Evaluation of the BioFire FilmArray Pneumonia Panel Plus to the Conventional Diagnostic Methods in Determining the Microbiological Etiology of Hospital-Acquired Pneumonia
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Standard Microbiological Examination
2.3. BFPP Microbiological Examination
2.4. Statistical Data Analysis
3. Results
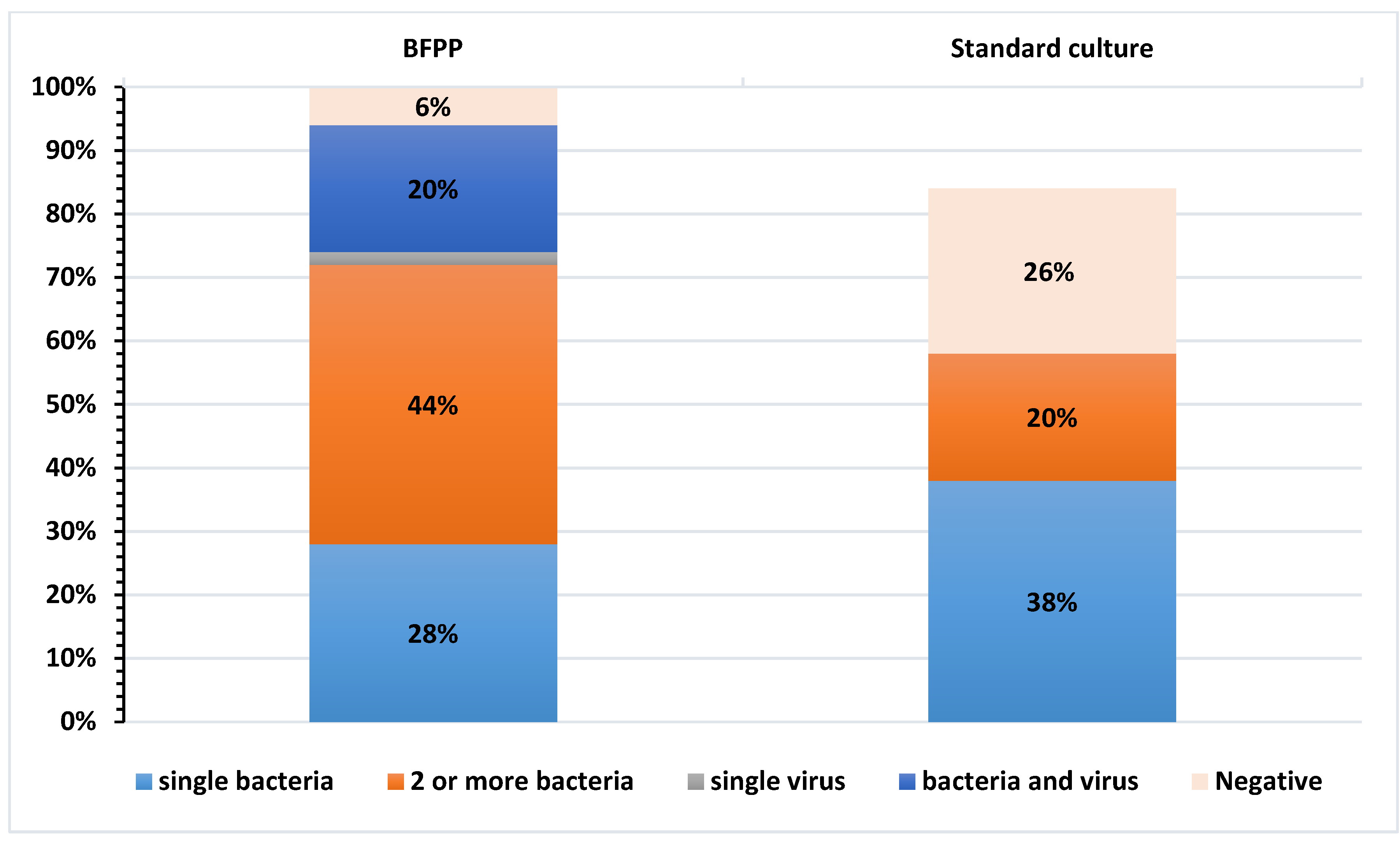
3.1. Evaluation of BFPP and Standard-of-Care Methods to Detect Most Relevant Respiratory Pathogens
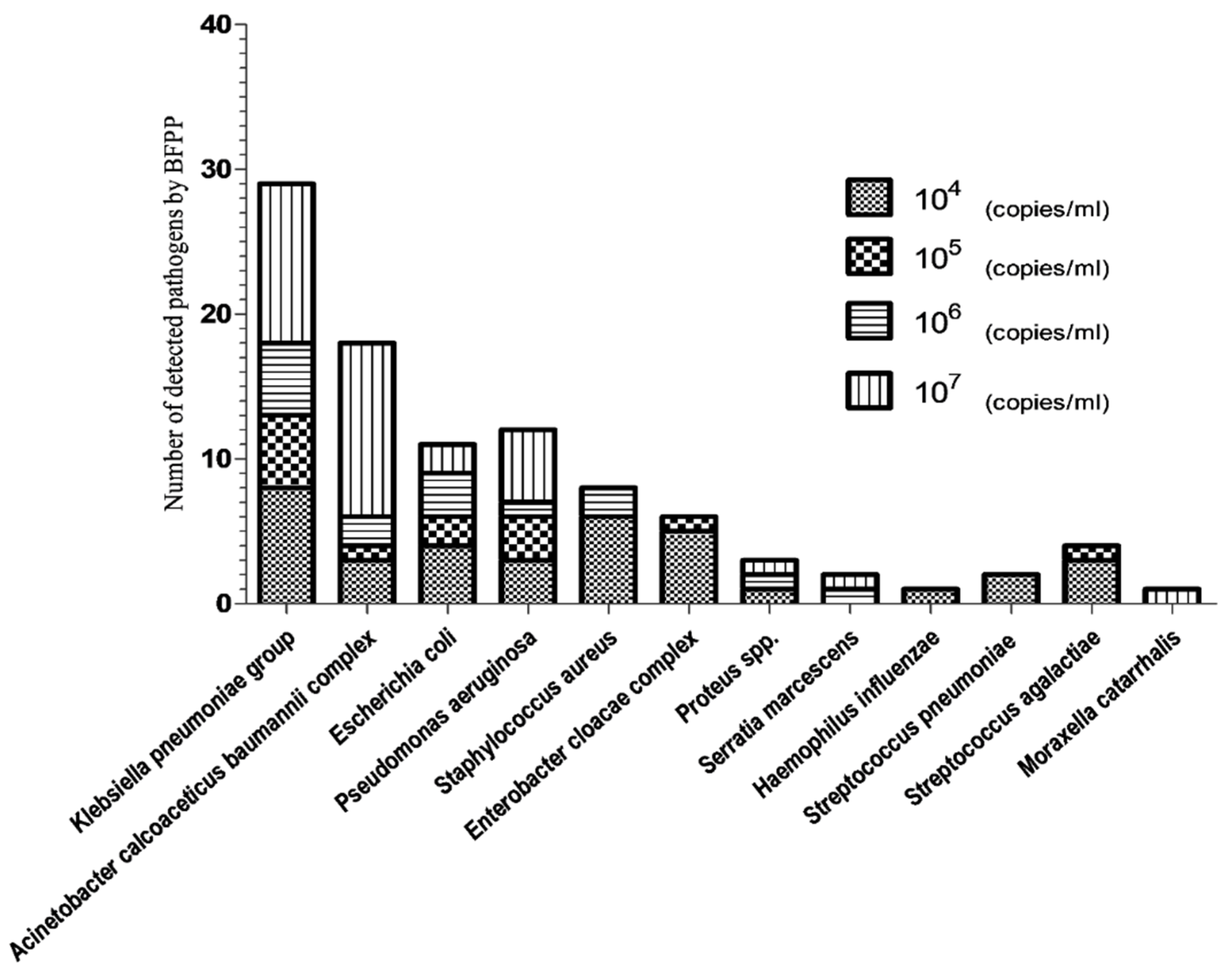
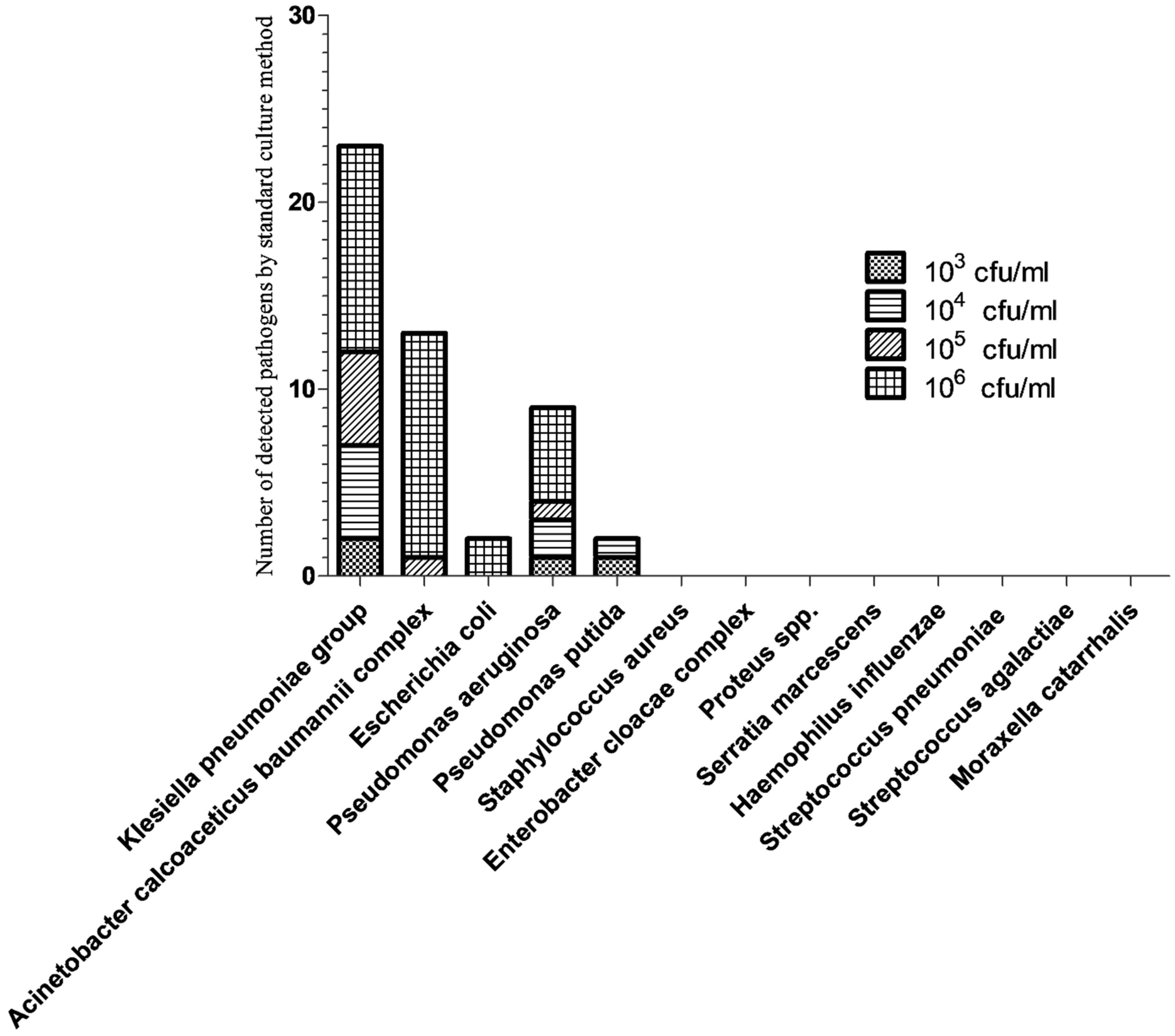
3.2. Semi-Quantitative Analysis of Bacterial Pathogens by BFPP and Standard Culture Methods
