Comparative Investigation of Gene Regulatory Processes Underlying Avian Influenza Viruses in Chicken and Duck
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
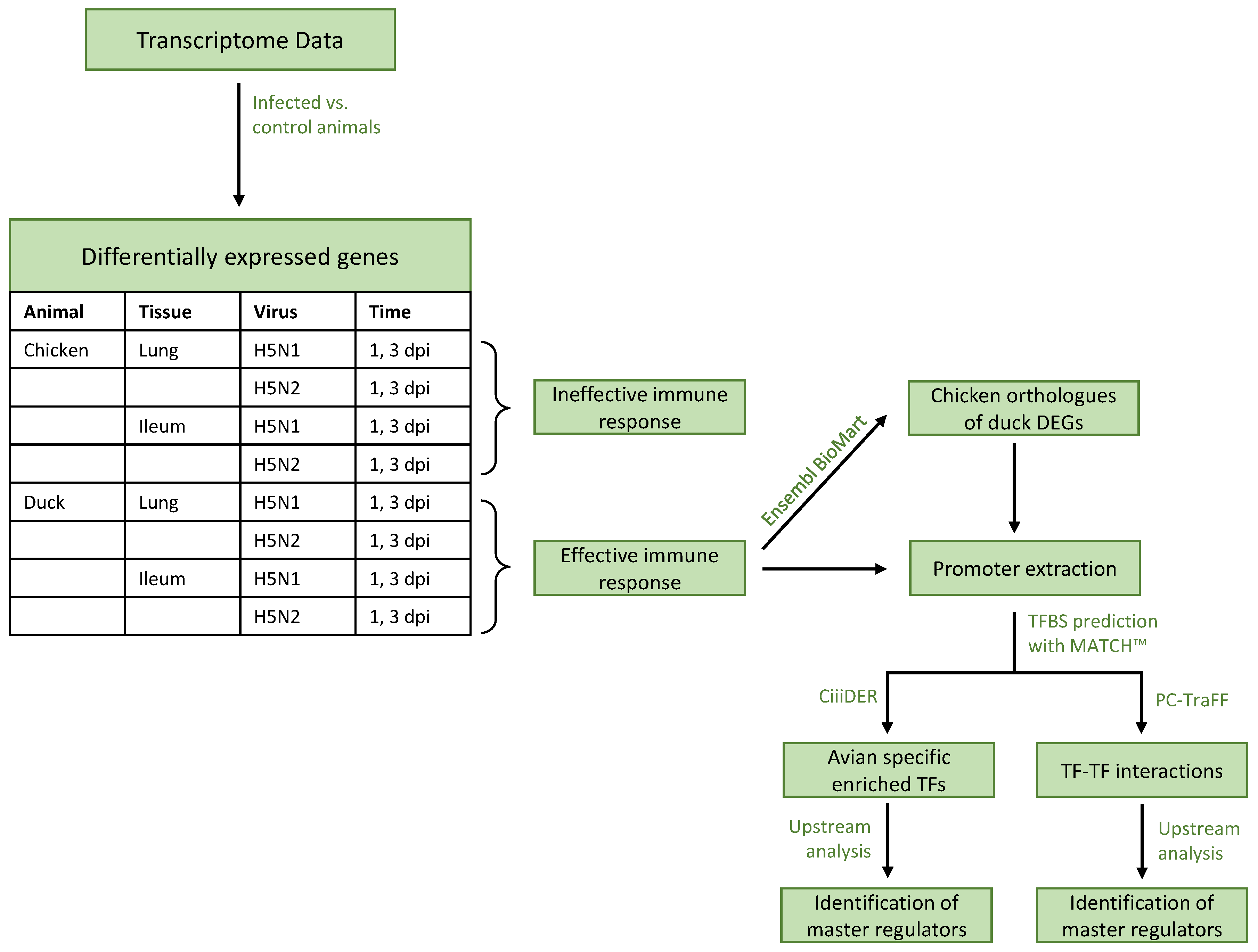
2.1. Transcriptome Data
2.2. Identification of Enriched TFs and TF-TF Cooperations
- Promoter sequences: Using the current versions of reference genomes GRCg6a and CAU_duck1.0, we extracted the promoter sequences ranging from −1000 base pairs (bp) to +100 bp relative to the transcription start site (TSS), similar to previous studies [35,40,41,42]. Sequences were rejected if the full promoter sequence could not be obtained, which was mostly the case for genes on scaffolds.
- Creation of the PWM profile and TFBS detection: Following our previous studies [31,32], we created a custom avian-specific PWM profile. For this, we first downloaded the TFs of avian species (chicken, duck, turkey, zebra finch, and flycatcher) from animalTFDB 3.0 [43] and selected those that were expressed in at least one RNA-Seq experimental condition. Second, we mapped the TFs to the PWMs stored in the TRANSFAC database (release 2018.1) [44]. Finally, we clustered the PWMs hierarchically based on their pairwise Pearson’s correlation coefficients and selected the representative with the highest information content for each cluster in order to create a non-redundant PWM profile with thresholds minimizing the sum of the false-positive and false-negative rates (“minSUM profile”). In total, the profile contains 553 PWMs, which are provided in File S1. We predicted the transcription factor binding sites by applying the MATCH tool [45], which obtains the custom avian-specific PWM profile and a matrix library provided by TRANSFAC [44] as input.
- TF enrichment: We performed a TFBS enrichment analysis by employing the CiiiDER tool [38] in order to identify over- and underrepresented TFBSs. In the following, we refer to a TF as over-/underrepresented in a condition if its corresponding TFBS is significantly over-/underrepresented in the set of promoter sequences of the respective DEGs compared to a custom background. The background set is composed of the promoter sequences of those genes that were not differentially expressed in any of the conditions. From this, the custom background was created as a subset of sequences of the same global GC distribution as the foreground sequences using BiasAway [46]. In a last step, a random sample of equal size was taken as the foreground gene set from the custom background for each gene set, which eventually led to individual background sets from the same distribution, thus making them comparable. Assessment of the distributions of TFBS predictions in foreground and background promoter sets is carried out by an FDR-adjusted p value threshold of 0.05.
