Gap Junction-Dependent and -Independent Functions of Connexin43 in Biology
Abstract
Simple Summary
Abstract
1. The Emergence of Connexin43 as a Gap Junction Protein in Evolution
2. Synthesis of Connexin43 and Forming of Gap Junctions on the Plasma Membrane
3. Connexin43 Gap Junction and Hemichannel Function in Physiology
| Cell Type | Promoter | Major Phenotype | References |
|---|---|---|---|
| Cardiomyocytes | Myh6 | Slow conduction and sudden arrhythmic death | [85] |
| Endothelial cells | Tek | Hypotension and bradycardia in mice | [74] |
| Smooth muscle | Myh11 | Defective in remodeling processes in response to vascular injury | [86] |
| Thermogenic adipocytes | Ucp1 | Impaired cold-induced adipose tissue beiging | [87] |
| Hepatocytes | Alb | Impaired glucose tolerance under high fat-diet feeding | [88] |
| Cardiac macrophage | Cx3cr1 | Delay in atrioventricular conduction | [89] |
| Bone cells (osteoblasts or osteocytes) | Twist2, Col1a1, Bglap, Dmp1 | Phenotype related to bone mineralization and homeostasis | Reviewed in [90] |
4. Channel-Independent Functions of Cx43 on Plasma Membrane
4.1. Transfer of Mitochondria
4.2. Regulator of Phagocytosis
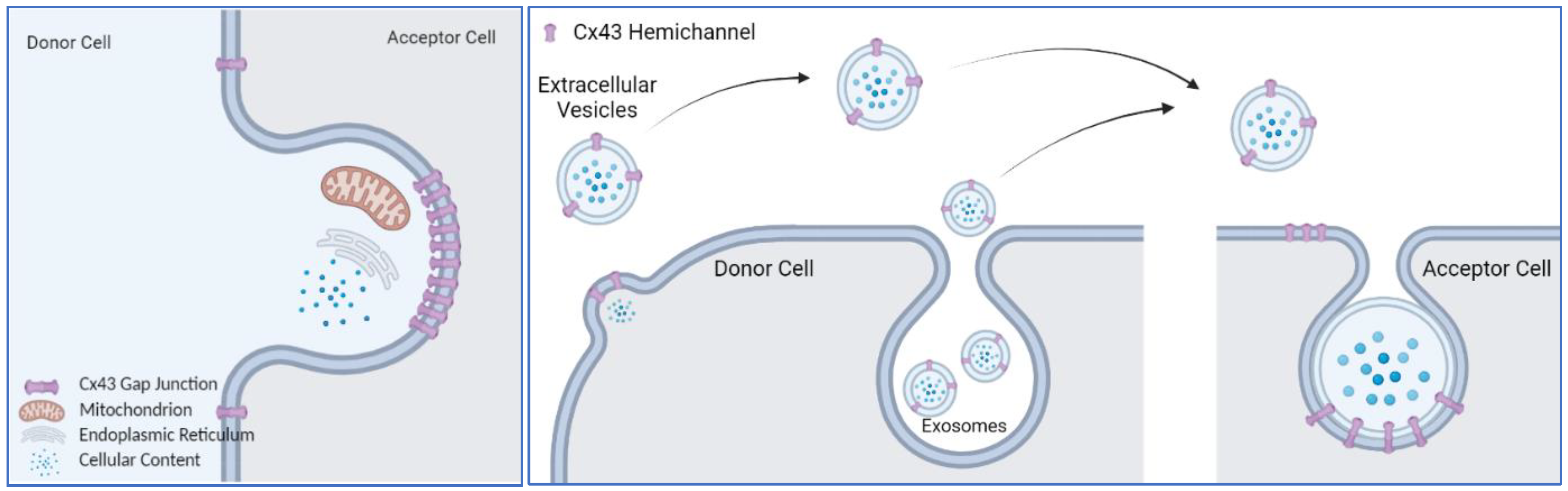
4.3. Cx43 on Extracellular Vesicles
4.4. Is Connexin43 an Active Driver of Membrane Trafficking or a Passenger in the Process?
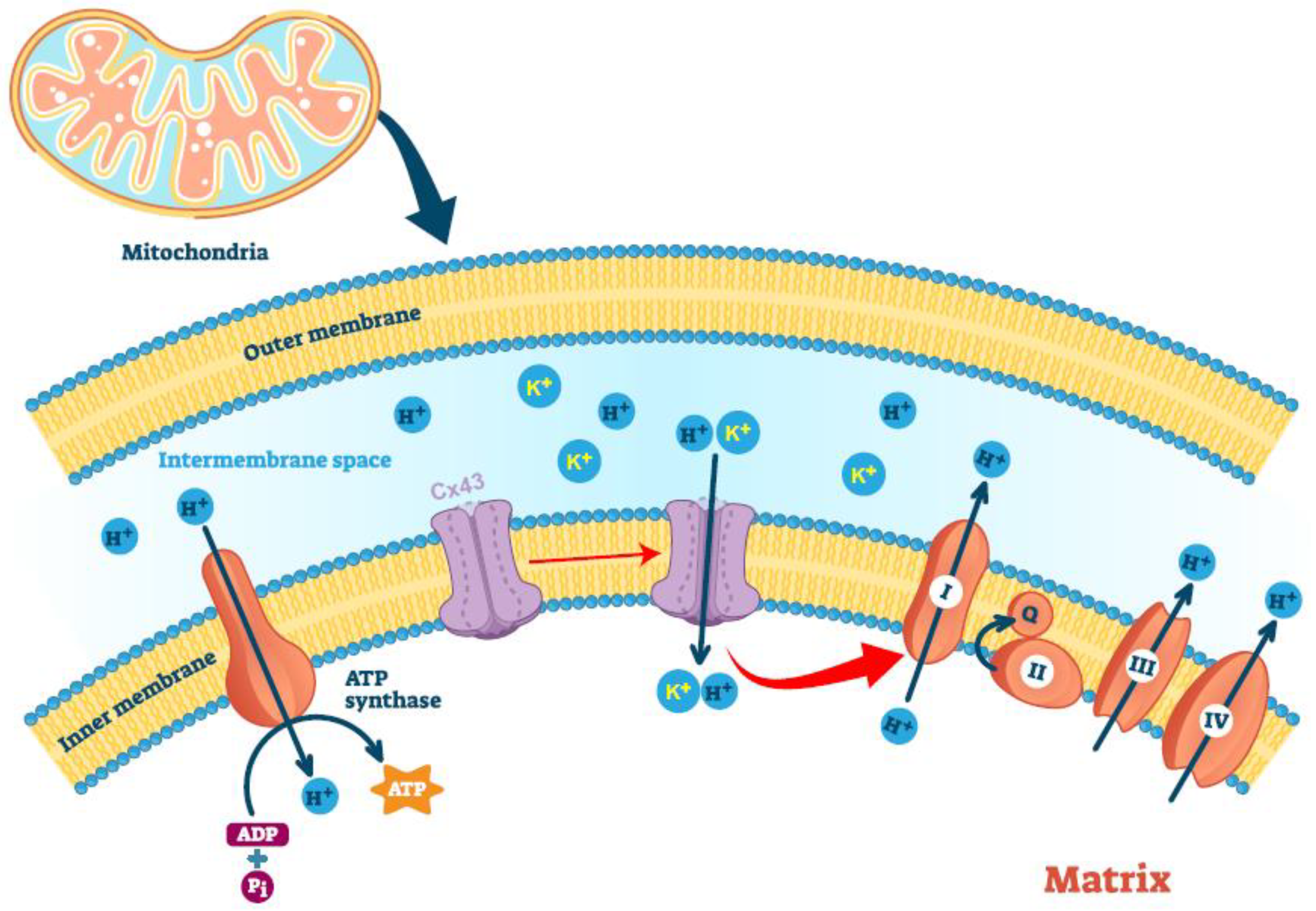
5. Mitochondria-Localized Connexin43
6. Alternative Translation of Connexin43 mRNA
7. Cx43 C-Terminal Fragment Generated by Proteomic Cleavage
8. Summary and Future Directions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Robertson, J.D. The occurrence of a subunit pattern in the unit membranes of club endings in mauthner cell synapses in goldfish brains. J. Cell Biol. 1963, 19, 201–221. [Google Scholar] [CrossRef] [PubMed]
- Revel, J.P.; Karnovsky, M.J. Hexagonal array of subunits in intercellular junctions of the mouse heart and liver. J. Cell Biol. 1967, 33, C7–C12. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulos, H.; Bottger, A.; Fischer, S.; Levin, A.; Wolf, A.; Fujisawa, T.; Hayakawa, S.; Gojobori, T.; Davies, J.A.; David, C.N.; et al. Evolution of gap junctions: The missing link? Curr. Biol. 2004, 14, R879–R880. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Baranova, A.; Ivanov, D.; Petrash, N.; Pestova, A.; Skoblov, M.; Kelmanson, I.; Shagin, D.; Nazarenko, S.; Geraymovych, E.; Litvin, O.; et al. The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics 2004, 83, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Penuela, S.; Bhalla, R.; Nag, K.; Laird, D.W. Glycosylation regulates pannexin intermixing and cellular localization. Mol. Biol. Cell 2009, 20, 4313–4323. [Google Scholar] [CrossRef]
- Sohl, G.; Willecke, K. Gap junctions and the connexin protein family. Cardiovasc. Res. 2004, 62, 228–232. [Google Scholar] [CrossRef]
- Saez, J.C.; Berthoud, V.M.; Branes, M.C.; Martinez, A.D.; Beyer, E.C. Plasma membrane channels formed by connexins: Their regulation and functions. Physiol. Rev. 2003, 83, 1359–1400. [Google Scholar] [CrossRef]
- Farquhar, M.G.; Palade, G.E. Junctional complexes in various epithelia. J. Cell Biol. 1963, 17, 375–412. [Google Scholar] [CrossRef]
- Mese, G.; Richard, G.; White, T.W. Gap junctions: Basic structure and function. J. Investig. Dermatol. 2007, 127, 2516–2524. [Google Scholar] [CrossRef]
- Pfenniger, A.; Wohlwend, A.; Kwak, B.R. Mutations in connexin genes and disease. Eur. J. Clin. Investig. 2011, 41, 103–116. [Google Scholar] [CrossRef]
- Srinivas, M.; Verselis, V.K.; White, T.W. Human diseases associated with connexin mutations. Biochim. Biophys. Acta Biomembr. 2018, 1860, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Beyer, E.C.; Berthoud, V.M. Gap junction structure: Unraveled, but not fully revealed. F1000 Res. 2017, 6, 568. [Google Scholar] [CrossRef] [PubMed]
- Epifantseva, I.; Shaw, R.M. Intracellular trafficking pathways of Cx43 gap junction channels. Biochim. Biophys. Acta Biomembr. 2018, 1860, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Johnstone, S.; Vidal-Brime, L.; Lynn, K.S.; Koval, M. Connexins: Synthesis, post-translational modifications, and trafficking in health and disease. Int. J. Mol. Sci. 2018, 19, 1296. [Google Scholar] [CrossRef]
- Pogoda, K.; Kameritsch, P. Molecular regulation of myoendothelial gap junctions. Curr. Opin. Pharmacol. 2019, 45, 16–22. [Google Scholar] [CrossRef]
- Willebrords, J.; Maes, M.; Crespo Yanguas, S.; Vinken, M. Inhibitors of connexin and pannexin channels as potential therapeutics. Pharmacol. Ther. 2017, 180, 144–160. [Google Scholar] [CrossRef]
- Laird, D.W.; Lampe, P.D. Therapeutic strategies targeting connexins. Nat. Rev. Drug Discov. 2018, 17, 905–921. [Google Scholar] [CrossRef]
- Leybaert, L.; Lampe, P.D.; Dhein, S.; Kwak, B.R.; Ferdinandy, P.; Beyer, E.C.; Laird, D.W.; Naus, C.C.; Green, C.R.; Schulz, R. Connexins in cardiovascular and neurovascular health and disease: Pharmacological implications. Pharmacol. Rev. 2017, 69, 396–478. [Google Scholar] [CrossRef]
- Bennett, M.V.; Barrio, L.C.; Bargiello, T.A.; Spray, D.C.; Hertzberg, E.; Saez, J.C. Gap junctions: New tools, new answers, new questions. Neuron 1991, 6, 305–320. [Google Scholar] [CrossRef]
- Beyer, E.C.; Berthoud, V.M. Gap junction gene and protein families: Connexins, innexins, and pannexins. Biochim. Biophys. Acta Biomembr. 2018, 1860, 5–8. [Google Scholar] [CrossRef]
