Simple Summary
Terpenoid phenazines generally produced in Streptomyces exhibit potential antitumor and antibacterial activities. In this study, we designed and constructed an artificial biosynthetic pathway for the synthesis of terpenoid phenazines in Pseudomonas chlororaphis P3. We successfully synthesized endophenazine A and endophenazine A1 for the first time in Pseudomonas by introducing the prenyltransferase PpzP. Moreover, we revealed the biosynthetic pathway of endophenazine A1 in P. chlororaphis P3. This study enriches the diversity of phenazines in P. chlororaphis P3 and provides a reference for the heterologous synthesis of terpenoid phenazines.
Abstract
Endophenazine A is a terpenoid phenazine with phenazine-1-carboxylic acid (PCA), and dimethylallyl diphosphate (DMAPP) derived from the 2-methyl-D-erythritol-4-phosphate (MEP) pathway as the precursor, which shows good antimicrobial activity against several Gram-positive bacteria and fungi. However, the highest yield of endophenazine A was about 20 mg/L in Streptomyces, limiting its large-scale industrial development. Pseudomonas chlororaphis P3, possessing an efficient PCA synthesis and MEP pathways, is a suitable chassis to synthesize endophenazine A. Herein, we designed an artificial biosynthetic pathway for the synthesis of endophenazine A in P. chlororaphis P3. Primarily, the prenyltransferase PpzP from Streptomyces anulatus 9663 was introduced into P. chlororaphis P3 and successfully synthesized endophenazine A. Another phenazine compound, endophenazine A1, was discovered and identified as a leakage of the intermediate 4-hydroxy-3-methyl-2-butene pyrophosphate (HMBPP). Finally, the yield of endophenazine A reached 279.43 mg/L, and the yield of endophenazine A1 reached 189.2 mg/L by metabolic engineering and medium optimization. In conclusion, we successfully synthesized endophenazine A and endophenazine A1 in P. chlororaphis P3 for the first time and achieved the highest titer, which provides a reference for the heterologous synthesis of terpenoid phenazines.
1. Introduction
Phenazines are a class of important nitrogen-containing heterocyclic compounds that are mainly secreted by Pseudomonas and Streptomyces [1]. Recently, they have attracted significant attention due to their widespread applications in agriculture, medicine, and industry [2,3,4,5]. Although phenazines can be chemically synthesized, these methods have many drawbacks, such as the generation of toxic by-products, low yields, and difficulty in purification [6,7]. Therefore, the biosynthesis of phenazines by green and environmentally friendly microbial fermentation is considered a promising choice.
Terpenoid phenazines show antitumor and antibacterial activities and are potential cancer therapeutic agents (Table S1) [3,8,9,10,11]. Endophenazine A is a C-prenylation phenazine of PCA, catalyzed by prenyltransferase PpzP, and possesses good antibacterial activity [12,13] (Figure 1). PpzP expresses a soluble protein and belongs to the ABBA prenyltransferase family due to its special α-β-β-α structure. In the absence of magnesium or other divalent cations, PpzP is able to accept the isopentenyl donor from dimethylallyl pyrophosphate (DMAPP), connecting C-1 of DMAPP and C-9 of 5,10-dihydrophenazine 1-carboxylate (dihydro-PCA) to form endophenazine A [12]. Endophenazine A is mainly occurred in Streptomyces, but the yield is quite low [14]. Generally, Streptomyces synthesized phenazine compounds using phenazine-1,6-dicarboxylic acid (PDC) as a precursor, such as griseolutein acid [15,16] and lomofungin [17], which is not a precursor for the synthesis of endophenazine A. Pseudomonas mainly use phenazine-1-carboxylic acid (PCA) as a precursor to synthesize phenazine derivatives, such as phenazine-1-carboxamide (PCN), 2-hydroxyphenazine (2-OH-PHZ), 1-hydroxyphenazine (1-OH-PHZ), and pyocyanin (PYO) [18,19,20,21,22]. Pseudomonas chlororaphis P3 is a high-yielding mutagenic strain for PCN production with complete phenazine synthetic gene cluster phzABCDEFG and phzH, encoding glutamine amidotransferase [18,23,24,25], and can be transformed into a good host for the synthesis of terpenoid phenazines.
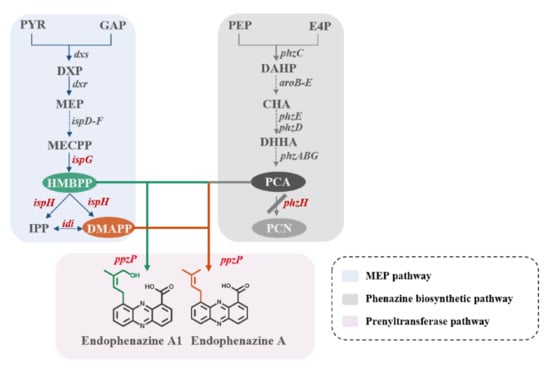
Figure 1.
Design of the biosynthetic pathway for endophenazines in Pseudomonas chlororaphis P3. The P. chlororaphis endogenous MEP pathway is shown in blue, the P. chlororaphis phenazine synthesis pathway is shown in grey, and the endophenazines pathway from S. anulatus 9663 is shown in pink. Genes: phzH, encoding glutamine amidotransferase; ispG, encoding 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase; ispH, encoding 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; idi, encoding isopentenyl diphosphate isomerase; and ppzP, encoding prenyltransferase.
