Simple Summary
Isoprene, a volatile hydrocarbon, is the second most abundantly produced climate-active gas, with largely indirect detrimental impacts, such as extending the residence time of the greenhouse gas methane. Isoprene is mainly emitted by plants and can be consumed by a range of microbes inhabiting diverse environments, including soil. Here, the ability of soil bacteria to degrade isoprene was investigated. Soil samples were taken from beneath wild Himalayan cherry trees in a tropical restored forest area, and an Alcaligenes sp. (strain 13f) was isolated. This isolate used isoprene as a sole source of carbon and energy (32.6% of isoprene was consumed in 18 days). A surprising finding from the genome analysis of Alcaligenes sp. strain 13f was that the well-characterised genes and genetic organisation typical of other isoprene-degrading bacteria were not observed. Thus, we propose that this strain uses a different metabolic pathway for isoprene degradation.
Abstract
Isoprene is a climate-active biogenic volatile organic compound (BVOC), emitted into the atmosphere in abundance, mainly from terrestrial plants. Soil is an important sink for isoprene due to its consumption by microbes. In this study, we report the ability of a soil bacterium to degrade isoprene. Strain 13f was isolated from soil beneath wild Himalayan cherry trees in a tropical restored forest. Based on phylogenomic analysis and an Average Nucleotide Identity score of >95%, it most probably belongs to the species Alcaligenes faecalis. Isoprene degradation by Alcaligenes sp. strain 13f was measured by using gas chromatography. When isoprene was supplied as the sole carbon and energy source at the concentration of 7.2 × 105 ppbv and 7.2 × 106 ppbv, 32.6% and 19.6% of isoprene was consumed after 18 days, respectively. Genome analysis of Alcaligenes sp. strain 13f revealed that the genes that are typically found as part of the isoprene monooxygenase gene cluster in other isoprene-degrading bacteria were absent. This discovery suggests that there may be alternative pathways for isoprene metabolism.
1. Introduction
Isoprene (2-methyl-1,3-butadiene; C5H8) is one of the primary biogenic volatile organic compounds [1]. It is emitted into the atmosphere by many organisms, especially plants [2]. It is estimated that global emissions of isoprene are in the range of 400 to 600 Tg per year, comparable in scale to methane [3,4]. Isoprene plays an important role in the Earth’s climate by reacting with free radicals in the atmosphere to produce greenhouse gases or by extending the residence time of other gases such as methane, thereby resulting in increased global temperatures [5].
Previous studies have shown that freshwater, marine, and soil ecosystems, and the phyllosphere, all operate as sinks for isoprene [6,7,8,9,10,11,12]. For example, it has been estimated that soil bacteria can consume up to 20.4 Tg of isoprene per year [7,10]. Many isoprene-degrading bacteria have been isolated from diverse environments, such as Arthrobacter, Nocardia, Nocardioides, Rhodococcus, Gordonia, Bacillus, Ramlibacter, Variovorax, Klebsiella, Mycobacterium, Pseudomonas, and Alcaligenes [7,8,9,13,14,15,16,17,18,19]. Some of these isoprene-degrading bacteria have been genetically characterised, including Gram-negative bacteria, such as Variovorax, Ramlibacter, and Sphingopyxis [18,19], and Gram-positive bacteria, such as Rhodococcus, Nocardioides, Gordonia, and Mycobacterium [15,16,17,20]. Among these, Rhodococcus AD45, a freshwater sediment strain, is probably the best-characterised isoprene-degrading bacterium and has been used as a model for isoprene metabolism studies [16,20].
Among those isoprene-degrading bacteria that have been analysed, there is a high level of genetic conservation. All of the genomes of isoprene degraders that have been sequenced harbour a gene cluster (iso cluster) that encodes the enzymes required for isoprene degradation. Some genes in the cluster (isoABCDEF) encode a multi-subunit isoprene monooxygenase that catalyses the oxidation of isoprene to epoxyisoprene, whereas others (isoGHIJ) encode a glutathione transferase and enzymes involved in subsequent stages of isoprene oxidation [16,21,22]. This genetic characterisation has led to the development of molecular methods to determine isoprene-degrading capability through PCR amplification of the isoA gene, the gene encoding the putative active site of isoprene monooxygenase which catalyses the first step in isoprene degradation. In all isoprene-degrading bacterial strains that were genetically examined, this gene has been found, thus making it a suitable molecular target to investigate the diversity and abundance of isoprene degraders [14,15,23]. However, since the methods were designed based on a relatively limited number of isoprene-degrading bacterial genera [14,23], it is not known if isoA is found in all isoprene degraders, or whether there are alternative mechanisms of isoprene degradation.
One of the most environmentally significant bacterial genera that has been the least studied in relation to isoprene degradation is Alcaligenes. It is a Gram-negative bacterium that is ubiquitous in the natural environment, such as soil, fresh waters, marine environments, and industrial effluent [24,25,26,27]. This genus was first described in 1919 [28] and has had a wide range of applications in bioremediation because of its ability to degrade many pollutants such as phenols, DDT insecticide, and hydrocarbons in crude oil [27,29,30]. In relation to isoprene, Alcaligenes was first reported as an isoprene degrader in 1990 [31]. Later, in 2015, a species of Alcaligenes was isolated from contaminated soil taken from a waste rubber dumping site and was confirmed for its ability to utilise isoprene as the sole source of carbon and energy [32]. However, these reports on isoprene-degrading Alcaligenes species did not provide characterisation at the biochemical or molecular biological level.
In this study, an Alcaligenes strain was isolated from tropical restored forest soil in Doi Suthep-Pui National Park in the north of Thailand. The strain was associated with wild Himalayan cherry, one of the indigenous tree species that has been recommended as a framework species for forest restoration [33]. Many isoprene-degrading bacteria, including Ochrobactrum, Arthrobacter, Bacillus, Friedmanniella, Klebsiella, Isoptericola, and Cellulosimicrobium, had been found in this location [34]. Here, we investigated the metabolism of isoprene by this Alcaligenes strain through its genome characterisation.