- TF-TF Cooperation: The PC-TraFF algorithm [35] and its extension PC-TraFF+ [39] are well-established, information-theory-based approaches to identify TF-TF cooperation pairs using the concept of pointwise mutual information. While PC-TraFF detects the co-occurring TFBSs of TF-pairs in the promoter sequences, PC-TraFF+ separates the highly sequence-set-specific TF-cooperations from the common ones by removing the background co-occurrences of TFBSs. The algorithm needs the predefined distance thresholds as input for the TFBSs. As in our previous studies [31,32], we used the recommended distances of ≥5 and ≤20 and defined a TF-pair as significant if its z-score ≥ 2.
2.3. Identification of Master Regulators
2.4. Annotations and Ortholog Mapping
3. Results and Discussion
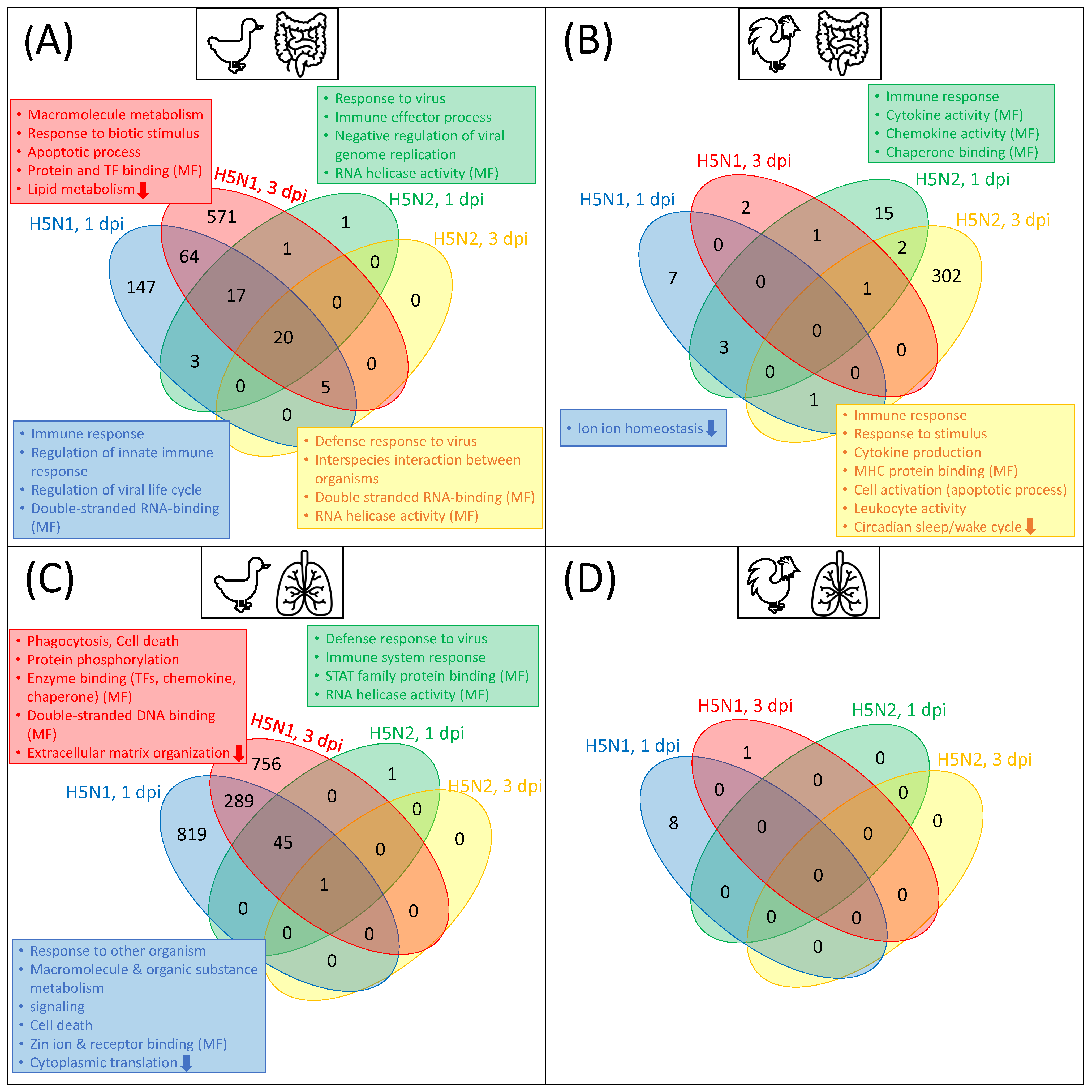
3.1. Transcription Factor Binding Site Enrichment
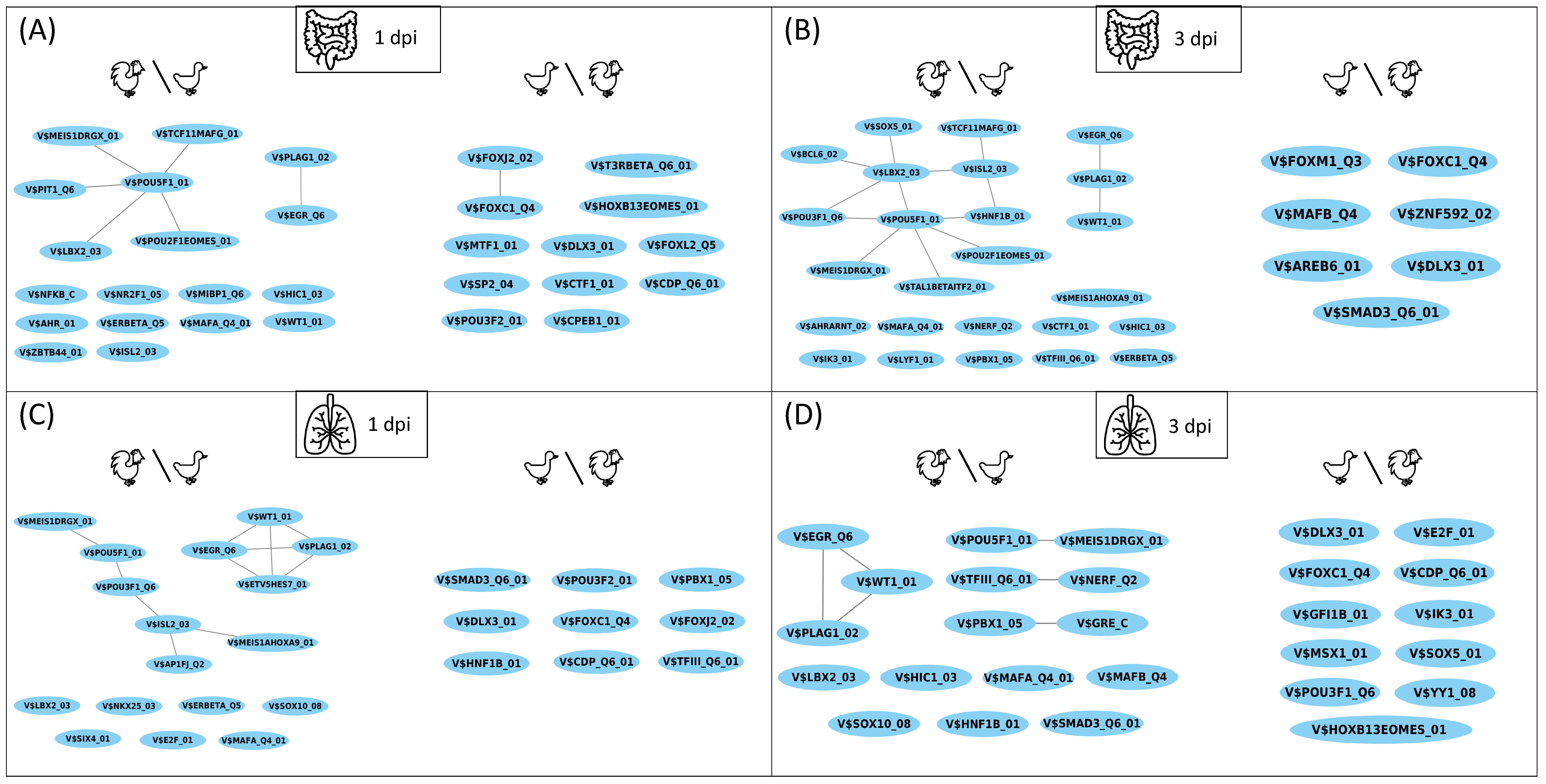
3.2. TF-TF Cooperations
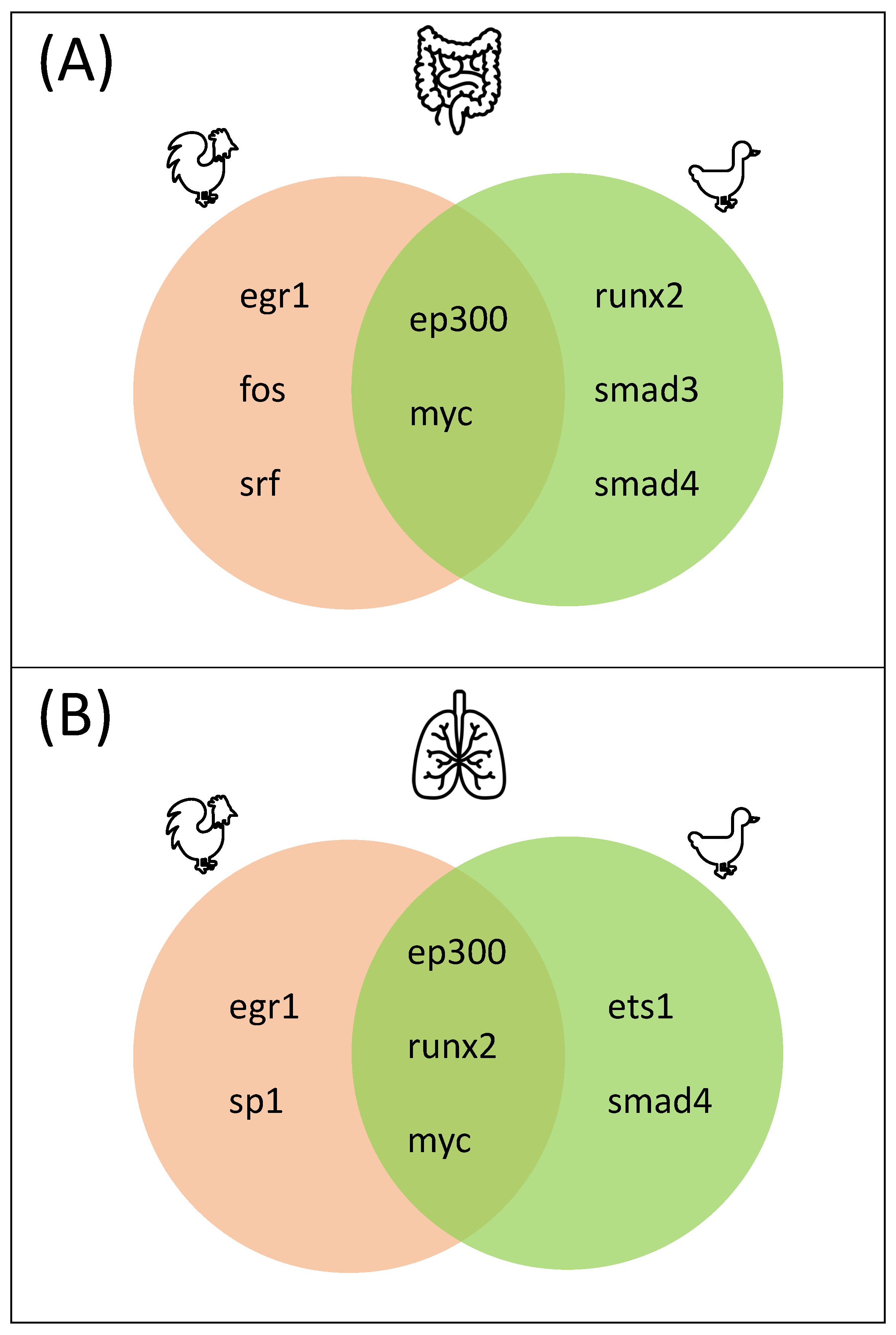
3.3. Master Regulators
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIV | Avian influenza virus |
| HPAI | High pathogenic avian influenza |
| LPAI | Low pathogenic avian influenza |
| dpi | Days post infection |
| DEG | Differentially expressed gene |
| TF | Transcription factor |
| TFBS | Transcription factor binding site |
| TSS | Transcription start site |
| BP | Base pairs |
| FDR | False discovery rate |
| LFC | Log fold change |
| GO | Gene Ontology |
| PMI | Pointwise mutual information |
| PWM | Position weight matrix |
| IFN | Interferon |
| IRF | Interferon regulatory factor |
| ISG | Interferon-stimulated gene |
| STAT | Signal transducer and activator of transcription |
| RLR | RIG-I-like receptor |
| FOX | Forkhead box |
| HOX | Homeobox |
| JAK | Janus–kinase |
References
- Smith, J.; Smith, N.; Yu, L.; Paton, I.R.; Gutowska, M.W.; Forrest, H.L.; Danner, A.F.; Seiler, J.P.; Digard, P.; Webster, R.G.; et al. A comparative analysis of host responses to avian influenza infection in ducks and chickens highlights a role for the interferon-induced transmembrane proteins in viral resistance. BMC Genom. 2015, 16, 574. [Google Scholar] [CrossRef] [PubMed]
- Swayne, D.E. Avian Influenza; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Ranaware, P.B.; Mishra, A.; Vijayakumar, P.; Gandhale, P.N.; Kumar, H.; Kulkarni, D.D.; Raut, A.A. Genome wide host gene expression analysis in chicken lungs infected with avian influenza viruses. PLoS ONE 2016, 11, e0153671. [Google Scholar] [CrossRef] [PubMed]
- WHO. Cumulative Number of Confirmed Human Cases for Avian Influenza A(H5N1) Reported to WHO, 2003–2020. 2020. Available online: https://www.who.int/influenza/human_animal_interface/2020_DEC_tableH5N1.pdf?ua=1 (accessed on 26 March 2021).
- Zou, A.; Nadeau, K.; Wang, P.W.; Lee, J.Y.; Guttman, D.S.; Sharif, S.; Korver, D.R.; Brumell, J.H.; Parkinson, J. Accumulation of genetic variants associated with immunity in the selective breeding of broilers. BMC Genet. 2020, 21, 5. [Google Scholar] [CrossRef] [PubMed]
- Barber, M.R.; Aldridge, J.R., Jr.; Fleming-Canepa, X.; Wang, Y.D.; Webster, R.G.; Magor, K.E. Identification of avian RIG-I responsive genes during influenza infection. Mol. Immunol. 2013, 54, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Evseev, D.; Magor, K.E. Innate immune responses to avian influenza viruses in ducks and chickens. Vet. Sci. 2019, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Looi, F.Y.; Baker, M.L.; Townson, T.; Richard, M.; Novak, B.; Doran, T.J.; Short, K.R. Creating disease resistant chickens: A viable solution to avian influenza? Viruses 2018, 10, 561. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Zhou, H. Overexpression of chicken IRF7 increased viral replication and programmed cell death to the avian influenza virus infection through TGF-Beta/FoxO signaling axis in DF-1. Front. Genet. 2018, 9, 415. [Google Scholar] [CrossRef] [PubMed]