In addition to PCA, DMAPP is also an essential precursor for the biosynthesis of endophenazine A, which is produced via two pathways: the 2-methyl-D-erythritol-4-phosphate (MEP) pathway and the mevalonate (MVA) pathway. Consistent with most bacteria, Pseudomonas only possesses the MEP pathway [26]. The MEP pathway, called the non-mevalonate (MVA) pathway, begins with pyruvate and glyceraldehyde 3-phosphate (GAP), and followed by continuous enzyme catalysis to synthesize 2-C-methyl-D-erythritol 2, 4-cyclodiphosphate (MECPP) [27]. Next, MECPP is catalyzed by 4-hydroxy-3-methyl-2-butene pyrophosphate (HMBPP) synthase (IspG) to HMBPP, which is reduced to isopentenyl pyrophosphate (IPP) and DMAPP by HMBPP reductase (IspH) [28]. In the last step, IPP and DMAPP can be reversibly converted by isopentenyl diphosphate isomerase (IDI) [29] (Figure 1). Due to the limitation of isoprene precursors, many studies have improved the concentration of isoprene by increasing the expression of key enzymes. For example, overexpression of idi enhanced the production of lycopene and β-carotene up to 4.5 and 2.7 folds, respectively [30]. In addition, the role of genes ispG and ispH in the MEP pathway have been elucidated and could be the important rate-limiting steps to synthesize secondary metabolites [31].
Based on the metabolic network of the phenazine biosynthesis pathway and MEP pathway, we designed an artificial biosynthetic pathway of endophenazines and successfully synthesized endophenazine A and endophenazine A1 in P. chlororaphis P3 for the first time (Figure 1). It was found that endophenazine A1 was a leakage of the intermediate HMBPP. Moreover, the yield of endophenazine A and endophenazine A1 were improved through metabolic engineering and medium optimization.
2. Materials and Methods
2.1. Strains, Plasmids, and Culture Conditions
All strains and plasmids are shown in Table 1, and primers are listed in Table S2. P. chlororaphis P3 was obtained by chemical mutagenesis of P. chlororaphis HT66 with a high production of PCN [24]. P. chlororaphis strains were grown in 5 mL Luria-Bertani (LB) broth and shake flask fermented in 60 mL King’s B medium (KB) and Y medium (37.08 mL/L glycerol, 20 g/L tryptone, 25.03 g/L soy peptone) at 28 °C, 200 rpm.

Table 1.
Strains and plasmids used in this study.
2.2. DNA Manipulation
Genes were deleted or integrated into the genome with a non-scar homologous recombination strategy in P. chlororaphis P3. First, the upstream and downstream fragment sequence of the target gene were ligated to the pK18mobsacB plasmid that contains the sucrose-sensitive sacB gene. Second, the recombinant plasmids were transformed into S17-1 (λplr) followed by P. chlororaphis P3. Finally, the resulting mutant strains were verified by DNA sequencing. The codon-optimized ppzP, ispG, ispH, and idi genes were ligated to the pBBR1MCS plasmid containing the lac promoter and transferred into P. chlororaphis by electroporation. The function and accession numbers of the genes are listed in Table S3.
2.3. Analytical Procedures for Phenazine Derivatives
During the fermentation process, 1 mL culture broth was taken every 12 h until 72 h. The extraction method of phenazine derivatives was consistent with that described by Deng et al. [32]. The mobile phase was 1‰ formic acid water and methanol. The compounds were separated by the gradient elution method (0−4 min, 20% methanol; 4−30 min, 50–95% methanol; 30−35 min, 20% methanol) with detection 254 nm.
2.4. Purification and Structural Identification of Phenazine Derivatives
Precisely, 5 L fermentation supernatant containing the target compound was adjusted to pH 2.0–3.0 with 6 M HCl, extracted with ethyl acetate and concentrated by evaporation at 40 °C. The crude phenazine extract from the supernatant was dissolved in 5-mL methanol and then passed through the Sephadex LH-20 column. The column was eluted with 10% methanol to remove impurities and eluted with 30% methanol to obtain the target compound. The 30% methanol eluate was collected separately in different tubes and analyzed by HPLC, and the eluate containing the target compound was mixed together. The filtered sample was separated by analytical HPLC (Agilent 1260 LC) with Agilent Eclipse XDB-C18. The purified compound was freeze-dried to form a solid powder. The purified compound was characterized by nuclear magnetic resonance (NMR) spectroscopy in the Instrumental Analysis Centre of Shanghai Jiao Tong University.
2.5. Statistical Analysis
All statistical figures were drawn using Prism software (GraphPad Software, La Jolla, CA, USA). All data presented were the average of three biological replicates and shown as the mean ± standard deviation.