2. Materials and Methods
2.1. Enrichment and Isolation of Isoprene-Degrading Bacteria
The isoprene-degrading bacterium Alcaligenes sp. strain 13f was isolated from topsoil (3 cm from the surface) beneath wild Himalayan cherry trees (Prunus cerasoides D. Don) in a restored forest area (18°51′51″ N 98°50′51″ E) located in Doi Suthep-Pui National Park, Chiang Mai, Thailand. A one-gram portion of the soil sample was suspended in 9 mL of minimal medium (composition described by Uttarotai et al. [34]) in a vial (125 cm3) and tightly closed with PTFE/silicone septum. Isoprene from Sigma-Aldrich (St Louis, MO, USA) (7.2 × 105 ppbv) was injected into the bottle containing the soil sample (isoprene for injection was prepared according to the method described by Acuña Alvarez et al. [13]). After incubation at 27 °C for 96 h, the enriched sample (1 mL) was drawn and subjected to serial dilution and spread-plating on minimal medium agar. The plates were incubated at 27 °C in a glass desiccator with the presence of isoprene (1 mL of isoprene was added to the 10.5 L desiccator). The representatives of colonies of the predominant bacteria grown on minimal medium agar were restreaked and incubated under the same condition until pure cultures were obtained.
2.2. Initial Identification of Isoprene-Degrading Bacteria by 16S rRNA Gene Sequencing
Genomic DNA was extracted by using a phenol-chloroform extraction method [35]. The bacterial universal primers (final concentration of 0.4 μM) 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-CGGTTACCTTGTTACGACTT-3′) [36], together with the AppTaq RedMix (Appleton, Birmingham, UK) reaction mixture, were used to amplify the 16S rRNA gene. The PCR was performed with an initial denaturation step at 95 °C for 3 min, 30 cycles of denaturation at 95 °C for 15 s, annealing at 55 °C for 15 s, extension at 72 °C for 30 s, and a 5-min final extension at 72 °C. The PCR products were purified by using a PCR purification kit (Sigma; GenElute, St Louis, MO, USA). Sanger sequencing was performed by Eurofins Genomics (Ebersberg, Germany). Chromas software (version 2.6.6; Technelysium, South Brisbane, Australia) was used to trim low-quality sequences and Bioedit (version 7.05.3) [37] was used to merge forward and reverse DNA sequences. The 16S rRNA sequence was subjected to BLASTn to identify the closest relatives. An isolate (13f), which was identified as Alcaligenes, was selected for further study. The sequence was deposited into the NCBI database (accession number MZ323998).
2.3. Test for Isoprene Degradation by Bacterial Isolates
The ability of Alcaligenes sp. strain 13f to degrade isoprene was investigated. Rhodococcus sp. strain bl28ba, a known isoprene degrader isolated by Murphy [38], was included as a positive control. A loopful of the bacterial culture, grown on minimal medium agar supplied with isoprene (as described in Section 2.1), was resuspended in 100 µL of minimal medium, and transferred to 9.9 mL of minimal medium without isoprene, minimal medium with isoprene, and glucose/yeast-extract broth [34], also with isoprene. The media were contained in 125 cm3 vials (as above). For each enrichment, isoprene was supplied at 7.2 × 105 ppbv and 7.2 × 106 ppbv (achieved by injecting 0.1 mL and 1 mL of headspace isoprene gas (prepared as described above) into the vials, respectively). Every three days, bacterial growth was determined by OD600, measured by using a Jenway 7300 spectrophotometer (Jenway, Stone, UK), and residual isoprene concentrations were measured by using a Unicam 610 gas chromatograph (Unicam, Lisbon, Portugal) with a flame ionisation detector (FID) and a 10% Apiezon L CW column, following the conditions described by Uttarotai et al. [34].
After 18 days of incubation, the cultures were diluted, spread on minimal medium agar, and incubated in a glass desiccator containing isoprene (as described in Section 2.1) for a further 18 days to confirm the presence of active bacteria and the purity of the bacterial cultures.
2.4. Amplification of the isoA Gene
PCR amplification of isoA was carried out by using two different protocols. The first protocol employed the isoA primers of El Khawand et al. [14] (5′-TGCATGGTCGARCAYATG-3′ and 5′-GRTCYTGYTCGAAGCACCACTT-3′), which yield an expected amplicon of 1015 bp. This protocol was used with a touchdown PCR, starting with an initial denaturation at 94 °C for 3 min, followed by 19 cycles of 94 °C for 30 s, 72 °C for 45 s with a reduction in temperatures of 1 °C per cycle until reaching 54 °C, 72 °C for 60 s, then 25 cycles of 94 °C for 30 s, 54 °C for 45 s, 72 °C for 60 s, and a final extension at 72 °C for 5 min. The other PCR was performed with primers isoA14F (5′-GVGACGAYTGGTAYGACA-3′) and isoA511R (5′-TCGTCRAAGAARTTCTTBAC-3′) of Carrión et al. [23], which yield an expected amplicon of 497 bp. The PCR consisted of an initial denaturation of 94 °C for 2 min, followed by 31 cycles of 95 °C for 15 s, 54 °C for 30 s, 72 °C for 1 min, and a final extension at 72 °C for 7 min. Both reactions were performed by using AppTaq RedMix (Appleton, Birmingham, UK). The PCR products were examined on an agarose gel stained with ethidium bromide.
2.5. Preparation of Genomic DNA for Genome Analysis
Genomic DNA from Alcaligenes sp. strain 13f was extracted from the culture grown in glucose/yeast-extract broth at 20 °C for 5 days by using the GenElute Bacterial Genomic DNA Extraction Kit (Sigma, St Louis, MO, USA). The DNA was examined on an agarose gel and quantified by using the Quant-iT PicoGreen dsDNA assay (Thermo Scientific, Loughborough, UK) in a Nanodrop spectrophotometer (NanoDrop 3300, Thermo Scientific, Loughborough, UK).