- Beyer, E.C.; Paul, D.L.; Goodenough, D.A. Connexin43: A protein from rat heart homologous to a gap junction protein from liver. J. Cell Biol. 1987, 105 Pt 1, 2621–2629. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, G.S.; Valiunas, V.; Brink, P.R. Selective permeability of gap junction channels. Biochim. Biophys. Acta 2004, 1662, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Gorlich, D.; Rapoport, T.A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 1993, 75, 615–630. [Google Scholar] [CrossRef]
- High, S.; Andersen, S.S.; Gorlich, D.; Hartmann, E.; Prehn, S.; Rapoport, T.A.; Dobberstein, B. Sec61p is adjacent to nascent type I and type II signal-anchor proteins during their membrane insertion. J. Cell Biol. 1993, 121, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Kalies, K.U.; Gorlich, D.; Rapoport, T.A. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J. Cell Biol. 1994, 126, 925–934. [Google Scholar] [CrossRef]
- Mothes, W.; Heinrich, S.U.; Graf, R.; Nilsson, I.; von Heijne, G.; Brunner, J.; Rapoport, T.A. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell 1997, 89, 523–533. [Google Scholar] [CrossRef]
- Falk, M.M.; Kumar, N.M.; Gilula, N.B. Membrane insertion of gap junction connexins: Polytopic channel forming membrane proteins. J. Cell Biol. 1994, 127, 343–355. [Google Scholar] [CrossRef]
- Zhang, J.T.; Chen, M.; Foote, C.I.; Nicholson, B.J. Membrane integration of in vitro-translated gap junctional proteins: Co- and post-translational mechanisms. Mol. Biol. Cell 1996, 7, 471–482. [Google Scholar] [CrossRef]
- Shivers, R.R.; Bowman, P.D. A freeze-fracture paradigm of the mechanism for delivery and insertion of gap junction particles into the plasma membrane. J. Submicrosc. Cytol. 1985, 17, 199–203. [Google Scholar]
- Segretain, D.; Falk, M.M. Regulation of connexin biosynthesis, assembly, gap junction formation, and removal. Biochim. Biophys. Acta 2004, 1662, 3–21. [Google Scholar] [CrossRef]
- Falk, M.M.; Buehler, L.K.; Kumar, N.M.; Gilula, N.B. Cell-free synthesis and assembly of connexins into functional gap junction membrane channels. EMBO J. 1997, 16, 2703–2716. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Diez, J.A.; George, C.H.; Evans, W.H. Synthesis and assembly of connexins in vitro into homomeric and heteromeric functional gap junction hemichannels. Biochem. J. 1999, 339 Pt 2, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Musil, L.S.; Goodenough, D.A. Multisubunit assembly of an integral plasma membrane channel protein, gap junction connexin43, occurs after exit from the ER. Cell 1993, 74, 1065–1077. [Google Scholar] [CrossRef]
- Das Sarma, J.; Wang, F.; Koval, M. Targeted gap junction protein constructs reveal connexin-specific differences in oligomerization. J. Biol. Chem. 2002, 277, 20911–20918. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Smith, T.D.; Sarma, J.D.; Ritzenthaler, J.D.; Maza, J.; Kaplan, B.E.; Cunningham, L.A.; Suaud, L.; Hubbard, M.J.; Rubenstein, R.C.; et al. ERp29 restricts Connexin43 oligomerization in the endoplasmic reticulum. Mol. Biol. Cell 2009, 20, 2593–2604. [Google Scholar] [CrossRef] [PubMed]
- Park, D.J.; Wallick, C.J.; Martyn, K.D.; Lau, A.F.; Jin, C.; Warn-Cramer, B.J. Akt phosphorylates connexin43 on Ser373, a “mode-1” binding site for 14-3-3. Cell Commun. Adhes. 2007, 14, 211–226. [Google Scholar] [CrossRef]
- Park, D.J.; Freitas, T.A.; Wallick, C.J.; Guyette, C.V.; Warn-Cramer, B.J. Molecular dynamics and in vitro analysis of Connexin43: A new 14-3-3 mode-1 interacting protein. Protein Sci. 2006, 15, 2344–2355. [Google Scholar] [CrossRef]
- Batra, N.; Riquelme, M.A.; Burra, S.; Jiang, J.X. 14-3-3theta facilitates plasma membrane delivery and function of mechanosensitive connexin 43 hemichannels. J. Cell Sci. 2014, 127 Pt 1, 137–146. [Google Scholar]
- Lin, D.; Zhou, J.; Zelenka, P.S.; Takemoto, D.J. Protein kinase Cgamma regulation of gap junction activity through caveolin-1-containing lipid rafts. Investig. Ophthalmol. Vis. Sci. 2003, 44, 5259–5268. [Google Scholar] [CrossRef]
- Langlois, S.; Cowan, K.N.; Shao, Q.; Cowan, B.J.; Laird, D.W. Caveolin-1 and -2 interact with connexin43 and regulate gap junctional intercellular communication in keratinocytes. Mol. Biol. Cell 2008, 19, 912–928. [Google Scholar] [CrossRef]
- Leithe, E.; Mesnil, M.; Aasen, T. The connexin 43 C-terminus: A tail of many tales. Biochim. Biophys. Acta Biomembr. 2018, 1860, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Goodenough, D.A.; Paul, D.L. Beyond the gap: Functions of unpaired connexon channels. Nat. Rev. Mol. Cell Biol. 2003, 4, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Ebihara, L. New roles for connexons. Am. Physiol. Sci. 2003, 18, 100–103. [Google Scholar] [CrossRef] [PubMed]
- D’Hondt, C.; Iyyathurai, J.; Himpens, B.; Leybaert, L.; Bultynck, G. Cx43-hemichannel function and regulation in physiology and pathophysiology: Insights from the bovine corneal endothelial cell system and beyond. Front. Physiol. 2014, 5, 348. [Google Scholar] [CrossRef]
- Weber, P.A.; Chang, H.C.; Spaeth, K.E.; Nitsche, J.M.; Nicholson, B.J. The permeability of gap junction channels to probes of different size is dependent on connexin composition and permeant-pore affinities. Biophys. J. 2004, 87, 958–973. [Google Scholar] [CrossRef]