3. Results
3.1. Construction of Biosynthetic Pathway for Endophenazine A in P. chlororaphis P3
PCA is a precursor for the synthesis of endophenazine A, thus gene phzH in P. chlororaphis P3 was knocked out to get the strain P3-A0. Then, ppzP was selected to connect the phenazine biosynthetic and MEP pathways and inserted into the original phzH locus of the chromosome in P3-A0 to construct the P3-A1 strain. To detect whether new phenazines were synthesized, we extracted the phenazines from the fermentation broth of the strains P3, P3-A0, and P3-A1, and analyzed them with HPLC. As shown in Figure 2A, the deletion of the phzH gene led to the accumulation of PCA instead of PCN in P3-A0, and the insertion of the ppzP gene resulted in a new compound A in strain P3-A1. The extract of the fermentation broth was then purified by the preparative HPLC, and purified compound A was obtained. Subsequently, the purified compound A was analyzed by UPLC-MS and NMR. The m/z value of compound A was 293.1289 [M + H]+, suggesting that the molecular formula of compound A is C18H16N2O2 and the calculated value is 293.1285 (Figure 2B). The proton and carbon chemical shifts of compound A are shown in Figure S1 and Table S4. The 1H NMR (400 MHz, DMSO-d6) spectra showed peaks at δ 8.61 (d, J = 7.0 Hz, H2), 8.11 (d, J = 7.9 Hz, H3), 8.51 (d, J = 8.7 Hz, H4), 7.87 (d, J = 6.8 Hz, H6), 7.98 (d, J = 7.8 Hz, H7), 8.17 (d, J = 8.1 Hz, H8), 3.37 (br, H12), 5.51 (t, J = 7.3 Hz, H13), 1.79 (s, H15), and 1.73 (s, H16). The 13C NMR (100 MHz, DMSO-d6) spectra showed peaks at δ 138.8 (C1), 132.1 (C2), 127.6 (C3), 134.8 (C4), 142.5 (C4a), 143.7 (5a), 130.5 (C6), 133.9 (C7), 131.1 (C8), 139.3 (C9), 139.4 (C9a), 127.5 (C10a), 166.1 (C11), 29.3 (C12), 121.6 (C13), 133.6(C14), 25.5 (C15), and 17.8 (C16). According to these results, the structure of compound A was determined to be endophenazine A (Figure 2C).
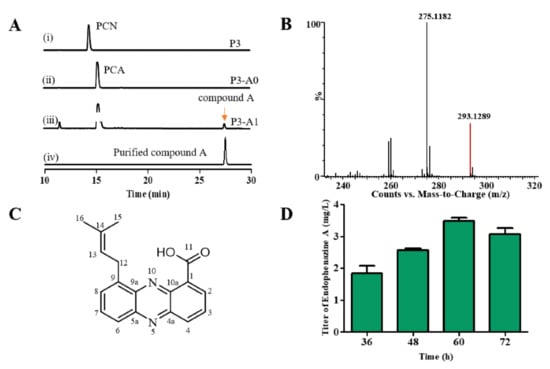
Figure 2.
Endophenazine A was synthesized by P. chlororaphis with inserted ppzP gene. (A) HPLC profiles of P3, P3-A0, and P3-A1 strains fermentation broth and purified compound A. (B) The UPLC-MS result of compound A. (C) The structure of compound A. (D) The concentration of compound A in P3-A1 in different culture time. P3: a mutant from P. chlororaphis HT66; P3-A0: deletion of phzH in P3; P3-A1: introduction of ppzP in the chromosome of P3.
3.2. Overexpression of ppzP Gene Enhanced the Production of Endophenazine A
Although the strain P3-A1 synthesized endophenazine A, its highest titer was only 3.5 mg/L at 60 h (Figure 2D). Since there was only a single copy of the ppzP gene in the genome, prenyltransferase PpzP might be a rate-limiting step in synthesizing endophenazine A. Therefore, the ppzP gene was overexpressed under the control of lac promoter in P3-A1 to get the strain P3-A2, resulting in the titer of 28.64 mg/L endophenazine A, which was 7.17-times higher than that in the control strain P3-A3, and the titer/DCW of endophenazine A was increased from 0.40 mg/g in P3-A3 to 2.96 mg/g in P3-A2 (Figure 3A). This result indicated that ppzP is a rate-limiting step, and increasing its expression level can promote the production of endophenazine A.
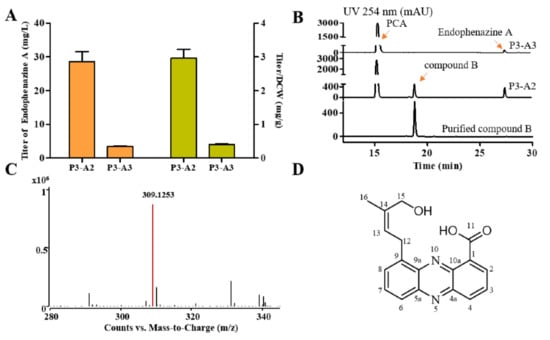
Figure 3.
Influence of overexpression of ppzP on the titer of endophenazines. (A) The concentration of endophenazine A in strain P3-A2 and P3-A3. (B) HPLC profiles of P3-A2 and P3-A3 strains fermentation broth and purified compound B. (C) The UPLC-MS result of compound B. (D) The structure of compound B. P3-A2: overexpression of ppzP in P3-A1; P3-A3: integration of pBBR1MCS plasmid in P3-A1.
3.3. Purification and Structural Identification of the Compound B
In addition to endophenazine A, another compound B in the strain P3-A2 was also detected (Figure 3B). Then, the purified compound B was analyzed by UPLC-MS and NMR. The m/z value of compound B is 309.1253 [M + H]+, suggesting that the molecular formula of compound B is C18H16N2O3 and the calculated value is 309.1234 (Figure 3C). The proton and carbon chemical shifts of compound B are shown in Figure S2 and Table S5. The 1H NMR (400 MHz, DMSO-d6) spectra showed peaks at δ 8.56 (dd, J = 7.0, 1.4 Hz, H2), 8.06 (dd, J = 8.7, 7.0 Hz, H3), 8.45 (dd, J = 8.7, 1.4 Hz, H4), 7.85 (dd, J = 6.8, 1.3 Hz, H6), 7.95 (dd, J = 8.8, 6.9 Hz, H7), 8.12 (dd, J = 8.7, 1.4 Hz, H8), 3.99 (d, J = 7.2 Hz H12), 5.71 (m, H13), 3.86 (s, H15), 4.68 (s, -OH15), and 1.76 (s, H16). The 13C NMR (100 MHz, DMSO-d6) spectra showed peaks at δ 137.9 (C1), 131.9 (C2), 127.4 (C3), 134.5 (C4), 142.2 (C4a), 144.3 (C5a), 130.4 (C6), 133.6 (C7), 130.9 (C8), 138.5 (C9), 139.1 (C9a), 127.4 (C10a), 166.0 (C11), 28.4 (C12), 120.2 (C13), 138.9(C14), 66.1 (C15), and 13.7 (C16). The structure of compound B was finally determined to be endophenazine A1 (Figure 3D).