- Pasick, J.; Diederich, S.; Berhane, Y.; Embury-Hyatt, C.; Xu, W. Imbalance between innate antiviral and pro-inflammatory immune responses may contribute to different outcomes involving low-and highly pathogenic avian influenza H5N3 infections in chickens. J. Gen. Virol. 2017, 98, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Giotis, E.S.; Robey, R.C.; Skinner, N.G.; Tomlinson, C.D.; Goodbourn, S.; Skinner, M.A. Chicken interferome: Avian interferon-stimulated genes identified by microarray and RNA-seq of primary chick embryo fibroblasts treated with a chicken type I interferon (IFN-α). Vet. Res. 2016, 47, 75. [Google Scholar] [CrossRef] [PubMed]
- Leymarie, O.; Embury-Hyatt, C.; Chevalier, C.; Jouneau, L.; Moroldo, M.; Da Costa, B.; Berhane, Y.; Delmas, B.; Weingartl, H.M.; Le Goffic, R. PB1-F2 attenuates virulence of highly pathogenic avian H5N1 influenza virus in chickens. PLoS ONE 2014, 9, e100679. [Google Scholar] [CrossRef]
- Abernathy, J.; Li, X.; Jia, X.; Chou, W.; Lamont, S.; Crooijmans, R.; Zhou, H. Copy number variation in F ayoumi and L eghorn chickens analyzed using array comparative genomic hybridization. Anim. Genet. 2014, 45, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lupiani, B.; Reddy, S.; Lamont, S.J.; Zhou, H. RNA-seq analysis revealed novel genes and signaling pathway associated with disease resistance to avian influenza virus infection in chickens. Poult. Sci. 2014, 93, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Sutejo, R.; Yeo, D.S.; Myaing, M.Z.; Hui, C.; Xia, J.; Ko, D.; Cheung, P.C.; Tan, B.H.; Sugrue, R.J. Activation of type I and III interferon signalling pathways occurs in lung epithelial cells infected with low pathogenic avian influenza viruses. PLoS ONE 2012, 7, e33732. [Google Scholar] [CrossRef] [PubMed]
- Uchida, Y.; Watanabe, C.; Takemae, N.; Hayashi, T.; Oka, T.; Ito, T.; Saito, T. Identification of host genes linked with the survivability of chickens infected with recombinant viruses possessing H5N1 surface antigens from a highly pathogenic avian influenza virus. J. Virol. 2012, 86, 2686–2695. [Google Scholar] [CrossRef] [PubMed]
- Reemers, S.S.; Jansen, C.; Koerkamp, M.J.G.; van Haarlem, D.; van de Haar, P.; Degen, W.G.; van Eden, W.; Vervelde, L. Reduced immune reaction prevents immunopathology after challenge with avian influenza virus: A transcriptomics analysis of adjuvanted vaccines. Vaccine 2010, 28, 6351–6360. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reemers, S.S.; van Leenen, D.; Koerkamp, M.J.G.; van Haarlem, D.; van de Haar, P.; van Eden, W.; Vervelde, L. Early host responses to avian influenza A virus are prolonged and enhanced at transcriptional level depending on maturation of the immune system. Mol. Immunol. 2010, 47, 1675–1685. [Google Scholar] [CrossRef] [PubMed]
- Reemers, S.S.; van Haarlem, D.A.; Koerkamp, M.J.G.; Vervelde, L. Differential gene-expression and host-response profiles against avian influenza virus within the chicken lung due to anatomy and airflow. J. Gen. Virol. 2009, 90, 2134–2146. [Google Scholar] [CrossRef]