- Wang, H.Z.; Veenstra, R.D. Monovalent ion selectivity sequences of the rat connexin43 gap junction channel. J. Gen. Physiol. 1997, 109, 491–507. [Google Scholar] [CrossRef]
- Contreras, J.E.; Saez, J.C.; Bukauskas, F.F.; Bennett, M.V. Gating and regulation of connexin 43 (Cx43) hemichannels. Proc. Natl. Acad. Sci. USA 2003, 100, 11388–11393. [Google Scholar] [CrossRef]
- Veenstra, R.D.; Wang, H.Z.; Beblo, D.A.; Chilton, M.G.; Harris, A.L.; Beyer, E.C.; Brink, P.R. Selectivity of connexin-specific gap junctions does not correlate with channel conductance. Circ. Res. 1995, 77, 1156–1165. [Google Scholar] [CrossRef]
- Cheng, K.; Haspel, H.C.; Vallano, M.L.; Osotimehin, B.; Sonenberg, M. Measurement of membrane potentials (psi) of erythrocytes and white adipocytes by the accumulation of triphenylmethylphosphonium cation. J. Membr. Biol. 1980, 56, 191–201. [Google Scholar] [CrossRef]
- Contreras, J.E.; Sanchez, H.A.; Eugenin, E.A.; Speidel, D.; Theis, M.; Willecke, K.; Bukauskas, F.F.; Bennett, M.V.; Saez, J.C. Metabolic inhibition induces opening of unapposed connexin 43 gap junction hemichannels and reduces gap junctional communication in cortical astrocytes in culture. Proc. Natl. Acad. Sci. USA 2002, 99, 495–500. [Google Scholar] [CrossRef]
- John, S.A.; Kondo, R.; Wang, S.Y.; Goldhaber, J.I.; Weiss, J.N. Connexin-43 hemichannels opened by metabolic inhibition. J. Biol. Chem. 1999, 274, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Sugishita, K.; Su, Z.; Ueda, I.; Barry, W.H. Activation of connexin-43 hemichannels can elevate [Ca(2+)]i and [Na(+)]i in rabbit ventricular myocytes during metabolic inhibition. J. Mol. Cell Cardiol. 2001, 33, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Kang, N.; Lovatt, D.; Torres, A.; Zhao, Z.; Lin, J.; Nedergaard, M. Connexin 43 hemichannels are permeable to ATP. J. Neurosci. 2008, 28, 4702–4711. [Google Scholar] [CrossRef] [PubMed]
- Swenson, K.I.; Jordan, J.R.; Beyer, E.C.; Paul, D.L. Formation of gap junctions by expression of connexins in Xenopus oocyte pairs. Cell 1989, 57, 145–155. [Google Scholar] [CrossRef]
- Johnson, R.G.; Reynhout, J.K.; TenBroek, E.M.; Quade, B.J.; Yasumura, T.; Davidson, K.G.; Sheridan, J.D.; Rash, J.E. Gap junction assembly: Roles for the formation plaque and regulation by the C-terminus of connexin43. Mol. Biol. Cell 2012, 23, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Yeh, H.I.; Rothery, S.; Dupont, E.; Coppen, S.R.; Severs, N.J. Individual gap junction plaques contain multiple connexins in arterial endothelium. Circ. Res. 1998, 83, 1248–1263. [Google Scholar] [CrossRef] [PubMed]
- McCutcheon, S.; Stout, R.F., Jr.; Spray, D.C. The dynamic Nexus: Gap junctions control protein localization and mobility in distinct and surprising ways. Sci. Rep. 2020, 10, 17011. [Google Scholar] [CrossRef]
- Xu, Q.; Kopp, R.F.; Chen, Y.; Yang, J.J.; Roe, M.W.; Veenstra, R.D. Gating of connexin 43 gap junctions by a cytoplasmic loop calmodulin binding domain. Am. J. Physiol. Cell Physiol. 2012, 302, C1548–C1556. [Google Scholar] [CrossRef]
- Morley, G.E.; Taffet, S.M.; Delmar, M. Intramolecular interactions mediate pH regulation of connexin43 channels. Biophys. J. 1996, 70, 1294–1302. [Google Scholar] [CrossRef]
- Hermans, M.M.; Kortekaas, P.; Jongsma, H.J.; Rook, M.B. pH sensitivity of the cardiac gap junction proteins, connexin 45 and 43. Pflug. Arch. 1995, 431, 138–140. [Google Scholar] [CrossRef]
- Delmar, M.; Coombs, W.; Sorgen, P.; Duffy, H.S.; Taffet, S.M. Structural bases for the chemical regulation of Connexin43 channels. Cardiovasc. Res. 2004, 62, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Oshima, A. Structure and closure of connexin gap junction channels. FEBS Lett. 2014, 588, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Musil, L.S.; Cunningham, B.A.; Edelman, G.M.; Goodenough, D.A. Differential phosphorylation of the gap junction protein connexin43 in junctional communication-competent and -deficient cell lines. J. Cell Biol. 1990, 111 Pt 1, 2077–2088. [Google Scholar] [CrossRef]
- Crow, D.S.; Beyer, E.C.; Paul, D.L.; Kobe, S.S.; Lau, A.F. Phosphorylation of connexin43 gap junction protein in uninfected and Rous sarcoma virus-transformed mammalian fibroblasts. Mol. Cell Biol. 1990, 10, 1754–1763. [Google Scholar] [PubMed]
- Beardslee, M.A.; Laing, J.G.; Beyer, E.C.; Saffitz, J.E. Rapid turnover of connexin43 in the adult rat heart. Circ. Res. 1998, 83, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Laing, J.G.; Beyer, E.C. The gap junction protein connexin43 is degraded via the ubiquitin proteasome pathway. J. Biol. Chem. 1995, 270, 26399–26403. [Google Scholar] [CrossRef]
- Berthoud, V.M.; Minogue, P.J.; Laing, J.G.; Beyer, E.C. Pathways for degradation of connexins and gap junctions. Cardiovasc. Res. 2004, 62, 256–267. [Google Scholar] [CrossRef]
- Bejarano, E.; Yuste, A.; Patel, B.; Stout, R.F., Jr.; Spray, D.C.; Cuervo, A.M. Connexins modulate autophagosome biogenesis. Nat. Cell Biol. 2014, 16, 401–414. [Google Scholar] [CrossRef]
- Reaume, A.G.; de Sousa, P.A.; Kulkarni, S.; Langille, B.L.; Zhu, D.; Davies, T.C.; Juneja, S.C.; Kidder, G.M.; Rossant, J. Cardiac malformation in neonatal mice lacking connexin43. Science 1995, 267, 1831–1834. [Google Scholar] [CrossRef]