3.4. HMBPP Is the Precursor for the Synthesis of Endophenazine A1
Endophenazine A1 was first isolated from Kitasatospora sp. HKI 714 [33], but its synthesis mechanism has not been elucidated. Since endophenazine A and endophenazine A1 are similar in structure, we first assumed that endophenazine A1 is an intermediate, which can be further reduced to endophenazine A, or endophenazine A might be unstable and can be oxidized to endophenazine A1. To test this hypothesis, we cultured strain P3-A0 in KB medium containing endophenazine A and endophenazine A1, respectively. However, the concentration of endophenazine A and endophenazine A1 did not decrease significantly after 60 h of incubation (Figure 4A,B). These results showed that endophenazine A1 and endophenazine A cannot interconvert in P. chlororaphis P3. Our other assumption was that the leakage of intermediate might cause endophenazine A1. In the MEP pathway, the intermediate HMBPP is almost the same as the isoprene unit of the endophenazine A1, so HMBPP may be the precursor of endophenazine A1. In order to investigate the effect of HMBPP concentration on the production of endophenazine A1, we co-expressed ispG and ispH with ppzP to obtain strains P3-A4 and P3-A5, respectively. Compared with strain P3-A2, the concentration of endophenazine A1 in strain P3-A4 increased by 62%, and the titer/DCW of endophenazine A1 was increased from 4.63 mg/g in P3-A2 to 7.32 mg/g in P3-A4. On the contrary, strain P3-A5 hardly synthesized endophenazine A1 (Figure 4C). The overexpression of the ispG gene had less effect on cell growth, while overexpression of the ispH gene had a significant inhibition on cell growth (Figure 4D). The above results revealed that HMBPP is the precursor for the synthesis of endophenazine A1, which provides different metabolic engineering strategies for subsequent improvement in the production of endophenazine A and endophenazine A1.
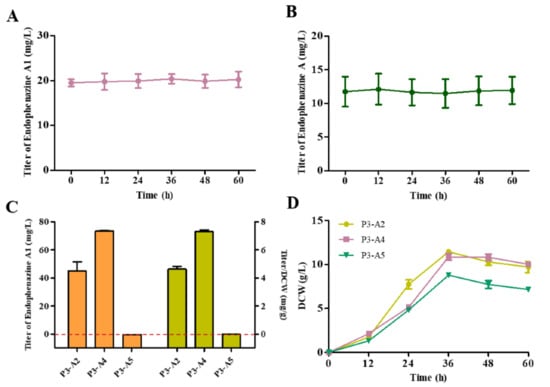
Figure 4.
The synthesis of endophenazine A1 in P. chlororaphis P3. (A) The stability of endophenazine A1 in P. chlororaphis P3. (B) The stability of endophenazine A in P. chlororaphis P3. (C) The production of endophenazine A1 in strain P3-A2, P3-A4, and P3-A5. (D) The growth curve of P3-A2, P3-A4, and P3-A5. P3-A2: overexpression of ppzP in P3-A1; P3-A4: co-overexpression of ppzP and ispG in P3-A1; P3-A5: co-overexpression of ppzP and ispH in P3-A1.
3.5. Modification of Metabolic Pathways to Increase the Production of Endophenazine A
To improve the production of endophenazine A, a rational strategy was designed to drive the carbon flux to the biosynthesis of endophenazine A. Firstly, the rate-limiting gene idi and ppzP was co-expressed in P3-A1 to get the strain P3-A6. As seen in Figure 5A, the production of endophenazine A in P3-A6 was 90.61 mg/L, which was 24.89 times higher than that in P3-A1, and the titer/DCW of endophenazine A was increased from 0.47 mg/g in P3-A1 to 10.04 mg/g in P3-A6. To minimize the leakage of HMBPP, ispH was co-expressed with ppzP and idi in strain P3-A1 obtaining the strain P3-A7. The production of endophenazine A was 94.14 mg/L in P3-A7, which is 3.9% higher than P3-A6 (Figure 5A). The overexpression of these genes had no significant effect on cell growth (Figure 5B). To further increase the production of endophenazine A, P3-A7 was cultured in the optimized Y medium, in which the concentration of endophenazine A was 279.43 mg/L, and the titer/DCW of endophenazine A was increased from 9.42 mg/g in KB medium to 13.75 mg/g in KB medium (Figure 5C). The reason may be that rich nutrients lead to high biomass (Figure 5D).
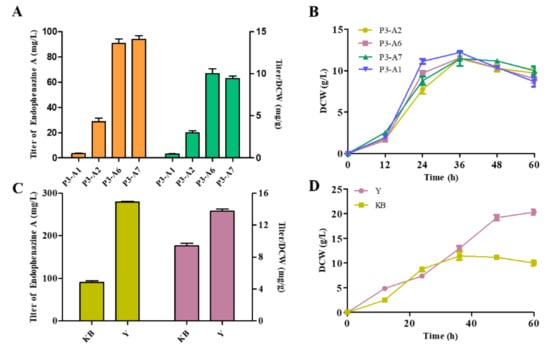
Figure 5.