- Reemers, S.S.; Koerkamp, M.J.G.; Holstege, F.C.; van Eden, W.; Vervelde, L. Cellular host transcriptional responses to influenza A virus in chicken tracheal organ cultures differ from responses in in vivo infected trachea. Vet. Immunol. Immunopathol. 2009, 132, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chiang, H.I.; Zhu, J.; Dowd, S.E.; Zhou, H. Characterization of a newly developed chicken 44K Agilent microarray. BMC Genom. 2008, 9, 60. [Google Scholar] [CrossRef]
- Degen, W.G.; Smith, J.; Simmelink, B.; Glass, E.J.; Burt, D.W.; Schijns, V.E. Molecular immunophenotyping of lungs and spleens in naive and vaccinated chickens early after pulmonary avian influenza A (H9N2) virus infection. Vaccine 2006, 24, 6096–6109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, Y.H.; Feng, H.P.; Huang, L.R.; Kang, Y.; Rong, E.G.; Chen, X.Y.; Li, J.W.; Zeng, W.; Zhu, P.Y.; Liu, X.J.; et al. Transcriptomic analyses reveal new genes and networks response to H5N1 influenza viruses in duck (Anas platyrhynchos). J. Integr. Agric. 2019, 18, 1460–1472. [Google Scholar] [CrossRef]
- Kumar, A.; Vijayakumar, P.; Gandhale, P.; Ranaware, P.; Kumar, H.; Kulkarni, D.; Raut, A.; Mishra, A. Genome-wide gene expression pattern underlying differential host response to high or low pathogenic H5N1 avian influenza virus in ducks. Acta Virol. 2017, 61, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, Y.; Burt, D.W.; Chen, H.; Zhang, Y.; Qian, W.; Kim, H.; Gan, S.; Zhao, Y.; Li, J.; et al. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nat. Genet. 2013, 45, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Maughan, M.N.; Dougherty, L.S.; Preskenis, L.A.; Ladman, B.S.; Gelb, J.; Spackman, E.V.; Keeler, C.L. Transcriptional analysis of the innate immune response of ducks to different species-of-origin low pathogenic H7 avian influenza viruses. Virol. J. 2013, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Mo, Y.; Wang, X.; Gu, M.; Hu, Z.; Zhong, L.; Wu, Q.; Hao, X.; Hu, S.; Liu, W.; et al. PA-X decreases the pathogenicity of highly pathogenic H5N1 influenza A virus in avian species by inhibiting virus replication and host response. J. Virol. 2015, 89, 4126–4142. [Google Scholar] [CrossRef] [PubMed]
- Kuchipudi, S.V.; Tellabati, M.; Sebastian, S.; Londt, B.Z.; Jansen, C.; Vervelde, L.; Brookes, S.M.; Brown, I.H.; Dunham, S.P.; Chang, K.C. Highly pathogenic avian influenza virus infection in chickens but not ducks is associated with elevated host immune and pro-inflammatory responses. Vet. Res. 2014, 45, 118. [Google Scholar] [CrossRef]
- Schat, K.A.; Bingham, J.; Butler, J.M.; Chen, L.M.; Lowther, S.; Crowley, T.M.; Moore, R.J.; Donis, R.O.; Lowenthal, J.W. Role of position 627 of PB2 and the multibasic cleavage site of the hemagglutinin in the virulence of H5N1 avian influenza virus in chickens and ducks. PLoS ONE 2012, 7, e30960. [Google Scholar]
- Liang, Q.l.; Luo, J.; Zhou, K.; Dong, J.X.; He, H.X. Immune-related gene expression in response to H5N1 avian influenza virus infection in chicken and duck embryonic fibroblasts. Mol. Immunol. 2011, 48, 924–930. [Google Scholar] [CrossRef]
- Rajavel, A.; Heinrich, F.; Schmitt, A.O.; Gültas, M. Identifying Cattle Breed-Specific Partner Choice of Transcription Factors during the African Trypanosomiasis Disease Progression Using Bioinformatics Analysis. Vaccines 2020, 8, 246. [Google Scholar] [CrossRef]