3.3. Detection of Antimicrobial Resistance Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Greenslade, L. World Pneumonia Day during a global pneumonia pandemic: 12 November 2020. Am. J. Physiol. Cell. Mol. Physiol. 2020, 319, L859–L860. [Google Scholar] [CrossRef]
- Leone, M.; Bouadma, L.; Bouhemad, B.; Brissaud, O.; Dauger, S.; Gibot, S.; Hraiech, S.; Jung, B.; Kipnis, E.; Launey, Y.; et al. Hospital-acquired pneumonia in ICU. Anaesth. Crit. Care Pain Med. 2018, 37, 83–98. [Google Scholar] [CrossRef]
- Papazian, L.; Klompas, M.; Luyt, C.-E. Ventilator-associated pneumonia in adults: A narrative review. Intensive Care Med. 2020, 46, 888–906. [Google Scholar] [CrossRef] [PubMed]
- Branco, A.; Lourençone, E.M.S.; Monteiro, A.B.; Fonseca, J.P.; Blatt, C.R.; Caregnato, R.C.A. Education to prevent ventilator-associated pneumonia in intensive care unit. Rev. Bras. Enferm. 2020, 73, e20190477. [Google Scholar] [CrossRef] [PubMed]
- Abdelrazik Othman, A.; Salah Abdelazim, M. Ventilator-associated pneumonia in adult intensive care unit prevalence and complications. Egypt. J. Crit. Care Med. 2017, 5, 61–63. [Google Scholar] [CrossRef]
- Feikin, D.R.; Hammitt, L.L.; Murdoch, D.R.; O’Brien, K.L.; Scott, J.A.G. The Enduring Challenge of Determining Pneumonia Etiology in Children: Considerations for Future Research Priorities. Clin. Infect. Dis. 2017, 64, S188–S196. [Google Scholar] [CrossRef] [PubMed]
- Sattar, S.B.A.; Sharma, S.; Headley, A.J.S. Bacterial Pneumonia (Nursing); StatPearls Publishing: Treasure Island, FL, USA, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK568697/ (accessed on 26 January 2022).