Effects of metabolic regulation and medium optimization on endophenazine A concentration (A,C) and cell growth (B,D) in P. chlororaphis P3. P3-A1: introduction of ppzP in the chromosome of P3; P3-A2: overexpression of ppzP in P3-A1; P3-A6: co-overexpression of ppzP and idi in P3-A1; P3-A7: co-overexpression of ppzP, idi, and ispH in P3-A1.
3.6. Modification of Metabolic Pathways to Increase the Production of Endophenazine A1
It has been proven that endophenazine A1 was a shunt pathway of endophenazine A. Therefore, we attempted to increase the production of endophenazine A1 by reducing the consumption of HMBPP and promoting the synthesis of HMBPP. The ispH gene is known to be necessary for bacterial growth and survival and cannot be deleted [34]. Therefore, we could not delete the ispH gene to block carbon flux from HMBPP to DMAPP. Previous results proved that overexpression of the idi gene promoted the flow of carbon flux to produce DMAPP, so we knocked out the idi gene in strain P3-A2 and obtained the strain P3-A8 to reduce the accumulation of DMAPP and increase the accumulation of HMBPP. As shown in Figure 6A, the production of endophenazine A1 in P3-A8 was 63.6 mg/L, which increased by 41% compared with P3-A2, and the titer/DCW of endophenazine A1 was increased from 4.63 mg/g in P3-A2 to 6.34 mg/g in P3-A8. Then, in order to increase the supply of precursor HMBPP, the ispG gene was co-overexpressed with ppzp in strain P3-A8 to get P3-A9, resulting in the production titer of 79.24 mg/L endophenazine A1, which was 24% higher than that of strain P3-A8, and the titer/DCW of endophenazine A1 was increased from 6.34 mg/g in P3-A8 to 7.45 mg/g in P3-A9 (Figure 6A). At the same time, these gene manipulations had no significant effect on cell growth (Figure 6B). Finally, the titer of endophenazine A1 in the Y medium of strain P3-A9 increased to 189.2 mg/L, which was 2.4 folds in the KB medium, and the titer/DCW of endophenazine A1 was increased from 7.45 mg/g in KB medium to 9.81 mg/g in KB medium (Figure 6C). The reason may be similar to the high production of endophenazine A (Figure 6D).
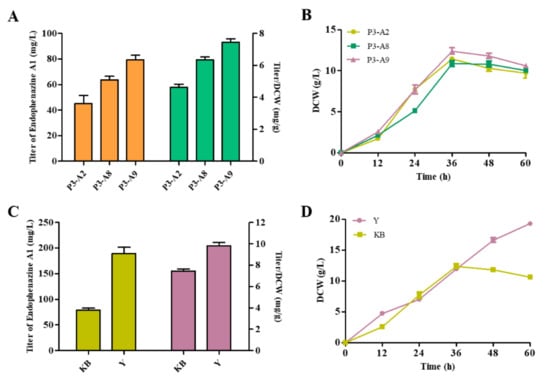
Figure 6.
Effects of metabolic regulation and medium optimization on endophenazine A1 production (A,C) and cell growth (B,D) in P. chlororaphis P3. P3-A2: overexpression of ppzP in P3-A1; P3-A8: deletion of idi in P3-A2; P3-A9: co-overexpression of ppzP and ispG in P3-A8.
4. Discussion
Endophenazine A is a C-prenylation phenazine catalyzed by prenyltransferases PpzP from S. anulatus 9663 or EpzP from S. cinnamonensis DSM 1042; the conversion rate of PpzP is higher than that of EpzP10 [35]. Although the production of endophenazine A was not reported in these two Streptomyces, the highest yield of endophenazine A was achieved in the engineered S. coelicolor M512, which heterologously expressed the whole gene cluster of endophenazine was only 20 mg/L [14]. Due to the fast growth and clear genetic background of P. chlororaphis P3 [36], the high yield of precursor compound PCA and endogenous MEP pathway, we designed an artificial biosynthetic pathway to produce endophenazine A in P. chlororaphis P3.
Besides endophenazine A, endophenazine A1 was synthesized (Figure 3B). Daniel et al. isolated endophenazine A1 in Kitasatospora sp. HKI 714, which showed antibacterial activity against Mycobacterium fortuitum and Mycobacterium aureus [33]. Later, it was reported that endophenazine A1 and endophenazine A were also detected in Kitasatospora sp. MBT66 [37,38]. However, the synthesis mechanism of endophenazine A1 was unclear. In this study, we found that endophenazine A1 and endophenazine A are relatively stable in the culture of P. chlororaphis P3 and cannot be converted to each other, indicating that endophenazine A1 is not an intermediate in the synthesis of endophenazine A and endophenazine A cannot be oxidized to form endophenazine A1 (Figure 4A,B). Then, we found that overexpression of the ispG gene enhanced the accumulation of endophenazine A1, while overexpression of the ispH gene reduced the production of endophenazine A1 (Figure 4C). These results are consistent with previous studies in which the overexpression of the ispG gene led to the enhancement of HMBPP, while overexpression of the ispH gene led to the reduction of HMBPP [31]. These results revealed that HMBPP is a key switch to control the metabolic flux toward endophenazine A and shunt products endophenazine A1.