- Steuernagel, L.; Meckbach, C.; Heinrich, F.; Zeidler, S.; Schmitt, A.O.; Gültas, M. Computational identification of tissue-specific transcription factor cooperation in ten cattle tissues. PLoS ONE 2019, 14, e0216475. [Google Scholar]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The human transcription factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Morgunova, E.; Taipale, J. Structural perspective of cooperative transcription factor binding. Curr. Opin. Struct. Biol. 2017, 47, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Meckbach, C.; Tacke, R.; Hua, X.; Waack, S.; Wingender, E.; Gültas, M. PC-TraFF: Identification of potentially collaborating transcription factors using pointwise mutual information. BMC Bioinform. 2015, 16, 400. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M. False discovery rates: A new deal. Biostatistics 2017, 18, 275–294. [Google Scholar] [CrossRef] [PubMed]
- Gearing, L.J.; Cumming, H.E.; Chapman, R.; Finkel, A.M.; Woodhouse, I.B.; Luu, K.; Gould, J.A.; Forster, S.C.; Hertzog, P.J. CiiiDER: A tool for predicting and analysing transcription factor binding sites. PLoS ONE 2019, 14, e0215495. [Google Scholar] [CrossRef] [PubMed]
- Meckbach, C.; Wingender, E.; Gültas, M. Removing Background Co-occurrences of Transcription Factor Binding Sites Greatly Improves the Prediction of Specific Transcription Factor Cooperations. Front. Genet. 2018, 9, 189. [Google Scholar] [CrossRef] [PubMed]
- Kel, A.; Boyarskikh, U.; Stegmaier, P.; Leskov, L.S.; Sokolov, A.V.; Yevshin, I.; Mandrik, N.; Stelmashenko, D.; Koschmann, J.; Kel-Margoulis, O.; et al. Walking pathways with positive feedback loops reveal DNA methylation biomarkers of colorectal cancer. BMC Bioinform. 2019, 20, 119. [Google Scholar] [CrossRef] [PubMed]
- Amlie-Wolf, A.; Tang, M.; Mlynarski, E.E.; Kuksa, P.P.; Valladares, O.; Katanic, Z.; Tsuang, D.; Brown, C.D.; Schellenberg, G.D.; Wang, L.S. INFERNO: Inferring the molecular mechanisms of noncoding genetic variants. Nucleic Acids Res. 2018, 46, 8740–8753. [Google Scholar] [CrossRef] [PubMed]
- Coetzee, S.G.; Rhie, S.K.; Berman, B.P.; Coetzee, G.A.; Noushmehr, H. FunciSNP: An R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res. 2012, 40, e139. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Miao, Y.R.; Jia, L.H.; Yu, Q.Y.; Zhang, Q.; Guo, A.Y. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019, 47, D33–D38. [Google Scholar] [CrossRef] [PubMed]
- Wingender, E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief. Bioinform. 2008, 9, 326–332. [Google Scholar] [CrossRef]
- Kel, A.E.; Gössling, E.; Reuter, I.; Cheremushkin, E.; Kel-Margoulis, O.V.; Wingender, E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003, 31, 3576–3579. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Puig, R.R.; Boddie, P.; Mathelier, A. BiasAway: Command-line and web server to generate nucleotide composition-matched DNA background sequences. Bioinformatics 2020, 37, 1607–1609. [Google Scholar] [CrossRef] [PubMed]
- Rajavel, A.; Schmitt, A.O.; Gültas, M. Computational Identification of Master Regulators Influencing Trypanotolerance in Cattle. Int. J. Mol. Sci. 2021, 22, 562. [Google Scholar] [CrossRef] [PubMed]
- Ramzan, F.; Klees, S.; Schmitt, A.O.; Cavero, D.; Gültas, M. Identification of Age-Specific and Common Key Regulatory Mechanisms Governing Eggshell Strength in Chicken Using Random Forests. Genes 2020, 11, 464. [Google Scholar] [CrossRef] [PubMed]
- Mekonnen, Y.A.; Gültas, M.; Effa, K.; Hanotte, O.; Schmitt, A.O. Identification of candidate signature genes and key regulators associated with Trypanotolerance in the Sheko Breed. Front. Genet. 2019, 10, 1095. [Google Scholar] [CrossRef] [PubMed]
- Wlochowitz, D.; Haubrock, M.; Arackal, J.; Bleckmann, A.; Wolff, A.; Beißbarth, T.; Wingender, E.; Gültas, M. Computational identification of key regulators in two different colorectal cancer cell lines. Front. Genet. 2016, 7, 42. [Google Scholar] [CrossRef]