- Britz-Cunningham, S.H.; Shah, M.M.; Zuppan, C.W.; Fletcher, W.H. Mutations of the Connexin43 gap-junction gene in patients with heart malformations and defects of laterality. N. Engl. J. Med. 1995, 332, 1323–1329. [Google Scholar] [CrossRef]
- Dasgupta, C.; Martinez, A.M.; Zuppan, C.W.; Shah, M.M.; Bailey, L.L.; Fletcher, W.H. Identification of connexin43 (alpha1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (DGGE). Mutat. Res. 2001, 479, 173–186. [Google Scholar] [CrossRef]
- Paznekas, W.A.; Boyadjiev, S.A.; Shapiro, R.E.; Daniels, O.; Wollnik, B.; Keegan, C.E.; Innis, J.W.; Dinulos, M.B.; Christian, C.; Hannibal, M.C.; et al. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am. J. Hum. Genet. 2003, 72, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Laird, D.W. Syndromic and non-syndromic disease-linked Cx43 mutations. FEBS Lett. 2014, 588, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Day, K.H.; Damon, D.N.; Duling, B.R. Endothelial cell-specific knockout of connexin 43 causes hypotension and bradycardia in mice. Proc. Natl. Acad. Sci. USA 2001, 98, 9989–9994. [Google Scholar] [CrossRef]
- Harris, A.L.; Bevans, C.G. Exploring hemichannel permeability in vitro. Methods Mol. Biol. 2001, 154, 357–377. [Google Scholar]
- Gomes, P.; Srinivas, S.P.; Van Driessche, W.; Vereecke, J.; Himpens, B. ATP release through connexin hemichannels in corneal endothelial cells. Investig. Ophthalmol. Vis. Sci. 2005, 46, 1208–1218. [Google Scholar] [CrossRef]
- Calder, B.W.; Matthew Rhett, J.; Bainbridge, H.; Fann, S.A.; Gourdie, R.G.; Yost, M.J. Inhibition of connexin 43 hemichannel-mediated ATP release attenuates early inflammation during the foreign body response. Tissue Eng. Part A 2015, 21, 1752–1762. [Google Scholar] [CrossRef]
- Xing, L.; Yang, T.; Cui, S.; Chen, G. Connexin hemichannels in astrocytes: Role in CNS disorders. Front. Mol. Neurosci. 2019, 12, 23. [Google Scholar] [CrossRef]
- Siller-Jackson, A.J.; Burra, S.; Gu, S.; Xia, X.; Bonewald, L.F.; Sprague, E.; Jiang, J.X. Adaptation of connexin 43-hemichannel prostaglandin release to mechanical loading. J. Biol. Chem. 2008, 283, 26374–26382. [Google Scholar] [CrossRef]
- Bruzzone, S.; Guida, L.; Zocchi, E.; Franco, L.; De Flora, A. Connexin 43 hemi channels mediate Ca2+-regulated transmembrane NAD+ fluxes in intact cells. FASEB J. 2001, 15, 10–12. [Google Scholar] [CrossRef]
- Shi, W.; Riquelme, M.A.; Gu, S.; Jiang, J.X. Connexin hemichannels mediate glutathione transport and protect lens fiber cells from oxidative stress. J. Cell Sci. 2018, 131, jcs212506. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; De Bock, M.; Decrock, E.; Bol, M.; Gadicherla, A.; Vinken, M.; Rogiers, V.; Bukauskas, F.F.; Bultynck, G.; Leybaert, L. Paracrine signaling through plasma membrane hemichannels. Biochim. Biophys. Acta 2013, 1828, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Dosch, M.; Zindel, J.; Jebbawi, F.; Melin, N.; Sanchez-Taltavull, D.; Stroka, D.; Candinas, D.; Beldi, G. Connexin-43-dependent ATP release mediates macrophage activation during sepsis. Elife 2019, 8, e42670. [Google Scholar] [CrossRef] [PubMed]
- Gutstein, D.E.; Morley, G.E.; Tamaddon, H.; Vaidya, D.; Schneider, M.D.; Chen, J.; Chien, K.R.; Stuhlmann, H.; Fishman, G.I. Conduction slowing and sudden arrhythmic death in mice with cardiac-restricted inactivation of connexin43. Circ. Res. 2001, 88, 333–339. [Google Scholar] [CrossRef]
- Liao, Y.; Regan, C.P.; Manabe, I.; Owens, G.K.; Day, K.H.; Damon, D.N.; Duling, B.R. Smooth muscle-targeted knockout of connexin43 enhances neointimal formation in response to vascular injury. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 1037–1042. [Google Scholar] [CrossRef]
- Zhu, Y.; Gao, Y.; Tao, C.; Shao, M.; Zhao, S.; Huang, W.; Yao, T.; Johnson, J.A.; Liu, T.; Cypess, A.M.; et al. Connexin 43 mediates white adipose tissue beiging by facilitating the propagation of sympathetic neuronal signals. Cell Metab. 2016, 24, 420–433. [Google Scholar] [CrossRef]
- Tirosh, A.; Tuncman, G.; Calay, E.S.; Rathaus, M.; Ron, I.; Tirosh, A.; Yalcin, A.; Lee, Y.G.; Livne, R.; Ron, S.; et al. Intercellular Transmission of hepatic ER Stress in obesity disrupts systemic metabolism. Cell Metab. 2021, 33, 1716. [Google Scholar] [CrossRef]
- Hulsmans, M.; Clauss, S.; Xiao, L.; Aguirre, A.D.; King, K.R.; Hanley, A.; Hucker, W.J.; Wulfers, E.M.; Seemann, G.; Courties, G.; et al. Macrophages facilitate electrical conduction in the heart. Cell 2017, 169, 510–522.e20. [Google Scholar] [CrossRef]
- Plotkin, L.I.; Bellido, T. Beyond gap junctions: Connexin43 and bone cell signaling. Bone 2013, 52, 157–166. [Google Scholar] [CrossRef]
- Khalil, A.A.; Ilina, O.; Vasaturo, A.; Venhuizen, J.H.; Vullings, M.; Venhuizen, V.; Bilos, A.; Figdor, C.G.; Span, P.N.; Friedl, P. Collective invasion induced by an autocrine purinergic loop through connexin-43 hemichannels. J. Cell Biol. 2020, 219, e201911120. [Google Scholar] [CrossRef]
- Shami, G.J.; Cheng, D.; Verhaegh, P.; Koek, G.; Wisse, E.; Braet, F. Three-dimensional ultrastructure of giant mitochondria in human non-alcoholic fatty liver disease. Sci. Rep. 2021, 11, 3319. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Gao, Y.; Liu, J.; Huang, Y.; Yin, J.; Feng, Y.; Shi, L.; Meloni, B.P.; Zhang, C.; Zheng, M.; et al. Intercellular mitochondrial transfer as a means of tissue revitalization. Signal Transduct. Target. Ther. 2021, 6, 65. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.N.; Das, S.R.; Emin, M.T.; Wei, M.; Sun, L.; Westphalen, K.; Rowlands, D.J.; Quadri, S.K.; Bhattacharya, S.; Bhattacharya, J. Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat. Med. 2012, 18, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Fan, X.L.; Jiang, D.; Zhang, Y.; Li, X.; Xu, Z.B.; Fang, S.B.; Chiu, S.; Tse, H.F.; Lian, Q.; et al. Connexin 43-mediated mitochondrial transfer of iPSC-MSCs alleviates asthma inflammation. Stem Cell Rep. 2018, 11, 1120–1135. [Google Scholar] [CrossRef]