Accordingly, we used metabolic engineering strategies to regulate the carbon flow of DMAPP and HMBP to achieve high production of endophenazine A and endophenazine A1, respectively. Firstly, we overexpressed the key genes of ppzP, idi, and ispH of the endophenazine A pathway in P3-A1, and the production of endophenazine A was significantly increased (Figure 5A). This result is consistent with the previous report, where the ispH gene catalyzed HMBPP into IPP and DMAPP at a ratio of 6.3:1, but it reduced up to 1:3.1 after treatment with gene idi [39]. Subsequently, the ispG and ppzP genes were co-overexpressed in P3-A8, and the concentration of endophenazine A1 increased by 80% (Figure 6A). This result is consistent with previous studies in which the overexpression of the ispG gene led to the enhancement of HMBPP. Finally, the concentration of endophenazine A and endophenazine A1 in the engineered P. chlororaphis P3 was 279.43 mg/L and 189.2 mg/L, respectively, which were remarkably enhanced with the stepwise metabolic engineering and medium optimization (Figure 7).
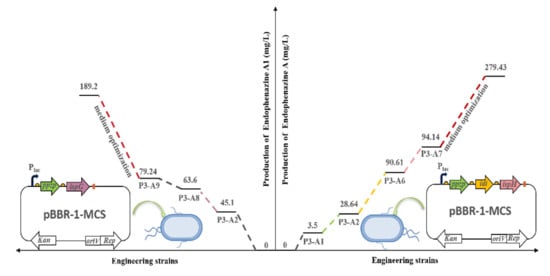
Figure 7.
Summary of metabolic engineering of P. chlororaphis P3 and medium optimization for stepwise increasing the titer of endophenazines. Left, concentration of endophenazine A1. Right, concentration of endophenazine A. P3-A1: introduction of ppzP in the chromosome of P3; P3-A2: overexpression of ppzP in P3-A1; P3-A3: integration of pBBR1MCS plasmid in P3-A1; P3-A4: co-overexpression of ppzP and ispG in P3-A1; P3-A5: co-overexpression of ppzP and ispH in P3-A1; P3-A6: co-overexpression of ppzP and idi in P3-A1; P3-A7: co-overexpression of ppzP, idi, and ispH in P3-A1; P3-A8: deletion of idi in P3-A2; P3-A9: co-overexpression of ppzP and ispG in P3-A8.
Protein engineering is a widely used effective strategy to improve enzyme-substrate specificity and catalytic efficiency. In the endophenazine A biosynthesis pathway, the catalytic efficiency of EpzP is 14 times higher than that of the wild-type enzyme through rational design [40]. Edward et al. realized the conversion of transketolase substrate specificity to fatty aldehyde through directed evolution [41]. This is a reference to increase the yield and diversity of terpenoid phenazines in the future.
5. Conclusions
In this study, two terpenoid phenazine pathways were designed and constructed in P. chlororaphis P3, resulting in the production of 279.43 mg/L endophenazine A and 189.2 mg/L endophenazine A1, respectively, through rational metabolic engineering strategies, which is the highest titers in microbial production of endophenazine A and endophenazine A1 up to now. This study enriched the diversity of phenazine compounds synthesized by P. chlororaphis P3 and laid the foundation for the future synthesis and large-scale production of other terpenoid phenazines.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11030363/s1, Figure S1: 1H NMR (a) and 13C NMR (b) spectra of compound A, Figure S2: 1H NMR (a) and 13C NMR (b) spectra of compound B; Table S1: Terpenoid phenazines, Table S2: The primers sequence used in this study, Table S3: The function and accession numbers of the genes, Table S4: 1H and 13C NMR spectral data of compound A in DMSO-d6, Table S5: 1H and 13C NMR spectral data of compound B in DMSO-d6. References in the supporting information are listed in the manuscript.
Author Contributions
Conceptualization, Y.L. and X.Z.; methodology, Y.L. and S.Y.; validation, Y.L.; formal analysis, Y.L.; investigation, Y.L.; data curation, Y.L.; writing—original draft preparation, Y.L.; writing—review and editing, X.Z., M.B. and M.J.; project administration, W.W. and H.H.; funding acquisition, X.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Scientific Research Projects (2019YFA0904300) and the National Natural Science Foundation of China (No. 32070051).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful to the Instrumental Analysis Center of Shanghai Jiao Tong University for their expert technical assistance in NMR.
Conflicts of Interest
The authors declare no competing financial interest.