- Chen, G.; Xu, K.; Sun, F.; Sun, Y.; Kong, Z.; Fang, B. Risk Factors of Multidrug-Resistant Bacteria in Lower Respiratory Tract Infections: A Systematic Review and Meta-Analysis. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 7268519. [Google Scholar] [CrossRef]
- Modi, A.R.; Kovacs, C.S. Hospital-acquired and ventilator-associated pneumonia: Diagnosis, management, and prevention. Clevel. Clin. J. Med. 2020, 87, 633–639. [Google Scholar] [CrossRef]
- Costa, M.I.; Cipriano, A.; Santos, F.V.; Valdoleiros, S.; Furtado, I.; Machado, A.; Abreu, M.; Bastos, H. Clinical profile and microbiological aetiology diagnosis in adult patients hospitalized with community-acquired pneumonia. Pulmonology 2020, S2531-0437(20)30246-4. [Google Scholar] [CrossRef]
- Murphy, C.N.; Fowler, R.; Balada-Llasat, J.M.; Carroll, A.; Stone, H.; Akerele, O.; Buchan, B.; Windham, S.; Hopp, A.; Ronen, S.; et al. Multicenter Evaluation of the BioFire FilmArray Pneumonia/Pneumonia Plus Panel for Detection and Quantification of Agents of Lower Respiratory Tract Infection. J. Clin. Microbiol. 2020, 58, e00128-20. [Google Scholar] [CrossRef]
- Behzadi, P.; Urbán, E.; Matuz, M.; Benkő, R.; Gajdács, M. The Role of Gram-Negative Bacteria in Urinary Tract Infections: Current Concepts and Therapeutic Options. Adv. Exp. Med. Biol. 2021, 1323, 35–69. [Google Scholar] [CrossRef] [PubMed]
- Shebl, E.; Gulick, P.G. Nosocomial Pneumonia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK535441/ (accessed on 18 January 2022).
- Yayan, J.; Ghebremedhin, B.; Rasche, K. Cefepime shows good efficacy and no antibiotic resistance in pneumonia caused by Serratia marcescens and Proteus mirabilis—An observational study. BMC Pharmacol. Toxicol. 2016, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Webber, D.M.; Wallace, M.A.; Burnham, C.-A.D.; Anderson, N.W. Evaluation of the BioFire FilmArray Pneumonia Panel for Detection of Viral and Bacterial Pathogens in Lower Respiratory Tract Specimens in the Setting of a Tertiary Care Academic Medical Center. J. Clin. Microbiol. 2020, 58, e00343-20. [Google Scholar] [CrossRef]
- Ranjbar, R.; Behzadi, P.; Mammina, C. Respiratory Tularemia: Francisella tularensis and Microarray Probe Designing. Open Microbiol. J. 2016, 10, 176–182. [Google Scholar] [CrossRef]
- Erden, V.; Basaranoglu, G.; Beycan, I.; Delatioglu, H.; Hamzaoglu, N.S. Reproducibility of mini-BAL culture results using 10 mL or 20 mL instilled fluid. Intensive Care Med. 2003, 29, 1856. [Google Scholar] [CrossRef] [PubMed]
- Leber, A.L. Respiratory Tract Cultures. In Clinical Microbiology Procedures Handbook; ASM Press: Washington, DC, USA, 2016; Available online: https://www.worldcat.org/title/clinical-microbiology-procedures-handbook/oclc/943710632 (accessed on 26 January 2022).