2.6. Genome Sequencing and Genome Assembly
The genome of Alcaligenes sp. strain 13f was sequenced by MicrobesNG (University of Birmingham, Birmingham, UK). Genomic DNA libraries were prepared by using a Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol with the following modifications: two nanograms of DNA were used, and the PCR elongation time was 1 min. DNA quantification and library preparation were carried out by using a Hamilton Microlab STAR automated liquid handler. Pooled libraries were quantified by using the Kapa Biosystems Library Quantification Kit for Illumina on a Roche LightCycler 96 qPCR machine. Libraries were sequenced on the Illumina HiSeq by using a 250-bp paired-end protocol. Reads were adapter-trimmed by using Trimmomatic (version 0.30) [39] with a sliding window quality cutoff of Q15. De-novo assembly was performed by using SPAdes (version 3.7) [40] with default parameters.
2.7. Phylogenomic Characterisation and Analysis of Average Nucleotide Identity (ANI)
Twelve publicly available genomes of Alcaligenes spp. were included for phylogenomic analysis. The Pathosystems Resource Integration Center (PATRIC) [41] was used to construct a codon-based tree built on 500 single-copy genes that are present in all genomes studied (Table S1), and aligned with the MAFFT (version 7.397) [42]. The maximum likelihood method (Jones-Taylor-Thornton (JTT) model) was performed by using RaxML (version 8.2.11) [43] with branch support values determined by 100 replicates of fast bootstrapping. The phylogenomic tree was visualised by using iTOL [44].
The similarities of eight genomes of closely related Alcaligenes species, including Alcaligenes sp. strain 13f, were analysed based on Average Nucleotide Identity (ANI) values, which were calculated and visualised by using OAT (version 0.93.1) [45].
2.8. Genome Analysis and Comparison
The quality of the assembly was assessed by using QUAST (version 5.0.2) [46] and sequences shorter than 200 bp were removed before the annotation process. The completeness of the genome assembly was determined by using Benchmarking Universal Single-Copy Orthologs (BUSCO) (version 4.1.3) [47]. The annotation was done by using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (version 5.2) (with the best-placed reference protein set and GeneMarkS-2+ methods) [48]. The amino acid sequences from Alcaligenes sp. strain 13f were compared with those of known bacterial isoprene degraders by using BLASTp (version 2.12.0) [49] to search for the occurrence of enzymes involved in isoprene degradation. To search for homologues of specific amino acid sequences, a local nucleotide BLAST database was queried with amino acid sequences of interest by using tBLASTn (version 2.2.10) [49]. The genomic data from this study have been deposited in the NCBI database under the accession number PRJNA734706.
3. Results and Discussion
3.1. Growth Characteristics and Isoprene Degradation of Alcaligenes sp. Strain 13f
Isoprene-degrading bacteria were isolated from isoprene-enriched soil samples collected beneath wild Himalayan cherry trees. A representative (isolate 13f) of the white, opaque colonies on minimal medium incubated with isoprene, which were the majority of colonies obtained, was further characterised and identified. It was Gram-negative, rod-shaped, identified as being related to Alcaligenes faecalis, according to 16S rRNA gene sequencing. Strain 13f used isoprene as the sole source of carbon and energy, as demonstrated by the growth on minimal medium agar incubated under isoprene as the sole carbon source.
Alcaligenes sp. strain 13f was then grown in minimal medium broth supplemented with isoprene at two concentrations (7.2 × 105 and 7.2 × 106 ppbv). The degradation of isoprene from the treatment with the lower isoprene concentration was 32.6% (Figure 1A, pink line), greater than that from the treatment with the higher isoprene concentration, which was 19.6% (Figure 1B, pink line) (Anova; p < 0.05).
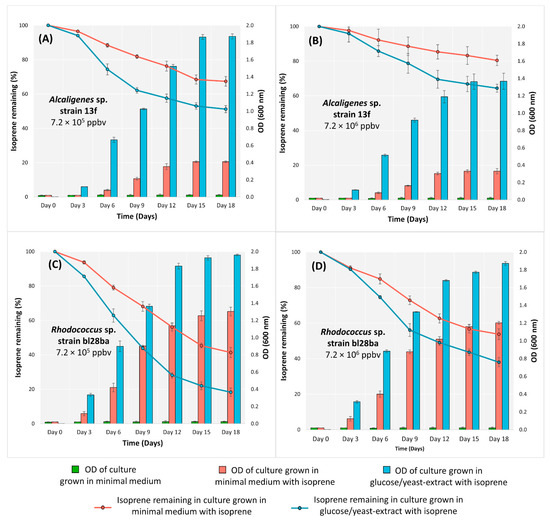
Figure 1.
Growth and isoprene degradation by Alcaligenes sp. strain 13f (A,B) and Rhodococcus sp. strain bl28ba (C,D) in minimal medium and glucose/yeast-extract medium incubated with isoprene at two different concentrations over 18 days. Growth of each isolate was demonstrated by an increase in OD600, and isoprene degradation is shown by the percentage of remaining isoprene in the culture, n = 3. The error bars show ± SD.
When Alcaligenes sp. strain 13f was additionally provided with glucose/yeast extract, it degraded isoprene more rapidly and to a greater extent than when isoprene was the only source of carbon and energy (Figure 1A,B, blue lines) (Anova; p < 0.05). It also grew more rapidly and extensively in the presence of glucose/yeast extract (Figure 1A,B, blue bars) (Anova; p < 0.05). These findings indicate that the genes for isoprene degradation are not repressed by the alternative organic growth substrates in the medium, as seen in other strains [34,50].
Similar to the cultures grown in minimal medium, in glucose yeast-extract broth, the extent of isoprene degradation was significantly lower (Anova; p < 0.05) when incubated at the higher isoprene concentration. Isoprene degradation in the glucose/yeast-extract broth after 18 days was 48.8% in the treatment supplied with the lower isoprene concentration (Figure 1A, blue line) and 35.6% with the higher isoprene concentration (Figure 1B, blue line).
An Alcaligenes sp. was previously described as an isoprene degrader by Ewers et al. [31]. Srivastva et al. [32] also observed the ability of an Alcaligenes sp. isolated from waste rubber dumping site soil to degrade isoprene and found that isoprene degradation by Alcaligenes strain ISO1 decreased with increasing isoprene concentrations, perhaps indicative of isoprene toxicity at higher concentrations.