- Golan, K.; Singh, A.K.; Kollet, O.; Bertagna, M.; Althoff, M.J.; Khatib-Massalha, E.; Petrovich-Kopitman, E.; Wellendorf, A.M.; Massalha, H.; Levin-Zaidman, S.; et al. Bone marrow regeneration requires mitochondrial transfer from donor Cx43-expressing hematopoietic progenitors to stroma. Blood 2020, 136, 2607–2619. [Google Scholar] [CrossRef]
- Osswald, M.; Jung, E.; Sahm, F.; Solecki, G.; Venkataramani, V.; Blaes, J.; Weil, S.; Horstmann, H.; Wiestler, B.; Syed, M.; et al. Brain tumour cells interconnect to a functional and resistant network. Nature 2015, 528, 93–98. [Google Scholar] [CrossRef]
- Otsu, K.; Das, S.; Houser, S.D.; Quadri, S.K.; Bhattacharya, S.; Bhattacharya, J. Concentration-dependent inhibition of angiogenesis by mesenchymal stem cells. Blood 2009, 113, 4197–4205. [Google Scholar] [CrossRef]
- Okafo, G.; Prevedel, L.; Eugenin, E. Tunneling nanotubes (TNT) mediate long-range gap junctional communication: Implications for HIV cell to cell spread. Sci. Rep. 2017, 7, 16660. [Google Scholar] [CrossRef]
- Anand, R.J.; Dai, S.; Gribar, S.C.; Richardson, W.; Kohler, J.W.; Hoffman, R.A.; Branca, M.F.; Li, J.; Shi, X.H.; Sodhi, C.P.; et al. A role for connexin43 in macrophage phagocytosis and host survival after bacterial peritoneal infection. J. Immunol. 2008, 181, 8534–8543. [Google Scholar] [CrossRef]
- Glass, A.M.; Wolf, B.J.; Schneider, K.M.; Princiotta, M.F.; Taffet, S.M. Connexin43 is dispensable for phagocytosis. J. Immunol. 2013, 190, 4830–4835. [Google Scholar] [CrossRef]
- Gemel, J.; Kilkus, J.; Dawson, G.; Beyer, E.C. Connecting Exosomes and Connexins. Cancers 2019, 11, 476. [Google Scholar] [CrossRef] [PubMed]
- Shimaoka, M.; Kawamoto, E.; Gaowa, A.; Okamoto, T.; Park, E.J. Connexins and integrins in exosomes. Cancers 2019, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.J.; Zhang, L.L.; Bi, Q.C.; Gan, L.J.; Wei, M.J.; Hong, T.; Tan, R.J.; Lan, X.M.; Liu, L.H.; Han, X.J.; et al. Exosomal connexin 43 regulates the resistance of glioma cells to temozolomide. Oncol. Rep. 2021, 45, 44. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.R.; Martins-Marques, T.; Ribeiro-Rodrigues, T.; Ferreira, J.V.; Catarino, S.; Pinho, M.J.; Zuzarte, M.; Isabel Anjo, S.; Manadas, B.; Pereira, P.; et al. Gap junctional protein Cx43 is involved in the communication between extracellular vesicles and mammalian cells. Sci. Rep. 2015, 5, 13243. [Google Scholar] [CrossRef] [PubMed]
- Martins-Marques, T.; Pinho, M.J.; Zuzarte, M.; Oliveira, C.; Pereira, P.; Sluijter, J.P.; Gomes, C.; Girao, H. Presence of Cx43 in extracellular vesicles reduces the cardiotoxicity of the anti-tumour therapeutic approach with doxorubicin. J. Extracell Vesicles 2016, 5, 32538. [Google Scholar] [CrossRef] [PubMed]
- Martins-Marques, T.; Ribeiro-Rodrigues, T.; de Jager, S.C.; Zuzarte, M.; Ferreira, C.; Cruz, P.; Reis, L.; Baptista, R.; Goncalves, L.; Slujiter, J.P.G.; et al. Myocardial infarction affects Cx43 content of extracellular vesicles secreted by cardiomyocytes. Life Sci. Alliance 2020, 3, e202000821. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Kojima, R.; Bojar, D.; Rizzi, G.; Hamri, G.C.; El-Baba, M.D.; Saxena, P.; Auslander, S.; Tan, K.R.; Fussenegger, M. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 2018, 9, 1305. [Google Scholar] [CrossRef]
- Ghoshroy, S.; Goodenough, D.A.; Sosinsky, G.E. Preparation, characterization, and structure of half gap junctional layers split with urea and EGTA. J. Membr. Biol. 1995, 146, 15–28. [Google Scholar] [CrossRef]
- Goodenough, D.A.; Gilula, N.B. The splitting of hepatocyte gap junctions and zonulae occludentes with hypertonic disaccharides. J. Cell Biol. 1974, 61, 575–590. [Google Scholar] [CrossRef] [PubMed]
- Jordan, K.; Chodock, R.; Hand, A.R.; Laird, D.W. The origin of annular junctions: A mechanism of gap junction internalization. J. Cell Sci. 2001, 114 Pt 4, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Spagnol, G.; Kieken, F.; Kopanic, J.L.; Li, H.; Zach, S.; Stauch, K.L.; Grosely, R.; Sorgen, P.L. Structural studies of the Nedd4 WW domains and their selectivity for the Connexin43 (Cx43) carboxyl terminus. J. Biol Chem. 2016, 291, 7637–7650. [Google Scholar] [CrossRef] [PubMed]
- Thevenin, A.F.; Kowal, T.J.; Fong, J.T.; Kells, R.M.; Fisher, C.G.; Falk, M.M. Proteins and mechanisms regulating gap-junction assembly, internalization, and degradation. Physiology 2013, 28, 93–116. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q.; et al. Progress, opportunity, and perspective on exosome isolation-efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707. [Google Scholar] [CrossRef] [PubMed]
- Adil, M.S.; Narayanan, S.P.; Somanath, P.R. Cell-cell junctions: Structure and regulation in physiology and pathology. Tissue Barriers 2021, 9, 1848212. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Brodsky, S.; Kumari, S.; Valiunas, V.; Brink, P.; Kaide, J.; Nasjletti, A.; Goligorsky, M.S. Paradoxical overexpression and translocation of connexin43 in homocysteine-treated endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 2002, 282, H2124–H2133. [Google Scholar] [CrossRef]