References
- Mavrodi, D.V.; Peever, T.L.; Mavrodi, O.V.; Parejko, J.A.; Raaijmakers, J.M.; Lemanceau, P.; Mazurier, S.; Heide, L.; Blankenfeldt, W.; Weller, D.M. Diversity and evolution of the phenazine biosynthesis pathway. Appl. Environ. Microbiol. 2010, 76, 866–879. [Google Scholar] [CrossRef] [PubMed]
- Pierson, L.S.; Pierson, E.A. Metabolism and function of phenazines in bacteria: Impacts on the behavior of bacteria in the environment and biotechnological processes. Appl. Microbiol. Biotechnol. 2010, 86, 1659–1670. [Google Scholar] [CrossRef] [PubMed]
- Laursen, J.B.; Nielsen, J. Phenazine natural products: Biosynthesis, synthetic analogues, and biological activity. Chem. Rev. 2004, 104, 1663–1686. [Google Scholar] [CrossRef] [PubMed]
- Ligon, J.M.; Hill, D.S.; Hammer, P.E.; Torkewitz, N.R.; Hofmann, D.; Kempf, H.J.; Pée, K.H.v. Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag. Sci. 2000, 56, 688–695. [Google Scholar] [CrossRef]
- Makgatho, M.E.; Anderson, R.; O’Sullivan, J.F.; Egan, T.J.; Freese, J.A.; Cornelius, N.; van Rensburg, C.E. Tetramethylpiperidine-substituted phenazines as novel anti-plasmodial agents. Drug Dev. Res. 2000, 50, 195–202. [Google Scholar] [CrossRef]
- Cheluvappa, R. Standardized chemical synthesis of Pseudomonas aeruginosa pyocyanin. MethodsX 2014, 1, 67–73. [Google Scholar] [CrossRef][Green Version]
- Elhady, H.A.; El-Mekawy, R.E.; Fadda, A. Valuable Chemistry of Phenazine Derivatives: Synthesis, Reactions and, Applications. Polycycl. Aromat. Compd. 2020, 1–33. [Google Scholar] [CrossRef]
- Kondratyuk, T.P.; Park, E.-J.; Yu, R.; Van Breemen, R.B.; Asolkar, R.N.; Murphy, B.T.; Fenical, W.; Pezzuto, J.M. Novel marine phenazines as potential cancer chemopreventive and anti-inflammatory agents. Mar. Drugs 2012, 10, 451–464. [Google Scholar] [CrossRef]
- Cimmino, A.; Evidente, A.; Mathieu, V.; Andolfi, A.; Lefranc, F.; Kornienko, A.; Kiss, R. Phenazines and cancer. Nat. Prod. Rep. 2012, 29, 487–501. [Google Scholar] [CrossRef]
- Gebhardt, K.; Schimana, J.; Krastel, P.; Dettner, K.; Rheinheimer, J.; Zeeck, A.; Fiedler, H.-P. Endophenazines A–D, New Phenazine Antibiotics from the Arthropod Associated Endosymbiont Streptomyces anulatus I. Taxonomy, Fermentation, Isolation and Biological Activities. J. Antibiot. 2002, 55, 794–800. [Google Scholar] [CrossRef]
- Omura, S.; Eda, S.; Funayama, S.; Komiyama, K.; Takahashi, Y.; Woodruff, H.B. Studies on a novel antitumor antibiotic, phenazinomycin: Taxonomy, fermentation, isolation, and physicochemical and biological characteristics. J. Antibiot. 1989, 42, 1037. [Google Scholar] [CrossRef]
- Saleh, O.; Gust, B.; Boll, B.; Fiedler, H.-P.; Heide, L. Aromatic prenylation in phenazine biosynthesis: Dihydrophenazine-1-carboxylate dimethylallyltransferase from Streptomyces anulatus. J. Biol. Chem. 2009, 284, 14439–14447. [Google Scholar] [CrossRef] [PubMed]
- Krastel, P.; Zeeck, A.; Gebhardt, K.; Fiedler, H.-P.; Rheinheimer, J. Endophenazines A–D, new phenazine antibiotics from the athropod associated endosymbiont Streptomyces anulatus II. Structure elucidation. J. Antibiot. 2002, 55, 801–806. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saleh, O.; Flinspach, K.; Westrich, L.; Kulik, A.; Gust, B.; Fiedler, H.-P.; Heide, L. Mutational analysis of a phenazine biosynthetic gene cluster in Streptomyces anulatus 9663. Beilstein J. Org. Chem. 2012, 8, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Giddens, S.R.; Feng, Y.; Mahanty, H.K. Characterization of a novel phenazine antibiotic gene cluster in Erwinia herbicola Eh1087. Mol. Microbiol. 2002, 45, 769–783. [Google Scholar] [CrossRef]
- Shi, Y.-M.; Brachmann, A.O.; Westphalen, M.A.; Neubacher, N.; Tobias, N.J.; Bode, H.B. Dual phenazine gene clusters enable diversification during biosynthesis. Nat. Chem. Biol. 2019, 15, 331–339. [Google Scholar] [CrossRef]
- Zhang, C.; Sheng, C.; Wang, W.; Hu, H.; Peng, H.; Zhang, X. Identification of the lomofungin biosynthesis gene cluster and associated flavin-dependent monooxygenase gene in Streptomyces lomondensis S015. PLoS ONE 2015, 10, e0136228. [Google Scholar] [CrossRef][Green Version]
- Chin-A-Woeng, T.F.; Thomas-Oates, J.E.; Lugtenberg, B.J.; Bloemberg, G.V. Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol. Plant-Microbe Interact. 2001, 14, 1006–1015. [Google Scholar] [CrossRef]
- Delaney, S.M.; Mavrodi, D.V.; Bonsall, R.F.; Thomashow, L.S. phzO, a gene for biosynthesis of 2-hydroxylated phenazine compounds in Pseudomonas aureofaciens 30-84. J. Bacteriol. 2001, 183, 318–327. [Google Scholar] [CrossRef][Green Version]
- Greenhagen, B.T.; Shi, K.; Robinson, H.; Gamage, S.; Bera, A.K.; Ladner, J.E.; Parsons, J.F. Crystal structure of the pyocyanin biosynthetic protein PhzS. Biochemistry 2008, 47, 5281–5289. [Google Scholar] [CrossRef]