- Doyle, D.; Peirano, G.; Lascols, C.; Lloyd, T.; Church, D.L.; Pitout, J.D.D. Laboratory Detection of Enterobacteriaceae That Produce Carbapenemases. J. Clin. Microbiol. 2012, 50, 3877–3880. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L.; Carrër, A.; Toleman, M.A.; Walsh, T.R. How To Detect NDM-1 Producers. J. Clin. Microbiol. 2011, 49, 718–721. [Google Scholar] [CrossRef]
- Bonnet, R.; Dutour, C.; Sampaio, J.L.; Chanal, C.; Sirot, D.; Labia, R.; De Champs, C.; Sirot, J. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240→Gly. Antimicrob. Agents Chemother. 2001, 45, 2269–2275. [Google Scholar] [CrossRef][Green Version]
- McHugh, L.C.; Snyder, K.; Yager, T.D. The effect of uncertainty in patient classification on diagnostic performance estimations. PLoS ONE 2019, 14, e0217146. [Google Scholar] [CrossRef]
- Rand, K.H.; Beal, S.G.; Cherabuddi, K.; Couturier, B.; Lingenfelter, B.; Rindlisbacher, C.; Jones, J.; Houck, H.J.; Lessard, K.J.; E Tremblay, E. Performance of a Semiquantitative Multiplex Bacterial and Viral PCR Panel Compared With Standard Microbiological Laboratory Results: 396 Patients Studied With the BioFire Pneumonia Panel. Open Forum Infect. Dis. 2020, 8, ofaa560. [Google Scholar] [CrossRef]
- Agarwal, A.; Malviya, D.; Harjai, M.; Tripathi, S.; Das, A. Comparative Evaluation of the Role of Nonbronchoscopic and Bronchoscopic Techniques of Distal Airway Sampling for the Diagnosis of Ventilator-Associated Pneumonia. Anesth. Essays Res. 2020, 14, 434–440. [Google Scholar]
- Afify, M.H.; Shaheen, E.A.; El-Dahdouh, S.S.; El-Feky, H.M. Comparison between bronchoscopic BAL and non-bronchoscopic BAL in patients with VAP. Egypt. J. Chest Dis. Tuberc. 2016, 65, 113–119. [Google Scholar] [CrossRef]
- Edin, A.; Eilers, H.; Allard, A.J. Evaluation of the Biofire Filmarray Pneumonia panel plus for lower respiratory tract infections. Infect. Dis. 2020, 52, 479–488. [Google Scholar] [CrossRef]
- Ginocchio, C.C.; Garcia-Mondragon, C.; Mauerhofer, B.; Rindlisbacher, C. Multinational evaluation of the BioFire® FilmArray® Pneumonia plus Panel as compared to standard of care testing. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1609–1622. [Google Scholar] [CrossRef]
- Lee, S.H.; Ruan, S.Y.; Pan, S.C.; Lee, T.F.; Chien, J.Y.; Hsueh, P.R. Performance of a multiplex PCR pneumonia panel for the identification of respiratory pathogens and the main determinants of resistance from the lower respiratory tract specimens of adult patients in intensive care units. J. Microbiol. Immunol. Infect. 2019, 52, 920–928. [Google Scholar] [CrossRef]
- Malhotra, R.K.; Indrayan, A. A simple nomogram for sample size for estimating sensitivity and specificity of medical tests. Indian J. Ophthalmol. 2010, 58, 519–522. [Google Scholar] [CrossRef]
- Hajian-Tilaki, K. Sample size estimation in diagnostic test studies of biomedical informatics. J. Biomed. Inform. 2014, 48, 193–204. [Google Scholar] [CrossRef]
- Ashurst, J.V.; Dawson, A. Klebsiella Pneumonia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK519004/ (accessed on 26 January 2022).