- Smith, A.C.; Robinson, A.J. MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res. 2016, 44, D1258–D1261. [Google Scholar] [CrossRef]
- Morgenstern, M.; Peikert, C.D.; Lubbert, P.; Suppanz, I.; Klemm, C.; Alka, O.; Steiert, C.; Naumenko, N.; Schendzielorz, A.; Melchionda, L.; et al. Quantitative high-confidence human mitochondrial proteome and its dynamics in cellular context. Cell Metab. 2021, 33, 2464–2483.e18. [Google Scholar] [CrossRef]
- Kazak, L.; Chouchani, E.T.; Jedrychowski, M.P.; Erickson, B.K.; Shinoda, K.; Cohen, P.; Vetrivelan, R.; Lu, G.Z.; Laznik-Bogoslavski, D.; Hasenfuss, S.C.; et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 2015, 163, 643–655. [Google Scholar] [CrossRef]
- Boengler, K.; Dodoni, G.; Rodriguez-Sinovas, A.; Cabestrero, A.; Ruiz-Meana, M.; Gres, P.; Konietzka, I.; Lopez-Iglesias, C.; Garcia-Dorado, D.; Di Lisa, F.; et al. Connexin 43 in cardiomyocyte mitochondria and its increase by ischemic preconditioning. Cardiovasc. Res. 2005, 67, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Kalogeris, T.; Baines, C.P.; Krenz, M.; Korthuis, R.J. Cell biology of ischemia/reperfusion injury. Int. Rev. Cell Mol. Biol. 2012, 298, 229–317. [Google Scholar] [PubMed]
- Halestrap, A.P. Mitochondria and preconditioning: A connexin connection? Circ. Res. 2006, 99, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Opie, L.H.; Sack, M.N. Metabolic plasticity and the promotion of cardiac protection in ischemia and ischemic preconditioning. J. Mol. Cell Cardiol. 2002, 34, 1077–1089. [Google Scholar] [CrossRef]
- Ferdinandy, P.; Schulz, R.; Baxter, G.F. Interaction of cardiovascular risk factors with myocardial ischemia/reperfusion injury, preconditioning, and postconditioning. Pharmacol. Rev. 2007, 59, 418–458. [Google Scholar] [CrossRef]
- Rodriguez-Sinovas, A.; Boengler, K.; Cabestrero, A.; Gres, P.; Morente, M.; Ruiz-Meana, M.; Konietzka, I.; Miro, E.; Totzeck, A.; Heusch, G.; et al. Translocation of connexin 43 to the inner mitochondrial membrane of cardiomyocytes through the heat shock protein 90-dependent TOM pathway and its importance for cardioprotection. Circ. Res. 2006, 99, 93–101. [Google Scholar] [CrossRef]
- Kavazis, A.N.; Alvarez, S.; Talbert, E.; Lee, Y.; Powers, S.K. Exercise training induces a cardioprotective phenotype and alterations in cardiac subsarcolemmal and intermyofibrillar mitochondrial proteins. Am. J. Physiol. Heart Circ. Physiol. 2009, 297, H144–H152. [Google Scholar] [CrossRef]
- Boengler, K.; Stahlhofen, S.; van de Sand, A.; Gres, P.; Ruiz-Meana, M.; Garcia-Dorado, D.; Heusch, G.; Schulz, R. Presence of connexin 43 in subsarcolemmal, but not in interfibrillar cardiomyocyte mitochondria. Basic Res. Cardiol. 2009, 104, 141–147. [Google Scholar] [CrossRef]
- Miro-Casas, E.; Ruiz-Meana, M.; Agullo, E.; Stahlhofen, S.; Rodriguez-Sinovas, A.; Cabestrero, A.; Jorge, I.; Torre, I.; Vazquez, J.; Boengler, K.; et al. Connexin43 in cardiomyocyte mitochondria contributes to mitochondrial potassium uptake. Cardiovasc. Res. 2009, 83, 747–756. [Google Scholar] [CrossRef]
- Soetkamp, D.; Nguyen, T.T.; Menazza, S.; Hirschhauser, C.; Hendgen-Cotta, U.B.; Rassaf, T.; Schluter, K.D.; Boengler, K.; Murphy, E.; Schulz, R. S-nitrosation of mitochondrial connexin 43 regulates mitochondrial function. Basic Res. Cardiol. 2014, 109, 433. [Google Scholar] [CrossRef]
- Cocozzelli, A.G.; White, T.W. Connexin 43 Mutations Lead to Increased Hemichannel Functionality in Skin Disease. Int. J. Mol. Sci. 2019, 20, 6186. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, M.; Augustynek, B.; Kulawiak, B.; Koprowski, P.; Bednarczyk, P.; Jarmuszkiewicz, W.; Szewczyk, A. What do we not know about mitochondrial potassium channels? Biochim. Biophys. Acta 2016, 1857, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Garlid, K.D.; Paucek, P. Mitochondrial potassium transport: The K(+) cycle. Biochim. Biophys. Acta 2003, 1606, 23–41. [Google Scholar] [CrossRef]
- Malinska, D.; Mirandola, S.R.; Kunz, W.S. Mitochondrial potassium channels and reactive oxygen species. FEBS Lett. 2010, 584, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
- Swietach, P.; Rossini, A.; Spitzer, K.W.; Vaughan-Jones, R.D. H+ ion activation and inactivation of the ventricular gap junction: A basis for spatial regulation of intracellular pH. Circ. Res. 2007, 100, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Boengler, K.; Ruiz-Meana, M.; Gent, S.; Ungefug, E.; Soetkamp, D.; Miro-Casas, E.; Cabestrero, A.; Fernandez-Sanz, C.; Semenzato, M.; Di Lisa, F.; et al. Mitochondrial connexin 43 impacts on respiratory complex I activity and mitochondrial oxygen consumption. J. Cell Mol. Med. 2012, 16, 1649–1655. [Google Scholar] [CrossRef]
- Kim, S.N.; Kwon, H.J.; Im, S.W.; Son, Y.H.; Akindehin, S.; Jung, Y.S.; Lee, S.J.; Rhyu, I.J.; Kim, I.Y.; Seong, J.K.; et al. Connexin 43 is required for the maintenance of mitochondrial integrity in brown adipose tissue. Sci. Rep. 2017, 7, 7159. [Google Scholar] [CrossRef]
- Trudeau, K.; Muto, T.; Roy, S. Downregulation of mitochondrial connexin 43 by high glucose triggers mitochondrial shape change and cytochrome C release in retinal endothelial cells. Investig. Ophthalmol. Vis. Sci. 2012, 53, 6675–6681. [Google Scholar] [CrossRef]