The isoprene degradation and the growth of Alcaligenes sp. strain 13f were higher than those of the bacteria that had previously been reported, which were Ochrobactrum, Arthrobacter, Bacillus, Friedmanniella, Klebsiella, Isoptericola, and Cellulosimicrobium genera [34]. However, the growth and isoprene degradation by Alcaligenes sp. strain 13f (Figure 1A,B) were lower than those of Rhodococcus sp. strain bl28ba in every treatment (Figure 1C,D). Rhodococcus is a commonly isolated isoprene-degrading species in soil enriched with isoprene [14,23].
3.2. Examination of isoA Gene in Alcaligenes sp. Strain 13f
The presence of the isoA gene, encoding the isoprene monooxygenase subunit and currently considered as an essential component of the pathway for isoprene degradation, was examined in Alcaligenes sp. strain 13f by using the primers designed by El Khawand et al. [14] and Carrión et al. [23]. Rhodococcus sp. strain bl28ba was used as a positive control. No isoA product was amplified by PCR from Alcaligenes sp. strain 13f, which led to the hypothesis that the isoA sequence from Alcaligenes sp. strain 13f was very different from the isoA sequences from diverse isoprene-degrading genera: Gordonia, Leifsonia, Loktanella, Micrococcus, Mycobacterium, Nocardioides, Rhodococcus, Shinella, Sphingopyxis, Stappia, and Variovorax [14,23]. Another hypothesis was that Alcaligenes sp. strain 13f lacks the isoA gene and has a novel mechanism of isoprene degradation. Therefore, to start to address these hypotheses, the genome of Alcaligenes sp. strain 13f was sequenced.
3.3. Overall Characteristics of the Genome of Alcaligenes sp. Strain 13f
The genome of Alcaligenes sp. strain 13f was 4,402,996 base pairs, which is within the size range for this genus (3.02–4.86 Mbp). The mol% GC content of 56.29% was also typical of this genus. Other genomic information is described in Table 1.

Table 1.
The characteristics of the genome of Alcaligenes sp. strain 13f.
According to its 16S rRNA gene sequence from the genome sequencing, strain 13f was identified as Alcaligenes faecalis, and the closest relative was strain NBRC 13111 with an identity of 99.73%.
To investigate the phylogenomic relationship of Alcaligenes sp. strain 13f, its genome sequence was uploaded to the PATRIC workspace [41]. In total, 500 single-copy genes that are commonly present in the genomes of 12 strains of Alcaligenes spp., which were selected based on previously described 16S rRNA phylogeny, were used to build a phylogenomic tree. Strain 13f was shown to most likely be a member of the species Alcaligenes faecalis (Figure 2). The closest strains were AU14, NBIB-017, BDB4, MB207, and MOR02 (Figure 2), and included members of the subspecies phenolicus, whereas 16S rRNA gene sequence analysis identified the closest strain as NBRC 13111, the type strain.
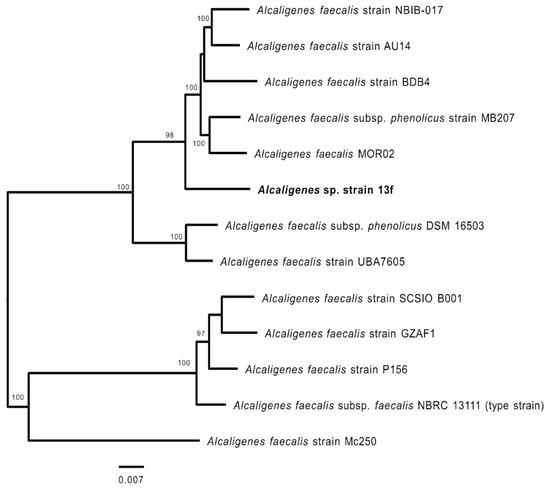
Figure 2.
Phylogenomic analysis of Alcaligenes spp. including Alcaligenes sp. strain 13f (bold font). The tree shown is a maximum likelihood phylogeny of 500 genes (presented in Table S1), based on 100 rounds of rapid bootstrapping. Only the bootstraps supporting values of >50% are shown.
To confirm the species assignment based on phylogenomic results, we applied one of the most effective measures for determining the relationships between bacterial species based on genome comparison, namely Average Nucleotide Identity (ANI) [51]. Analysis of the genome of Alcaligenes sp. strain 13f compared with the genomes of six of the most closely related strains and the type strain (Alcaligenes faecalis subsp. faecalis NBRC 13111) revealed that all of the ANI values were higher than 92% (Figure 3). If the type strain (NBRC 13111) is excluded, Alcaligenes sp. strain 13f and each of the other six strains had ANI values of more than 95% (the threshold for shared species identity [52,53]). The highest ANI value was 96.99% identity with Alcaligenes faecalis strain AU14, a wheat root strain with the ability to reduce nitrous oxide [54]. Thus, the phylogenomic circumscription of strain 13f by strains of Alcaligenes faecalis and the high genome relatedness based on ANI suggest that it is probably a member of this species.
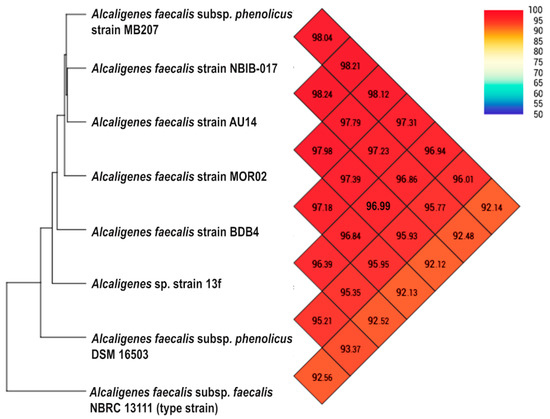
Figure 3.
A heatmap presenting the ANI values of Alcaligenes sp. strain 13f and seven closely related strains, including the type strain (Alcaligenes faecalis subsp. faecalis NBRC 13111). The heatmap was calculated by using the Orthologous Average Nucleotide Identity Tool (OAT) software.