- Koschmann, J.; Bhar, A.; Stegmaier, P.; Kel, A.E.; Wingender, E. “Upstream analysis”: An integrated promoter-pathway analysis approach to causal interpretation of microarray data. Microarrays 2015, 4, 270–286. [Google Scholar] [CrossRef]
- Wingender, E.; Kel, A. geneXplain—Eine integrierte Bioinformatik-Plattform. BIOspektrum 2012, 18, 554–556. [Google Scholar] [CrossRef]
- Rzhetsky, A.; Iossifov, I.; Koike, T.; Krauthammer, M.; Kra, P.; Morris, M.; Yu, H.; Duboué, P.A.; Weng, W.; Wilbur, W.J.; et al. GeneWays: A system for extracting, analyzing, visualizing, and integrating molecular pathway data. J. Biomed. Inform. 2004, 37, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, 2011, bar030. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Bowden, T.A.; Wilson, I.A.; Crispin, M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta-(Bba)-Gen. Subj. 2019, 1863, 1480–1497. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Perez, J.T.; Chen, C.; Li, Y.; Benitez, A.; Kandasamy, M.; Lee, Y.; Andrade, J.; tenOever, B.; Manicassamy, B. Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication. Cell Rep. 2018, 23, 596–607. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Schulman, J.; Itamura, S.; Palese, P. Glycosylation of neuraminidase determines the neurovirulence of influenza A/WSN/33 virus. J. Virol. 1993, 67, 6667–6673. [Google Scholar] [CrossRef]
- Wingender, E.; Schoeps, T.; Dönitz, J. TFClass: An expandable hierarchical classification of human transcription factors. Nucleic Acids Res. 2013, 41, D165–D170. [Google Scholar] [CrossRef]
- Shatskaya, E.; Kovner, A.; Potapova, O.; Cherdantseva, L.; Shkurupy, V.; Shestopalov, A. Study of SMAD-dependent signal pathway in the development of early pulmonary fibrosis in mice infected with influenza A/H1N1 virus. Bull. Exp. Biol. Med. 2017, 162, 647. [Google Scholar] [CrossRef]
- Pokharel, S.M.; Shil, N.K.; Bose, S. Autophagy, TGF-β and SMAD-2/3 signaling regulates interferon-β response in respiratory syncytial virus infected macrophages. Front. Cell. Infect. Microbiol. 2016, 6, 174. [Google Scholar] [CrossRef]
- Xu, P.; Bailey-Bucktrout, S.; Xi, Y.; Xu, D.; Du, D.; Zhang, Q.; Xiang, W.; Liu, J.; Melton, A.; Sheppard, D.; et al. Innate antiviral host defense attenuates TGF-β function through IRF3-mediated suppression of Smad signaling. Mol. Cell 2014, 56, 723–737. [Google Scholar] [CrossRef]
- Jang, C.W.; Chen, C.H.; Chen, C.C.; Chen, J.Y.; Su, Y.H.; Chen, R.H. TGF-β induces apoptosis through Smad-mediated expression of DAP-kinase. Nat. Cell Biol. 2002, 4, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Gallant, S.; Gilkeson, G. ETS transcription factors and regulation of immunity. Arch. Immunol. Ther. Exp. 2006, 54, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Froggatt, H.M.; Harding, A.T.; Chaparian, R.R.; Heaton, N.S. ETV7 limits antiviral gene expression and control of influenza viruses. Sci. Signal. 2021, 14, eabe1194. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, G.; Kaestner, K.H. SnapShot: Forkhead transcription factors I. Cell 2007, 130, 1160. [Google Scholar] [CrossRef]
- van der Horst, A.; Burgering, B.M. Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell. Biol. 2007, 8, 440–450. [Google Scholar] [CrossRef]