- Mohammad, G.; Kowluru, R.A. Novel role of mitochondrial matrix metalloproteinase-2 in the development of diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3832–3841. [Google Scholar] [CrossRef]
- Joshi-Mukherjee, R.; Coombs, W.; Burrer, C.; de Mora, I.A.; Delmar, M.; Taffet, S.M. Evidence for the presence of a free C-terminal fragment of cx43 in cultured cells. Cell Commun. Adhes. 2007, 14, 75–84. [Google Scholar] [CrossRef]
- Salat-Canela, C.; Sese, M.; Peula, C.; Ramon y Cajal, S.; Aasen, T. Internal translation of the connexin 43 transcript. Cell Commun. Signal. 2014, 12, 31. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Initiation of translation in prokaryotes and eukaryotes. Gene 1999, 234, 187–208. [Google Scholar] [CrossRef]
- Kozak, M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 2005, 361, 13–37. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, C.; Peixeiro, I.; Romao, L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013, 9, e1003529. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988, 334, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Schiavi, A.; Hudder, A.; Werner, R. Connexin43 mRNA contains a functional internal ribosome entry site. FEBS Lett. 1999, 464, 118–122. [Google Scholar] [CrossRef]
- Ul-Hussain, M.; Olk, S.; Schoenebeck, B.; Wasielewski, B.; Meier, C.; Prochnow, N.; May, C.; Galozzi, S.; Marcus, K.; Zoidl, G.; et al. Internal ribosomal entry site (IRES) activity generates endogenous carboxyl-terminal domains of Cx43 and is responsive to hypoxic conditions. J. Biol. Chem. 2014, 289, 20979–20990. [Google Scholar] [CrossRef]
- Jackson, R.J. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005, 33 Pt 6, 1231–1241. [Google Scholar] [CrossRef]
- Zeitz, M.J.; Calhoun, P.J.; James, C.C.; Taetzsch, T.; George, K.K.; Robel, S.; Valdez, G.; Smyth, J.W. Dynamic UTR usage regulates alternative translation to modulate gap junction formation during stress and aging. Cell Rep. 2019, 27, 2737–2747.e5. [Google Scholar] [CrossRef]
- James, C.C.; Zeitz, M.J.; Calhoun, P.J.; Lamouille, S.; Smyth, J.W. Altered translation initiation of Gja1 limits gap junction formation during epithelial-mesenchymal transition. Mol. Biol. Cell 2018, 29, 797–808. [Google Scholar] [CrossRef]
- Basheer, W.A.; Xiao, S.; Epifantseva, I.; Fu, Y.; Kleber, A.G.; Hong, T.; Shaw, R.M. GJA1-20k arranges actin to guide Cx43 delivery to cardiac intercalated discs. Circ. Res. 2017, 121, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Shimura, D.; Baum, R.; Hernandez, D.M.; Agvanian, S.; Nagaoka, Y.; Katsumata, M.; Lampe, P.D.; Kleber, A.G.; Hong, T.; et al. Auxiliary trafficking subunit GJA1-20k protects connexin-43 from degradation and limits ventricular arrhythmias. J. Clin. Investig. 2020, 130, 4858–4870. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhang, S.S.; Xiao, S.; Basheer, W.A.; Baum, R.; Epifantseva, I.; Hong, T.; Shaw, R.M. Cx43 isoform GJA1-20k promotes microtubule dependent mitochondrial transport. Front. Physiol. 2017, 8, 905. [Google Scholar] [CrossRef] [PubMed]
- Shimura, D.; Nuebel, E.; Baum, R.; Valdez, S.E.; Xiao, S.; Warren, J.S.; Palatinus, J.A.; Hong, T.; Rutter, J.; Shaw, R.M. Protective mitochondrial fission induced by stress-responsive protein GJA1-20k. Elife 2021, 10, e69207. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.; Doble, B.W.; Kardami, E. The carboxy-tail of connexin-43 localizes to the nucleus and inhibits cell growth. Mol. Cell Biochem. 2003, 242, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Moorby, C.; Patel, M. Dual functions for connexins: Cx43 regulates growth independently of gap junction formation. Exp. Cell Res. 2001, 271, 238–248. [Google Scholar] [CrossRef]
- Crespin, S.; Fromont, G.; Wager, M.; Levillain, P.; Cronier, L.; Monvoisin, A.; Defamie, N.; Mesnil, M. Expression of a gap junction protein, connexin43, in a large panel of human gliomas: New insights. Cancer Med. 2016, 5, 1742–1752. [Google Scholar] [CrossRef]
- Silver, P.A. How proteins enter the nucleus. Cell 1991, 64, 489–497. [Google Scholar] [CrossRef]
- Bernhofer, M.; Goldberg, T.; Wolf, S.; Ahmed, M.; Zaugg, J.; Boden, M.; Rost, B. NLSdb-major update for database of nuclear localization signals and nuclear export signals. Nucleic Acids Res. 2018, 46, D503–D508. [Google Scholar] [CrossRef]
- Lindsey, M.L.; Escobar, G.P.; Mukherjee, R.; Goshorn, D.K.; Sheats, N.J.; Bruce, J.A.; Mains, I.M.; Hendrick, J.K.; Hewett, K.W.; Gourdie, R.G.; et al. Matrix metalloproteinase-7 affects connexin-43 levels, electrical conduction, and survival after myocardial infarction. Circulation 2006, 113, 2919–2928. [Google Scholar] [CrossRef]
- Verma, R.P.; Hansch, C. Matrix metalloproteinases (MMPs): Chemical-biological functions and (Q)SARs. Bioorg. Med. Chem. 2007, 15, 2223–2268. [Google Scholar] [CrossRef] [PubMed]
- Cauwe, B.; Opdenakker, G. Intracellular substrate cleavage: A novel dimension in the biochemistry, biology and pathology of matrix metalloproteinases. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 351–423. [Google Scholar] [CrossRef] [PubMed]
- De Bock, M.; Wang, N.; Decrock, E.; Bultynck, G.; Leybaert, L. Intracellular cleavage of the Cx43 C-terminal domain by matrix-metalloproteases: A novel contributor to inflammation? Mediat. Inflamm. 2015, 2015, 257471. [Google Scholar] [CrossRef]