3.4. Genome Comparison between Alcaligenes sp. Strain 13f and Other Isoprene-Degrading Bacteria
In order to identify the sequences that might code for genes involved in isoprene degradation, the genes from the genome of Alcaligenes sp. strain 13f were submitted to the NCBI database for annotation by using the prokaryotic genome annotation pipeline (PGAP). Out of the 3942 gene products analysed, seven were found to have some similarities (ca. 20–30%) to proteins previously known to be involved in isoprene degradation in other species (Table 2).

Table 2.
Percentages of protein sequence identity derived from Alcaligenes sp. strain 13f compared with proteins in the Iso metabolic cluster of isoprene-degrading bacteria.
Specifically, the amino acid sequences of Alcaligenes sp. strain 13f were compared with the translated sequences from the iso gene cluster of Rhodococcus AD45, Sphingopyxis OPL5, and Variovorax WS11, which are isoprene degraders that have been well characterised genetically [18,19,20]. The translated amino acid sequences of Alcaligenes sp. strain 13f had some similarities with several proteins encoded in the iso metabolic gene cluster found in these strains. However, the percent identities to the Iso sequences were not as high as expected, in the range of approximately 20 to 30% (Table 2). These sequences from Alcaligenes sp. strain 13f were identified as other proteins, with more than 98% identities (Table 2). For example, the sequence that had approximately 28% identity to the IsoA sequences in Rhodococcus AD45, Sphingopyxis OPL5, and Variovorax WS11 was identified as having 99.80% identity to an aromatic/alkene/methane monooxygenase hydroxylase/oxygenase subunit alpha from Alcaligenes (Table 2). Moreover, some genes typically found together in the iso cluster were missing (Figure 4), including isoB (encoding isoprene monooxygenase gamma subunit), isoG (encoding a putative coenzyme-A transferase), isoI, and isoJ (encoding a glutathione S-transferase). The absence of these genes indicated that Alcaligenes sp. strain 13f might have had a different oxygenase from the previously known isoprene monooxygenase, which enabled it to catabolise isoprene, albeit less rapidly than most well-characterised isoprene-degrading strains [15,16,17,18,19,20].
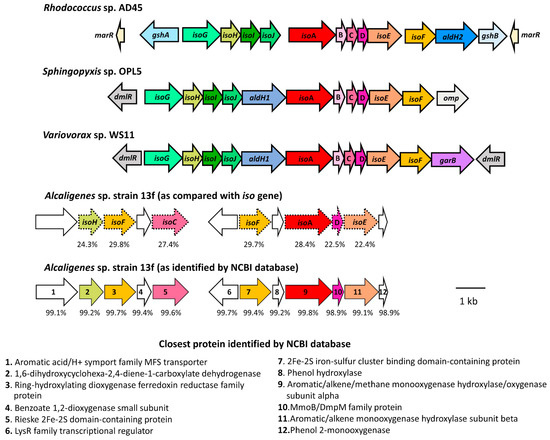
Figure 4.
The organisation of the genes putatively involved in isoprene degradation in Alcaligenes sp. strain 13f, presented in comparison with the iso gene organisation in the reference isoprene degraders: Rhodococcus AD45, Sphingopyxis OPL5, and Variovorax WS11 [18,19,20]. The arrows represent the orientation of the genes. The dotted frames surrounding the gene names are to remind the reader of the putative functional homologues involved in isoprene degradation, with the percent identity to the protein sequences of Variovorax WS11 shown below. The bottom-row diagram shows the gene arrangement in Alcaligenes sp. strain 13f according to the closest proteins identified by NCBI protein database.
Because a monooxygenase (aromatic/alkene/methane monooxygenase hydroxylase/oxygenase subunit alpha) that had some degrees of similarity to IsoA was found in Alcaligenes sp. strain 13f (as seen in Table 2), its amino acid sequence was compared with other isoA sequences of selected known isoprene degraders and other monooxygenases, such as methane monooxygenase, toluene monooxygenase, and phenol monooxygenase, by using Bioedit sequence alignments [37]. From the comparison, a phylogenetic tree was constructed using MEGA-X [55] (Figure 5).
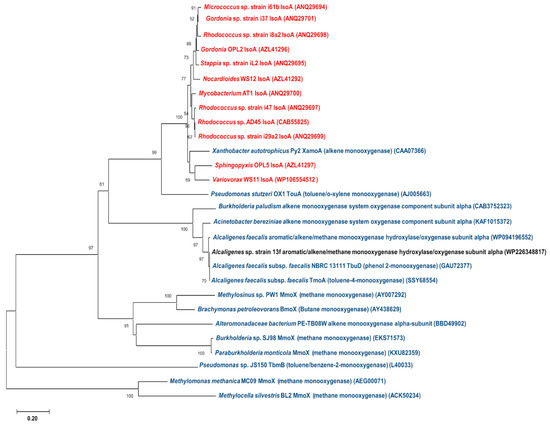
Figure 5.
A phylogenetic tree demonstrating the similarities among the IsoA amino acid sequences of known isoprene-degrading strains (red), monooxygenase of Alcaligenes sp. strain 13f (black), and other hydrocarbon monooxygenases of other bacteria (blue). A tree was constructed through the maximum composite likelihood model of neighbour-joining method with 1000 bootstrap replicates.
The phylogenetic tree revealed notable distances between the monooxygenase found in Alcaligenes sp. strain 13f and the isoA gene of other isoprene degraders (Figure 5). It can also be seen that the IsoA sequences of Gram-positive bacteria (Micrococcus, Gordonia, Rhodococcus, Stappia, Nocardioides, and Mycobacterium) were phylogenetically grouped, and separate from the group of IsoA sequences of Gram-negative bacteria (Sphingopyxis and Variovorax), which were closely related to the alkene monooxygenase alpha subunit (xamoA) of the Gram-negative Xanthobacter strain Py2 (Figure 5), as previously reported by Dawson et al. [18]. The aromatic/alkene/methane monooxygenase found in Alcaligenes sp. strain 13f, however, was in a different cluster from both of these groups, and was much more similar to other hydrocarbon monooxygenases (e.g., phenol monooxygenase, alkene monooxygenase, and toluene monooxygenase) from other Alcaligenes strains and some other Gram-negative bacteria. The low similarities between the monooxygenases of Alcaligenes sp. strain 13f and the proteins in the Iso cluster found in other isoprene degraders are consistent with the lack of detection of the isoA gene in Alcaligenes sp. strain 13f by PCR (see Section 3.2).