- Majoros, A.; Platanitis, E.; Kernbauer-Hölzl, E.; Rosebrock, F.; Müller, M.; Decker, T. Canonical and non-canonical aspects of JAK–STAT signaling: Lessons from interferons for cytokine responses. Front. Immunol. 2017, 8, 29. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Chiang, H.S.; Liu, H.M. The Molecular Basis of Viral Inhibition of IRF- and STAT-Dependent Immune Responses. Front. Immunol. 2018, 9, 3086. [Google Scholar] [CrossRef]
- Harrison, D.A. The jak/stat pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011205. [Google Scholar] [CrossRef]
- Strutt, T.M.; McKinstry, K.K.; Marshall, N.B.; Vong, A.M.; Dutton, R.W.; Swain, S.L. Multipronged CD4(+) T-cell effector and memory responses cooperate to provide potent immunity against respiratory virus. Immunol. Rev. 2013, 255, 149–164. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Robertson, D.L.; Van de Peer, Y.; Oliver, S.G. Choose your partners: Dimerization in eukaryotic transcription factors. Trends Biochem. Sci. 2008, 33, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kel, A.; Konovalova, T.; Waleev, T.; Cheremushkin, E.; Kel-Margoulis, O.; Wingender, E. Composite Module Analyst: A fitness-based tool for identification of transcription factor binding site combinations. Bioinformatics 2006, 22, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Tan, M.; Xu, X.; Cui, M.Z. Histamine induces Egr-1 expression in human aortic endothelial cells via the H1 receptor-mediated protein kinase Cδ-dependent ERK activation pathway. J. Biol. Chem. 2008, 283, 26928–26936. [Google Scholar] [CrossRef] [PubMed]
- Khachigian, L.M. Early growth response-1 in cardiovascular pathobiology. Circ. Res. 2006, 98, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.F.; Fujita, T.; Lu, J.; Okada, K.; Zou, Y.S.; Mackman, N.; Pinsky, D.J.; Stern, D.M. Egr-1, a master switch coordinating upregulation of divergent gene families underlying ischemic stress. Nat. Med. 2000, 6, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Grogan, L.; Nau, M.M.; Allegra, C.J.; Chu, E.; Wright, J.J. Physical interaction between p53 and primary response gene Egr-1. Int. J. Oncol. 2001, 18, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Tatebe, K.; Zeytun, A.; Ribeiro, R.M.; Hoffmann, R.; Harrod, K.S.; Forst, C.V. Response network analysis of differential gene expression in human epithelial lung cells during avian influenza infections. BMC Bioinform. 2010, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Zhao, M.; Liu, H.; Su, R.; Mao, Q.; Gong, L.; Cao, X.; Hao, Y. Uncovering the pharmacological mechanisms of Xijiao Dihuang decoction combined with Yinqiao powder in treating influenza viral pneumonia by an integrative pharmacology strategy. Biomed. Pharmacother. 2021, 141, 111676. [Google Scholar] [CrossRef]
- Roy, A.L. Biochemistry and biology of the inducible multifunctional transcription factor TFII-I: 10 years later. Gene 2012, 492, 32–41. [Google Scholar] [CrossRef]
- Treisman, R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995, 14, 4905–4913. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fu, W.; Cao, K.; He, Q.; Ding, X.; Chen, J.; Zhu, L.; Chen, T.; Ding, L.; Yang, Y.; et al. IFN-κ suppresses the replication of influenza A viruses through the IFNAR-MAPK-Fos-CHD6 axis. Sci. Signal. 2020, 13, eaaz3381. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Saitoh, K.; Koike, C.; Narita, T.; Yasugi, S.; Iba, H. Differential expression of fos and jun family members in the developing chicken gastrointestinal tract. Oncogene 1998, 16, 1611–1616. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Li, X.; Shi, Q.; Zhang, Z.; Xu, S. Hydrogen sulfide exposure triggers chicken trachea inflammatory injury through oxidative stress-mediated FOS/IL8 signaling. J. Hazard. Mater. 2019, 368, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Kern, C.; Zhou, H. Knockout of IRF7 Highlights its Modulator Function of Host Response Against Avian Influenza Virus and the Involvement of MAPK and TOR Signaling Pathways in Chicken. Genes 2020, 11, 385. [Google Scholar] [CrossRef]