- Wilson, C.L.; Ouellette, A.J.; Satchell, D.P.; Ayabe, T.; Lopez-Boado, Y.S.; Stratman, J.L.; Hultgren, S.J.; Matrisian, L.M.; Parks, W.C. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 1999, 286, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Shirafuji, Y.; Tanabe, H.; Satchell, D.P.; Henschen-Edman, A.; Wilson, C.L.; Ouellette, A.J. Structural determinants of procryptdin recognition and cleavage by matrix metalloproteinase-7. J. Biol. Chem. 2003, 278, 7910–7919. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, A.; Oyler, G.; McKay, R.; Gerfen, C.; Conant, K. Cleavage of neuronal synaptosomal-associated protein of 25 kDa by exogenous matrix metalloproteinase-7. J. Neurochem. 2007, 102, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Varela-Eirin, M.; Varela-Vazquez, A.; Rodriguez-Candela Mateos, M.; Vila-Sanjurjo, A.; Fonseca, E.; Mascarenas, J.L.; Eugenio Vazquez, M.; Mayan, M.D. Recruitment of RNA molecules by connexin RNA-binding motifs: Implication in RNA and DNA transport through microvesicles and exosomes. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 728–736. [Google Scholar] [CrossRef]
- Kotini, M.; Barriga, E.H.; Leslie, J.; Gentzel, M.; Rauschenberger, V.; Schambony, A.; Mayor, R. Gap junction protein Connexin-43 is a direct transcriptional regulator of N-cadherin in vivo. Nat. Commun. 2018, 9, 3846. [Google Scholar] [CrossRef]
- Seibel, N.M.; Eljouni, J.; Nalaskowski, M.M.; Hampe, W. Nuclear localization of enhanced green fluorescent protein homomultimers. Anal. Biochem. 2007, 368, 95–99. [Google Scholar] [CrossRef]
- Mouse Genome Sequencing, C.; Waterston, R.H.; Lindblad-Toh, K.; Birney, E.; Rogers, J.; Abril, J.F.; Agarwal, P.; Agarwala, R.; Ainscough, R.; Alexandersson, M.; et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [CrossRef]
- Pujar, S.; O’Leary, N.A.; Farrell, C.M.; Loveland, J.E.; Mudge, J.M.; Wallin, C.; Giron, C.G.; Diekhans, M.; Barnes, I.; Bennett, R.; et al. Consensus coding sequence (CCDS) database: A standardized set of human and mouse protein-coding regions supported by expert curation. Nucleic Acids Res. 2018, 46, D221–D228. [Google Scholar] [CrossRef] [PubMed]
- Yura, K.; Shionyu, M.; Hagino, K.; Hijikata, A.; Hirashima, Y.; Nakahara, T.; Eguchi, T.; Shinoda, K.; Yamaguchi, A.; Takahashi, K.; et al. Alternative splicing in human transcriptome: Functional and structural influence on proteins. Gene 2006, 380, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, O.; Convertini, P.; Zhang, Z.; Wen, Y.; Shen, M.; Falaleeva, M.; Stamm, S. Function of alternative splicing. Gene 2013, 514, 1–30. [Google Scholar] [CrossRef] [PubMed]
- James, C.C.; Smyth, J.W. Alternative mechanisms of translation initiation: An emerging dynamic regulator of the proteome in health and disease. Life Sci. 2018, 212, 138–144. [Google Scholar] [CrossRef]
- Seo, J.; Lee, K.J. Post-translational modifications and their biological functions: Proteomic analysis and systematic approaches. J. Biochem. Mol. Biol. 2004, 37, 35–44. [Google Scholar] [CrossRef]
- Rogers, L.D.; Overall, C.M. Proteolytic post-translational modification of proteins: Proteomic tools and methodology. Mol. Cell Proteom. 2013, 12, 3532–3542. [Google Scholar] [CrossRef]
- Kameritsch, P.; Khandoga, N.; Pohl, U.; Pogoda, K. Gap junctional communication promotes apoptosis in a connexin-type-dependent manner. Cell Death Dis. 2013, 4, e584. [Google Scholar] [CrossRef]
- Ma, J.W.; Ji, D.D.; Li, Q.Q.; Zhang, T.; Luo, L. Inhibition of connexin 43 attenuates oxidative stress and apoptosis in human umbilical vein endothelial cells. BMC Pulm. Med. 2020, 20, 19. [Google Scholar] [CrossRef]
- Johnstone, S.R.; Best, A.K.; Wright, C.S.; Isakson, B.E.; Errington, R.J.; Martin, P.E. Enhanced connexin 43 expression delays intra-mitotic duration and cell cycle traverse independently of gap junction channel function. J. Cell Biochem. 2010, 110, 772–782. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Morita, I.; Ikeda, M.; Ma, K.W.; Murota, S. Connexin43 suppresses proliferation of osteosarcoma U2OS cells through post-transcriptional regulation of p27. Oncogene 2001, 20, 4138–4149. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Nakayama, K.; Nakayama, K.; Morita, I. A novel route for connexin 43 to inhibit cell proliferation: Negative regulation of S-phase kinase-associated protein (Skp 2). Cancer Res. 2003, 63, 1623–1630. [Google Scholar] [PubMed]
- King, D.R.; Sedovy, M.W.; Leng, X.; Xue, J.; Lamouille, S.; Koval, M.; Isakson, B.E.; Johnstone, S.R. Mechanisms of connexin regulating peptides. Int. J. Mol. Sci. 2021, 22, 10186. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y. Gap Junction-Dependent and -Independent Functions of Connexin43 in Biology. Biology 2022, 11, 283. https://doi.org/10.3390/biology11020283
Zhu Y. Gap Junction-Dependent and -Independent Functions of Connexin43 in Biology. Biology. 2022; 11(2):283. https://doi.org/10.3390/biology11020283
Chicago/Turabian StyleZhu, Yi. 2022. "Gap Junction-Dependent and -Independent Functions of Connexin43 in Biology" Biology 11, no. 2: 283. https://doi.org/10.3390/biology11020283
APA StyleZhu, Y. (2022). Gap Junction-Dependent and -Independent Functions of Connexin43 in Biology. Biology, 11(2), 283. https://doi.org/10.3390/biology11020283