Due to the lack of close homologues of isoprene monooxygenase, and the possibility that a different class of monooxygenase might catalyse the initial oxidation of isoprene, the genome of Alcaligenes sp. strain 13f was searched for another key gene of isoprene degradation, isoI, encoding the gluthathione-S-transferase which catalyses the second step of the isoprene degradation pathway. This gene is universally present in all previously sequenced isoprene degraders [6]. Since recently, an analogous pathway of styrene degradation has been identified in Gordonia, whereby styrene oxide, formed by the action of a flavin-dependent monooxygenase (StyAB), is similarly conjugated with glutathione by StyI [56], the closely-related styI gene that was included in the search. When the Alcaligenes sp. strain 13f genome was searched for isoI or styI sequences, no close homologues were found (no hits at E-value < 0.00001). Because the styrene degradation pathway may be non-specific, the Alcaligenes sp. strain 13f genome was also searched for homologues of the gene encoding styrene monooxygenase large subunit, styA, using as query the amino acid sequences from styrene degraders Pseudomonas sp. VLB120 and Gordonia rubripertincta CWB2 [56,57]. An FAD-binding oxidoreductase (WP_226349106.1) was identified at a relatively low level of amino acid identity (29% for each query sequence). Because close homologues of WP_226349106.1 are present in many other Alcaligenes and related bacteria not known for isoprene degradation, and downstream genes involved in isoprene or styrene degradation were not located nearby in the genome (phenylacetaldehyde dehydrogenase (styD), styrene oxide isomerase (styC), or glutathione-S-transferase (isoI, styI)), this was not considered a likely candidate enabling growth on isoprene in Alcaligenes sp. strain 13f.
Interestingly, the growth and isoprene degradation of Alcaligenes sp. strain 13f were lower than those of Rhodococcus sp. strain bl28ba (a representative Rhodococcus strain harbouring the isoA gene [38], which was used as a positive control) (Figure 1). This was also another indication that the genetics and the mechanisms involved in isoprene metabolism of Alcaligenes sp. strain 13f might be different from the typical iso-driven mechanisms found in Rhodococcus and other genera such as Gordonia and Variovorax [15,16,18,20], which can contribute to the differences in its pattern of growth and isoprene degradation. However, the mechanism of isoprene catabolism by Alcaligenes sp. strain 13f is not yet known but certainly warrants further investigation. The aromatic/alkene monooxygenase found in Alcaligenes sp. strain 13f might catalyse the initial oxidation of isoprene, resulting in the decrease of isoprene from the headspace gaseous content. It has been shown previously that aromatic monooxygenases can catalyse the oxidation of alkenes [58]. The resulting products of this catalysis could enter the central metabolic pathway through beta-oxidation, similar to the known isoprene degradation pathway of Rhodococcus AD45. Further studies on the intermediate metabolites, as well as transcriptomic/proteomic responses to growth on isoprene compared with other carbon and energy sources, are therefore crucial in deciphering the isoprene degradation pathway of this Alcaligenes isolate and Alcaligenes spp. in general.
4. Conclusions
In this study, Alcaligenes sp. strain 13f obtained from soil associated with wild Himalayan cherry was investigated for its isoprene-degrading ability, and its genome was sequenced. Overall, our study showed the capacity for isoprene degradation of an Alcaligenes species and raised the intriguing possibility that the isoprene metabolism pathway of this bacterium might be different from other isoprene-degrading bacteria found in previous studies, as revealed by the genome analysis. The incompleteness of the isoprene monooxygenase gene cluster in isoprene-degrading bacteria has never been reported previously. This study, therefore, provides a basis for further exploration into isoprene degradation mechanisms in Alcaligenes to gain a better understanding of isoprene metabolism in this environmentally significant isoprene-degrading bacterium.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11040519/s1, Table S1: Genes of Alcaligenes sp. strain 13f and the other twelve A. faecalis strains included in phylogenomic analysis.
Author Contributions
Conceptualisation, T.U., J.C.M., S.B., T.J.M. and T.C.; data curation, T.U.; formal analysis, T.U., S.S., A.T.C., J.C.M., W.M., N.N., T.J.M. and T.C.; Funding acquisition, J.C.M., T.J.M., S.B. and T.C.; Investigation, T.U., T.J.M. and T.C.; Project administration, T.J.M., S.B. and T.C.; Supervision, J.C.M., T.J.M., S.B. and T.C.; Writing—original draft, T.U., T.J.M. and T.C.; Writing—review & editing, T.U., S.S., A.T.C., J.C.M., W.M., N.N., S.W., S.B., T.C. and T.J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Science Achievement Scholarship of Thailand (granted to T.U), Biodiversity-based Economy Development Office (Public Organisation) (BEDO) grant to T.C. and S.B (36/2562), Natural Environment Research Council (NERC) grant to T.J.M. (NE/J009555/1)), European Research Council (ERC) Advanced Grant (IsoMet 694578) to J.C.M. and partially supported by Chiang Mai University, Thailand and the University of Essex, UK.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The 16S rRNA sequence and the genomic data are available on the NCBI database (accession number MZ323998 and PRJNA734706).