- Hauber, H.P.; Foley, S.C.; Hamid, Q. Mucin overproduction in chronic inflammatory lung disease. Can. Respir. J. 2006, 13, 327–335. [Google Scholar] [CrossRef]
- Feng, X.H.; Lin, X.; Derynck, R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15Ink4B transcription in response to TGF-β. EMBO J. 2000, 19, 5178–5193. [Google Scholar] [CrossRef]
- Boxer, L.M.; Dang, C.V. Translocations involving c-myc and c-myc function. Oncogene 2001, 20, 5595–5610. [Google Scholar] [CrossRef]
- Pastorcic, M.; Das, H.K. Regulation of transcription of the human presenilin-1 gene by ets transcription factors and the p53 protooncogene. J. Biol. Chem. 2000, 275, 34938–34945. [Google Scholar] [CrossRef]
- Paulson, M.; Pisharody, S.; Pan, L.; Levy, D.E.; Guadagno, S.; Mui, A.L. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 1999, 274, 25343–25349. [Google Scholar] [CrossRef]
- Leymarie, O.; Meyer, L.; Hervé, P.L.; Da Costa, B.; Delmas, B.; Chevalier, C.; Le Goffic, R. Host Response Comparison of H1N1- and H5N1-Infected Mice Identifies Two Potential Death Mechanisms. Int. J. Mol. Sci. 2017, 18, 1631. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.J.; Lee, H.J.; Kang, K.S.; Song, K.D.; Kim, T.H.; Song, C.S.; Park, M.N. Molecular responses to the influenza A virus in chicken trachea-derived cells. Poult. Sci. 2015, 94, 1190–1201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Feng, X.H.; Derynck, R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature 1998, 394, 909–913. [Google Scholar] [CrossRef] [PubMed]

| Virus | Time | Tissue | Duck DEGs | Chicken DEGs | ||
|---|---|---|---|---|---|---|
| Upregulated | Downregulated | Upregulated | Downregulated | |||
| H5N1 | 1 dpi | lung | 804 | 350 | 1 | 7 |
| ileum | 193 | 63 | 5 | 6 | ||
| 3 dpi | lung | 605 | 486 | 1 | 0 | |
| ileum | 332 | 346 | 3 | 1 | ||
| H5N2 | 1 dpi | lung | 47 | 0 | 0 | 0 |
| ileum | 42 | 1 | 20 | 2 | ||
| 3 dpi | lung | 1 | 0 | 0 | 0 | |
| ileum | 25 | 0 | 286 | 20 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klees, S.; Schlüter, J.-S.; Schellhorn, J.; Bertram, H.; Kurzweg, A.C.; Ramzan, F.; Schmitt, A.O.; Gültas, M. Comparative Investigation of Gene Regulatory Processes Underlying Avian Influenza Viruses in Chicken and Duck. Biology 2022, 11, 219. https://doi.org/10.3390/biology11020219
Klees S, Schlüter J-S, Schellhorn J, Bertram H, Kurzweg AC, Ramzan F, Schmitt AO, Gültas M. Comparative Investigation of Gene Regulatory Processes Underlying Avian Influenza Viruses in Chicken and Duck. Biology. 2022; 11(2):219. https://doi.org/10.3390/biology11020219
Chicago/Turabian StyleKlees, Selina, Johanna-Sophie Schlüter, Jendrik Schellhorn, Hendrik Bertram, Antje Christine Kurzweg, Faisal Ramzan, Armin Otto Schmitt, and Mehmet Gültas. 2022. "Comparative Investigation of Gene Regulatory Processes Underlying Avian Influenza Viruses in Chicken and Duck" Biology 11, no. 2: 219. https://doi.org/10.3390/biology11020219
APA StyleKlees, S., Schlüter, J.-S., Schellhorn, J., Bertram, H., Kurzweg, A. C., Ramzan, F., Schmitt, A. O., & Gültas, M. (2022). Comparative Investigation of Gene Regulatory Processes Underlying Avian Influenza Viruses in Chicken and Duck. Biology, 11(2), 219. https://doi.org/10.3390/biology11020219






