friendly interface. SpringerPlus 2014, 3, 431. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Dupanloup, I.; Schneider, S.; Excoffier, L.A. Simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 2002, 11, 2571–2581. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Pew, J.; Muir, P.H.; Wang, J.; Frasier, T.R. Related: An R Package for Analysing Pairwise Relatedness from Codominant Molecular Markers. Mol. Ecol. Resour. 2015, 15, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. An Estimator for Pairwise Relatedness Using Molecular Markers. Genetics 2002, 160, 1203–1215. [Google Scholar] [CrossRef] [PubMed]
- Dyer, R.J.; Nason, J.D. Population Graphs: The Graph Theoretic Shape of Genetic Structure. Mol. Ecol. 2004, 13, 1713–1727. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising How the Computer Program Cervus Accommodates Genotyping Error Increases Success in Paternity Assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Kitada, S.; Nakajima, K.; Hamasaki, K.; Shishidou, H.; Waples, R.S.; Kishino, H. Rigorous Monitoring of a Large-Scale Marine Stock Enhancement Program Demonstrates the Need for Comprehensive Management of Fisheries and Nursery Habitat. Sci. Rep. 2019, 9, 5290. [Google Scholar] [CrossRef]
- Hsu, T.-H.; Gwo, J.-C. Fine-Scale Genetic Structure of Rabbitfish, Siganus Fuscescens, in Penghu Archipelago Following a Mass Mortality Event Caused by Extreme Cold Winter Weather. Genes Genom. 2017, 39, 645–652. [Google Scholar] [CrossRef]
- Hansen, M.M.; Fraser, D.J.; Meier, K.; Mensberg, K.-L.D. Sixty Years of Anthropogenic Pressure: A Spatio-Temporal Genetic Analysis of Brown Trout Populations Subject to Stocking and Population Declines. Mol. Ecol. 2009, 18, 2549–2562. [Google Scholar] [CrossRef]
- Araki, H.; Schmid, C. Is Hatchery Stocking a Help or Harm?: Evidence, Limitations and Future Directions in Ecological and Genetic Surveys. Aquaculture 2010, 308, S2–S11. [Google Scholar] [CrossRef]
- Weng, Z.; Yang, Y.; Wang, X.; Wu, L.; Hua, S.; Zhang, H.; Meng, Z. Parentage Analysis in Giant Grouper (Epinephelus lanceolatus) Using Microsatellite and SNP Markers from Genotyping-by-Sequencing Data. Genes 2021, 12, 1042. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, E.B.; Umino, T.; Nagasawa, K. Stock Enhancement Programme for Black Sea Bream, Acanthopagrus Schlegelii (Bleeker), in Hiroshima Bay, Japan: A Review. Aquac. Res. 2008, 39, 1307–1315. [Google Scholar] [CrossRef]
- Gonzalez, E.B.; Nagasawa, K.; Umino, T. Stock Enhancement Program for Black Sea Bream (Acanthopagrus schlegelii) in Hiroshima Bay: Monitoring the Genetic Effects. Aquaculture 2008, 276, 36–43. [Google Scholar] [CrossRef]
- Wang, X.; Weng, Z.; Yang, Y.; Hua, S.; Zhang, H.; Meng, Z. Genetic Evaluation of Black Sea Bream (Acanthopagrus schlegelii) Stock Enhancement in the South China Sea Based on Microsatellite DNA Markers. Fishes 2021, 6, 47. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Chang, P.-H.; Shih, C.-H.; Shiao, J.-C.; Tzeng, T.-D.; Chang, W.-C. The Impact of Religious Release Fish on Conservation. Glob. Ecol. Conserv. 2021, 27, e01556. [Google Scholar] [CrossRef]
- Kawai, K.; Fujita, H.; Sanchez, G.; Umino, T. Oyster Farms Are the Main Spawning Grounds of the Black Sea Bream Acanthopagrus Schlegelii in Hiroshima Bay, Japan. PeerJ 2021, 9, e11475. [Google Scholar] [CrossRef]
- Saito, H.; Nakanishi, Y.; Shigeta, T.; Umino, T.; Kawai, K.; Imabayashi, H. Effect of predation of fishes on oyster spats in Hiroshima Bay. Nippon. Suisan Gakkaishi 2008, 74, 809–815. [Google Scholar] [CrossRef]
- Laikre, L.; Schwartz, M.K.; Waples, R.S.; Ryman, N. Compromising Genetic Diversity in the Wild: Unmonitored Large-Scale Release of Plants and Animals. Trends Ecol. Evol. 2010, 25, 520–529. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).