Acknowledgments
We would like to thank Farid Benyahia and John Green (University of Essex, UK) and the staff of the Department of Biology, Chiang Mai University for providing technical support. We also would like to express our gratitude to the academic and technical staff members of the Forest Restoration Research Unit of Chiang Mai University (FORRU-CMU) for providing information on framework species and for their assistance with soil sampling in the restored forest area.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Müller, J.-F.; Stavrakou, T.; Wallens, S.; De Smedt, I.; Van Roozendael, M.; Potosnak, M.J.; Rinne, J.; Munger, B.; Goldstein, A.; Guenther, A.B. Global isoprene emissions estimated using MEGAN, ECMWF analyses and a detailed canopy environment model. Atmos. Chem. Phys. 2008, 8, 1329–1341. [Google Scholar] [CrossRef] [Green Version]
- Sharkey, T.D.; Wiberley, A.E.; Donohue, A.R. isoprene emission from plants: Why and how. Ann. Bot. 2008, 101, 5–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arneth, A.; Monson, R.K.; Schurgers, G.; Niinemets, Ü.; Palmer, P.I. Why are estimates of global terrestrial isoprene emissions so similar (and why is this not so for monoterpenes)? Atmos. Chem. Phys. 2008, 8, 4605–4620. [Google Scholar] [CrossRef] [Green Version]
- Guenther, A.B.; Jiang, X.; Heald, C.L.; Sakulyanontvittaya, T.; Duhl, T.; Emmons, L.K.; Wang, X. The model of emissions of gases and aerosols from Nature version 2.1 (MEGAN2.1): An extended and updated framework for modeling biogenic emissions. Geosci. Model Dev. 2012, 5, 1471–1492. [Google Scholar] [CrossRef] [Green Version]
- Pacifico, F.; Harrison, S.P.; Jones, C.D.; Sitch, S. Isoprene emissions and climate. Atmos. Environ. 2009, 43, 6121–6135. [Google Scholar] [CrossRef]
- McGenity, T.J.; Crombie, A.T.; Murrell, J.C. Microbial cycling of Isoprene, the most abundantly produced biological volatile organic compound on earth. ISME J. 2018, 12, 931–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cleveland, C.C.; Yavitt, J.B. Consumption of atmospheric isoprene in soil. Geophys. Res. Lett. 1997, 24, 2379–2382. [Google Scholar] [CrossRef] [Green Version]
- Murrell, J.C.; McGenity, T.J.; Crombie, A.T. Microbial metabolism of isoprene: A much-neglected climate-active gas. Microbiology 2020, 166, 600–613. [Google Scholar] [CrossRef] [PubMed]
- Gray, C.M.; Helmig, D.; Fierer, N. Bacteria and fungi associated with isoprene consumption in soil. Elem. Sci. Anth. 2015, 3, 000053. [Google Scholar] [CrossRef] [Green Version]
- Cleveland, C.C.; Yavitt, J.B. Microbial consumption of atmospheric isoprene in a temperate forest soil. Appl. Environ. Microbiol. 1998, 64, 172–177. [Google Scholar] [CrossRef] [Green Version]
- Pegoraro, E.; Abrell, L.; van Haren, J.; Barron-Gafford, G.; Grieve, K.; Malhi, Y.; Murthy, R.; Lin, G. The effect of elevated atmospheric CO2 and drought on sources and sinks of isoprene in a temperate and tropical rainforest mesocosm. Glob. Change Biol. 2005, 11, 1234–1246. [Google Scholar] [CrossRef]
- Pegoraro, E.; Rey, A.; Abrell, L.; Haren, J.V.; Lin, G. Drought effect on isoprene production and consumption in biosphere 2 tropical rainforest. Glob. Chang. Biol. 2006, 12, 456–469. [Google Scholar] [CrossRef]
- Acuña Alvarez, L.A.; Exton, D.A.; Timmis, K.N.; Suggett, D.J.; McGenity, T.J. Characterization of marine isoprene-degrading communities. Environ. Microbiol. 2009, 11, 3280–3291. [Google Scholar] [CrossRef] [PubMed]
- El Khawand, M.; Crombie, A.T.; Johnston, A.; Vavlline, D.V.; McAuliffe, J.C.; Latone, J.A.; Primak, Y.A.; Lee, S.-K.; Whited, G.M.; McGenity, T.J.; et al. Isolation of isoprene degrading bacteria from soils, development of IsoA gene probes and identification of the active isoprene-degrading soil community using DNA-stable isotope probing: Isoprene-degrading bacteria. Env. Microbiol. 2016, 18, 2743–2753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, A.; Crombie, A.T.; Khawand, M.E.; Sims, L.; Whited, G.M.; McGenity, T.J.; Murrell, J.C. Identification and characterisation of isoprene-degrading bacteria in an estuarine environment. Env. Microbiol. 2017, 19, 3526–3537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Hylckama Vlieg, J.E.T.; Leemhuis, H.; Spelberg, J.H.L.; Janssen, D.B. Characterization of the gene cluster involved in isoprene metabolism in Rhodococcus sp. strain AD45. J. Bacteriol. 2000, 182, 1956–1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, L.; Larke-Mej, N.L. Complete genome of isoprene degrading Nocardioides sp. WS12. Microorganisms 2020, 8, 889. [Google Scholar] [CrossRef] [PubMed]
- Dawson, R.A.; Larke-Mejía, N.L.; Crombie, A.T.; Ul Haque, M.F.; Murrell, J.C. Isoprene oxidation by the gram-negative model bacterium Variovorax sp. WS11. Microorganisms 2020, 8, 349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larke-Mejía, N.L.; Carrión, O.; Crombie, A.T.; McGenity, T.J.; Murrell, J.C. Sphingopyxis sp. strain OPL5, an isoprene-degrading bacterium from the Sphingomonadaceae family isolated from oil palm leaves. Microorganisms 2020, 8, 1557. [Google Scholar] [CrossRef] [PubMed]
- Crombie, A.T.; Khawand, M.E.; Rhodius, V.A.; Fengler, K.A.; Miller, M.C.; Whited, G.M.; McGenity, T.J.; Murrell, J.C. Regulation of plasmid-encoded isoprene metabolism in Rhodococcus, a representative of an important link in the global isoprene cycle. Env. Microbiol. 2015, 17, 3314–3329. [Google Scholar] [CrossRef] [Green Version]