- Ferreira, R.L.; da Silva, B.C.M.; Rezende, G.S.; Nakamura-Silva, R.; Pitondo-Silva, A.; Campanini, E.B.; Brito, M.C.A.; Da Silva, E.M.L.; Freire, C.C.D.M.; Cunha, A.; et al. High Prevalence of Multidrug-Resistant Klebsiella pneumoniae Harboring Several Virulence and β-Lactamase Encoding Genes in a Brazilian Intensive Care Unit. Front. Microbiol. 2019, 9, 3198. [Google Scholar] [CrossRef]
- Welp, A.L.; Bomberger, J.M. Bacterial Community Interactions During Chronic Respiratory Disease. Front. Cell. Infect. Microbiol. 2020, 10, 213. [Google Scholar] [CrossRef]
- Koulenti, D.; Tsigou, E.; Rello, J. Nosocomial pneumonia in 27 ICUs in Europe: Perspectives from the EU-VAP/CAP study. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1999–2006. [Google Scholar] [CrossRef]
- Cilloniz, C.; Martin-Loeches, I.; Garcia-Vidal, C.; San Jose, A.; Torres, A. Microbial Etiology of Pneumonia: Epidemiology, Diagnosis and Resistance Patterns. Int. J. Mol. Sci. 2016, 17, 2120. [Google Scholar] [CrossRef]
- Bakaletz, L.O. Viral–bacterial co-infections in the respiratory tract. Curr. Opin. Microbiol. 2017, 35, 30–35. [Google Scholar] [CrossRef]
- Gaibani, P.; Viciani, E.; Bartoletti, M.; Lewis, R.E.; Tonetti, T.; Lombardo, D.; Castagnetti, A.; Bovo, F.; Horna, C.S.; Ranieri, M.; et al. The lower respiratory tract microbiome of critically ill patients with COVID-19. Sci. Rep. 2021, 11, 10103. [Google Scholar] [CrossRef]
- Buchan, B.W.; Windham, S.; Balada-Llasat, J.; Leber, A.; Harrington, A.; Relich, R.; Murphy, C.; Bard, J.D.; Naccache, S.; Ronen, S.; et al. Practical Comparison of the BioFire FilmArray Pneumonia Panel to Routine Diagnostic Methods and Potential Impact on Antimicrobial Stewardship in Adult Hospitalized Patients with Lower Respiratory Tract Infections. J. Clin. Microbiol. 2020, 58, e00135-20. [Google Scholar] [CrossRef]
- Kumari, M.; Verma, S.; Venkatesh, V.; Gupta, P.; Tripathi, P.; Agarwal, A.; Siddiqui, S.S.; Arshad, Z.; Prakash, V. Emergence of blaNDM-1 and blaVIM producing Gram-negative bacilli in ventilator-associated pneumonia at AMR Surveillance Regional Reference Laboratory in India. PLoS ONE 2021, 16, e0256308. [Google Scholar] [CrossRef]