- Parsons, J.F.; Greenhagen, B.T.; Shi, K.; Calabrese, K.; Robinson, H.; Ladner, J.E. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry 2007, 46, 1821–1828. [Google Scholar] [CrossRef] [PubMed]
- Mavrodi, D.V.; Blankenfeldt, W.; Thomashow, L.S. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 2006, 44, 417–445. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Zhang, P.; Bilal, M.; Wang, W.; Hu, H.; Zhang, X. Enhanced biosynthesis of phenazine-1-carboxamide by engineered Pseudomonas chlororaphis HT66. Microb. Cell Fact. 2018, 17, 117. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.-J.; Peng, H.-S.; Hu, H.-B.; Huang, X.-Q.; Wang, W.; Zhang, X.-H. iTRAQ-based quantitative proteomic analysis reveals potential factors associated with the enhancement of phenazine-1-carboxamide production in Pseudomonas chlororaphis P3. Sci. Rep. 2016, 6, 27393. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Ksenzenko, V.N.; Bonsall, R.F.; Cook, R.J.; Boronin, A.M.; Thomashow, L.S. A seven-gene locus for synthesis of phenazine1-carboxylic acid by Pseudomonas fluorescens 2-79. J. Bacteriol. 1998, 180, 2541–2548. [Google Scholar] [CrossRef]
- Hernandez-Arranz, S.; Perez-Gil, J.; Marshall-Sabey, D.; Rodriguez-Concepcion, M. Engineering Pseudomonas putida for isoprenoid production by manipulating endogenous and shunt pathways supplying precursors. Microb. Cell Fact. 2019, 18, 152. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Wiberley, A.E.; Donohue, A.R. Isoprene emission from plants: Why and how. Ann. Bot. 2008, 101, 5–18. [Google Scholar] [CrossRef]
- Zhao, L.; Chang, W.-C.; Xiao, Y.; Liu, H.-W.; Liu, P. Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu. Rev. Biochem. 2013, 82, 497–530. [Google Scholar] [CrossRef]
- Banerjee, A.; Sharkey, T. Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat. Prod. Rep. 2014, 31, 1043–1055. [Google Scholar] [CrossRef]
- Albrecht, M.; Misawa, N.; Sandmann, G. Metabolic engineering of the terpenoid biosynthetic pathway of Escherichia coli for production of the carotenoids β-carotene and zeaxanthin. Biotechnol. Lett. 1999, 21, 791–795. [Google Scholar] [CrossRef]
- Li, Q.; Fan, F.; Gao, X.; Yang, C.; Bi, C.; Tang, J.; Liu, T.; Zhang, X. Balanced activation of IspG and IspH to eliminate MEP intermediate accumulation and improve isoprenoids production in Escherichia coli. Metab. Eng. 2017, 44, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.X.; Zhang, Z.; Li, H.L.; Wang, W.; Hu, H.B.; Zhang, X.H. Identification of a Novel Bioactive Phenazine Derivative and Regulation of phoP on Its Production in Streptomyces lomondensis S015. J. Agric. Food Chem. 2021, 69, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Heine, D.; Martin, K.; Hertweck, C. Genomics-guided discovery of endophenazines from Kitasatospora sp. HKI 714. J. Nat. Prod. 2014, 77, 1083–1087. [Google Scholar] [CrossRef] [PubMed]
- McAteer, S.; Coulson, A.; McLennan, N.; Masters, M. The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J. Bacteriol. 2001, 183, 7403–7407. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seeger, K.; Flinspach, K.; Haug-Schifferdecker, E.; Kulik, A.; Gust, B.; Fiedler, H.P.; Heide, L. The biosynthetic genes for prenylated phenazines are located at two different chromosomal loci of Streptomyces cinnamonensis DSM 1042. Microb. Biotechnol. 2011, 4, 252–262. [Google Scholar] [CrossRef]
- Wang, S.; Fu, C.; Bilal, M.; Hu, H.; Wang, W.; Zhang, X. Enhanced biosynthesis of arbutin by engineering shikimate pathway in Pseudomonas chlororaphis P3. Microb. Cell Fact. 2018, 17, 174. [Google Scholar] [CrossRef]
- Wu, C.; Van Wezel, G.P.; Choi, Y.H. Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66. J. Antibiot. 2015, 68, 445–452. [Google Scholar] [CrossRef]
- Wu, C.; Medema, M.H.; Läkamp, R.M.; Zhang, L.; Dorrestein, P.C.; Choi, Y.H.; Van Wezel, G.P. Leucanicidin and endophenasides result from methyl-rhamnosylation by the same tailoring enzymes in Kitasatospora sp. MBT66. ACS Chem. Biol. 2016, 11, 478–490. [Google Scholar] [CrossRef]
- Gräwert, T.; Kaiser, J.; Zepeck, F.; Laupitz, R.; Hecht, S.; Amslinger, S.; Schramek, N.; Schleicher, E.; Weber, S.; Haslbeck, M. IspH protein of Escherichia coli: Studies on Iron− Sulfur cluster implementation and catalysis. J. Am. Chem. Soc. 2004, 126, 12847–12855. [Google Scholar] [CrossRef]
- Zocher, G.; Saleh, O.; Heim, J.B.; Herbst, D.A.; Heide, L.; Stehle, T. Structure-based engineering increased the catalytic turnover rate of a novel phenazine prenyltransferase. PLoS ONE 2012, 7, e48427. [Google Scholar] [CrossRef]
- Hibbert, E.G.; Senussi, T.; Smith, M.E.; Costelloe, S.J.; Ward, J.M.; Hailes, H.C.; Dalby, P.A. Directed evolution of transketolase substrate specificity towards an aliphatic aldehyde. J. Biotechnol. 2008, 134, 240–245. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).