- van Hylckama Vlieg, J.E.T.; Kingma, J.; van den Wijngaard, A.J.; Janssen, D.B. A glutathione S-transferase with activity towards cis-1,2-dichloroepoxyethane is involved in isoprene utilization by Rhodococcus sp. strain AD45. Appl. Environ. Microbiol. 1998, 64, 2800–2805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Hylckama Vlieg, J.E.T.; Kingma, J.; Kruizinga, W.; Janssen, D.B. Purification of a glutathione S-transferase and a glutathione conjugate-specific dehydrogenase involved in isoprene metabolism in Rhodococcus sp. strain AD45. J. Bacteriol. 1999, 181, 2094–2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrión, O.; Larke-Mejía, N.L.; Gibson, L.; Farhan Ul Haque, M.; Ramiro-García, J.; McGenity, T.J.; Murrell, J.C. Gene probing reveals the widespread distribution, diversity and abundance of isoprene-degrading bacteria in the environment. Microbiome 2018, 6, 219. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.-S.; Hirai, M.; Shoda, M. Piggery wastewater treatment using Alcaligenes Faecalis strain No. 4 with heterotrophic nitrification and aerobic denitrification. Water Res. 2006, 40, 3029–3036. [Google Scholar] [CrossRef] [PubMed]
- Van Trappen, S.; Tan, T.-L.; Samyn, E.; Vandamme, P. Alcaligenes aquatilis sp. nov., a novel bacterium from sediments of the Weser Estuary, Germany, and a salt marsh on Shem Creek in Charleston Harbor, USA. Int. J. Syst. Evol. Microbiol. 2005, 55, 2571–2575. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.; Ahmed, I.; Iida, T.; Lee, Y.-J.; Busse, H.-J.; Fujiwara, T.; Ohkuma, M. A heavy-metal tolerant novel bacterium, Alcaligenes pakistanensis sp. nov., isolated from industrial effluent in Pakistan. Antonie Van Leeuwenhoek 2015, 108, 859–870. [Google Scholar] [CrossRef]
- Mahjoubi, M.; Aliyu, H.; Cappello, S.; Naifer, M.; Souissi, Y.; Cowan, D.A.; Cherif, A. The genome of Alcaligenes aquatilis strain BU33N: Insights into hydrocarbon degradation capacity. PLoS ONE 2019, 14, e0221574. [Google Scholar] [CrossRef] [PubMed]
- Castellani, A.; Chalmers, A.J. Manual of Tropical Medicine, 3rd ed.; Williams Wood and Co.: New York, NK, USA, 1919; pp. 959–960. [Google Scholar]
- Rehfuss, M.; Urban, J. Alcaligenes faecalis subsp. phenolicus subsp. nov. a phenol-degrading, denitrifying bacterium isolated from a graywater bioprocessor. Syst. Appl. Microbiol. 2005, 28, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Lin, D.; Zheng, Y.; Zhang, Q.; Yin, Y.; Cai, L.; Fang, H.; Yu, Y. Biodegradation of DDT by Stenotrophomonas sp. DDT-1: Characterization and genome functional analysis. Sci. Rep. 2016, 6, 21332. [Google Scholar] [CrossRef] [Green Version]
- Ewers, J.; Freier-Schröder, D.; Knackmuss, H.J. Selection of trichloroethene (TCE) degrading bacteria that resist inactivation by TCE. Arch. Microbiol. 1990, 154, 410–413. [Google Scholar] [CrossRef]
- Srivastva, N.; Shukla, A.K.; Singh, R.S.; Upadhyay, S.N.; Dubey, S.K. Characterization of bacterial isolates from rubber dump site and their use in biodegradation of isoprene in batch and continuous bioreactors. Bioresour. Technol. 2015, 188, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Elliott, S.; Navakitbumrung, P.; Kuarak, C.; Zangkum, S.; Anusarnsunthorn, V.; Blakesley, D. Selecting framework tree species for restoring seasonally dry tropical forests in northern Thailand based on field performance. For. Ecol. Manag. 2003, 184, 177–191. [Google Scholar] [CrossRef]
- Uttarotai, T.; McKew, B.A.; Benyahia, F.; Murrell, J.C.; Mhuantong, W.; Wangkarn, S.; Chitov, T.; Bovonsombut, S.; McGenity, T.J. Isoprene-degrading bacteria from soils associated with tropical economic crops and framework forest trees. Microorganisms 2021, 9, 1024. [Google Scholar] [CrossRef]
- Griffiths, R.I.; Whiteley, A.S.; O’Donnell, A.G.; Bailey, M.J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 2000, 66, 5488–5491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Murphy, G. Isoprene Degradation in the Terrestrial Environment. Ph.D. Thesis, University of Essex, Colchester, UK, 2017. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014, 42, D581–D591. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (ITOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Srivastva, N.; Vishwakarma, P.; Bhardwaj, Y.; Singh, A.; Manjunath, K.; Dubey, S.K. Kinetic and molecular analyses reveal isoprene degradation potential of Methylobacterium sp. Bioresour. Technol. 2017, 242, 87–91. [Google Scholar] [CrossRef]
- Lalucat, J.; Mulet, M.; Gomila, M.; García-Valdés, E. Genomics in bacterial taxonomy: Impact on the genus Pseudomonas. Genes 2020, 11, 139. [Google Scholar] [CrossRef] [Green Version]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M.Y. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef] [Green Version]
- Usyskin-Tonne, A.; Hadar, Y.; Minz, D. Altering N2O emissions by manipulating wheat root bacterial community. Sci. Rep. 2019, 9, 7613. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Heine, T.; Zimmerling, J.; Ballmann, A.; Kleeberg, S.B.; Rückert, C.; Busche, T.; Winkler, A.; Kalinowski, J.; Poetsch, A.; Scholtissek, A.; et al. On the enigma of glutathione-dependent styrene degradation in Gordonia rubripertincta CWB2. Appl. Environ. Microbiol. 2018, 84, e00154-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panke, S.; Witholt, B.; Schmid, A.; Wubbolts, M.G. Towards a biocatalyst for (S)-styrene oxide production: Characterization of the styrene degradation pathway of Pseudomonas sp. strain VLB120. Appl. Environ. Microbiol. 1998, 64, 2032–2043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClay, K.; Fox, B.G.; Steffan, R.J. Toluene monooxygenase-catalyzed epoxidation of alkenes. Appl. Environ. Microbiol. 2000, 66, 1877–1882. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).