- Kamel, N.A.; Elsayed, K.M.; Awad, M.F.; Aboshanab, K.; El Borhamy, M. Multimodal Interventions to Prevent and Control Carbapenem-Resistant Enterobacteriaceae and Extended-Spectrum β-Lactamase Producer-Associated Infections at a Tertiary Care Hospital in Egypt. Antibiotics 2021, 10, 509. [Google Scholar] [CrossRef]
- Mabrouk, S.S.; Abdellatif, G.R.; El-Ansary, M.R.; Aboshanab, K.M.; Ragab, Y.M. Carbapenemase Producers Among Extensive Drug-Resistant Gram-Negative Pathogens Recovered from Febrile Neutrophilic Patients in Egypt. Infect. Drug Resist. 2020, 13, 3113–3124. [Google Scholar] [CrossRef]
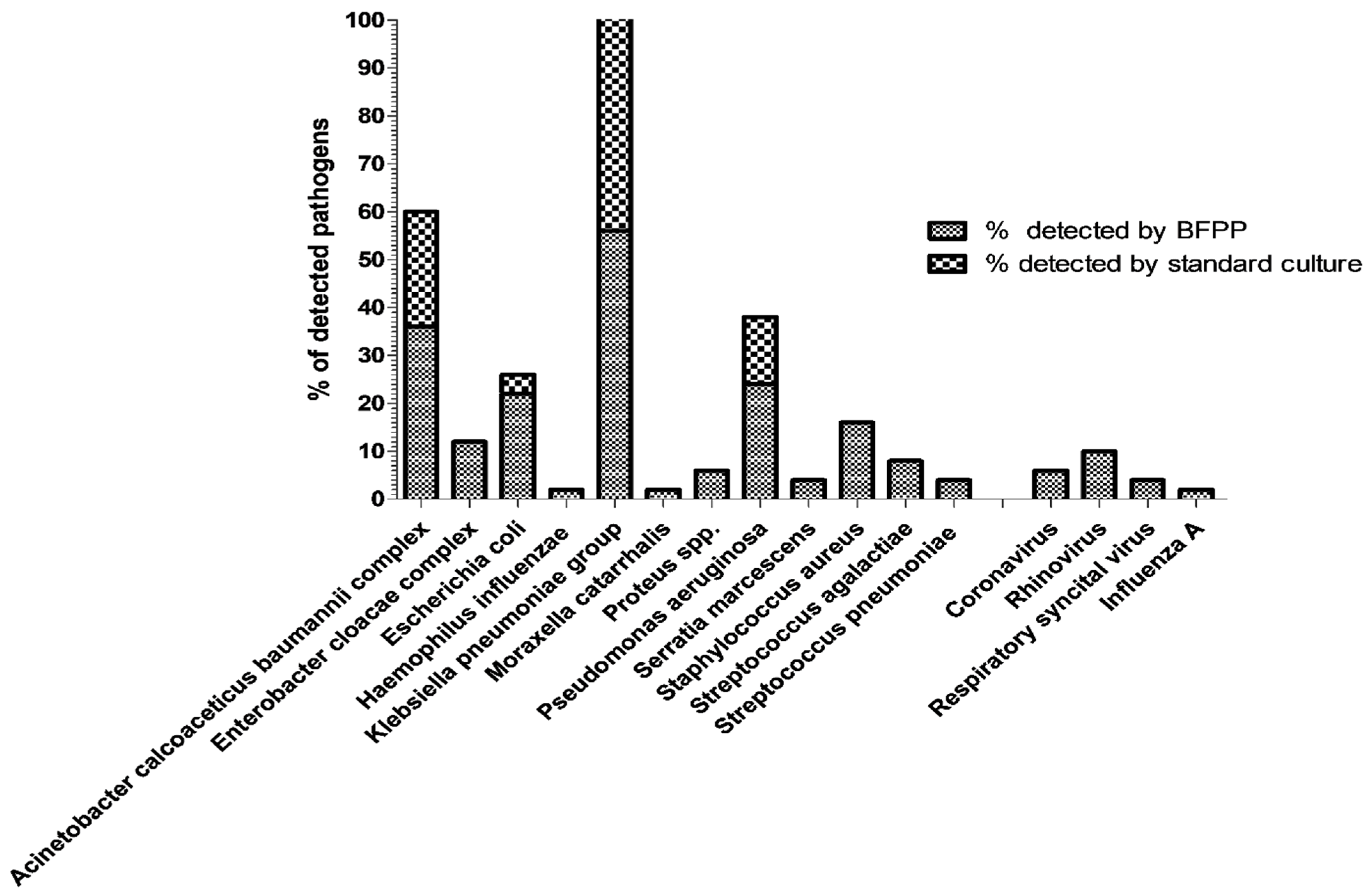
| BFPP Target Organisms (*) | mBAL Specimens [No of BFPP Detections/No of Standard Culture Detections] | |||||||
|---|---|---|---|---|---|---|---|---|
| [+/+]; True Positive | [+/−]; False Positive | [−/+]; False Negative | [−/−]; True Negative | PPA a %; [95%CI] | NPA b %; [95%CI] | OPA%; [95%CI] | ||
| Bacterial targets | Acinetobacter calcoaceticus baumannii complex | 12 | 6 | 0 | 32 | 100%; [72–100] | 84.2%; [69–93] | 88%; [76–95] |
| Enterobacter cloacae complex | 0 | 6 | 0 | 44 | NA | 88%; [76–95] | 88%; [76–95] | |
| Escherichia coli | 2 | 9 | 0 | 39 | 100%; [29–100] | 81%; [68–90] | 82%; [69–90] | |
| Haemophilus influenzae | 0 | 1 | 0 | 49 | NA | 98%; [88.5–100] | 98%; [88.5–100] | |
| Klebsiella pneumoniae group | 23 | 5 | 0 | 22 | 100%; [83–100] | 81.4%; [63–92] | 50%; [37–63] | |
| Moraxella catarrhalis | 0 | 1 | 0 | 49 | NA | 98%; [88.5–100] | 98%; [88.5–100] | |
| Proteus spp. | 0 | 3 | 0 | 47 | NA | 94%; [83–99] | 94%; [83–99] | |
| Pseudomonas aeruginosa | 7 | 5 | 0 | 38 | 100%; [60–100] | 88.3%; [75–95.3] | 90%; [78.2–96] | |
| Serratia marcescens | 0 | 2 | 0 | 48 | NA | 96%; [86–100] | 96%; [86–100] | |
| Staphylococcus aureus | 0 | 8 | 0 | 42 | NA | 84%; [71.2–92] | 84%; [71.2–92] | |
| Streptococcus agalactiae | 0 | 4 | 0 | 46 | NA | 92%; [81–97] | 92%; [81–97] | |
| Streptococcus pneumoniae | 0 | 2 | 0 | 48 | NA | 96%; [86–100] | 96%; [86–100] | |
| Total bacterial pathogens | 44 | 52 | 0* | 484 | 100; [90–100] | 90%; [87.4–92.5] | 91%; [88.4–93.1] | |
| BFPP Target Organisms Antimicrobial Resistance Genes | mBAL Specimens [No of Results for BFPP/VITEK2 Antimicrobial Breakpoints] | |||||||
|---|---|---|---|---|---|---|---|---|
| [+/+] | [+/−] | [−/+] | [−/−] | PPA %; [95%CI] | NPA%; [95%CI] | OPA%; [95%CI] | ||
| Carbapenemase producing Gram negative bacilli A | IMP | 0 | 1 | 0 | 49 | NA | 98%; [88.5–100] | 98%; [88.5–100] |
| KPC | 5 | 0 | 0 | 45 | 100%; [51–100] | 100%; [91–100] | 100%; [91.4–100] | |
| NDM | 21 | 3 | 0 | 26 | 100%; [82–100] | 90%; [73–97] | 94%; [83–99] | |
| OXA-48 | 16 | 1 | 0 | 33 | 100%; [77–100] | 97%; [84–100] | 98%; [88.5–100] | |
| VIM | 4 | 1 | 0 | 45 | 100%; [45–100] | 98%; [88–100] | 98%; [88.5–100] | |
| Total carbapenamase producers | 46 | 6 | 0 | 198 | 100%; [91–100] | 97%; [94–99] | 98%; [95–99] | |
| ESBL producing bacteria B | CTX-M | 28 | 3 | 2 | 17 | 93%; [78–99] | 85%; [63–96] | 90%; [78.2–96] |
| Methicillin resistant Staphylococcus aureus C | MecA/C and MREJ | 0 | 5 | 0 | 45 | NA | 90%; [78–96] | 90%; [78.2–96] |
| Total no of detected resistant genes | 74 | 14 | 2 | 260 | 97%; [90–100] | 95%; [91.5–97] | 95%; [93–97] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamel, N.A.; Alshahrani, M.Y.; Aboshanab, K.M.; El Borhamy, M.I. Evaluation of the BioFire FilmArray Pneumonia Panel Plus to the Conventional Diagnostic Methods in Determining the Microbiological Etiology of Hospital-Acquired Pneumonia. Biology 2022, 11, 377. https://doi.org/10.3390/biology11030377
Kamel NA, Alshahrani MY, Aboshanab KM, El Borhamy MI. Evaluation of the BioFire FilmArray Pneumonia Panel Plus to the Conventional Diagnostic Methods in Determining the Microbiological Etiology of Hospital-Acquired Pneumonia. Biology. 2022; 11(3):377. https://doi.org/10.3390/biology11030377
Chicago/Turabian StyleKamel, Noha A., Mohammad Y. Alshahrani, Khaled M. Aboshanab, and Mervat I. El Borhamy. 2022. "Evaluation of the BioFire FilmArray Pneumonia Panel Plus to the Conventional Diagnostic Methods in Determining the Microbiological Etiology of Hospital-Acquired Pneumonia" Biology 11, no. 3: 377. https://doi.org/10.3390/biology11030377
APA StyleKamel, N. A., Alshahrani, M. Y., Aboshanab, K. M., & El Borhamy, M. I. (2022). Evaluation of the BioFire FilmArray Pneumonia Panel Plus to the Conventional Diagnostic Methods in Determining the Microbiological Etiology of Hospital-Acquired Pneumonia. Biology, 11(3), 377. https://doi.org/10.3390/biology11030377







