Genome-Wide Identification of DNA Binding with One Finger (Dof) Gene Family in Tartary Buckwheat (Fagopyrum tataricum) and Analysis of Its Expression Pattern after Exogenous Hormone Stimulation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Dof Family Genes in Tartary buckwheat
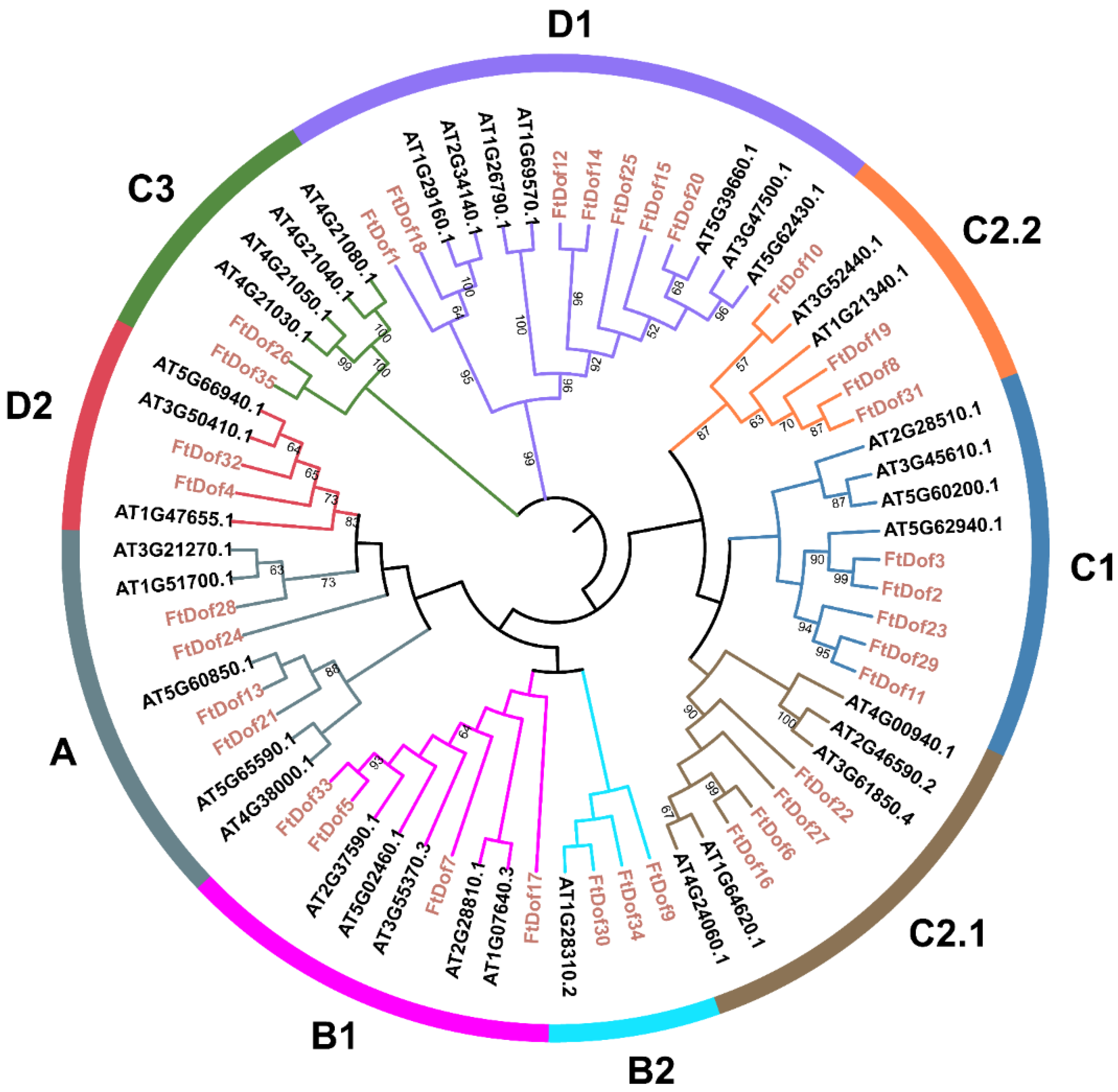
2.2. Phylogenetic Analyses and Classification of FtDof Family Members
2.3. Sequence Characteristic Analysis
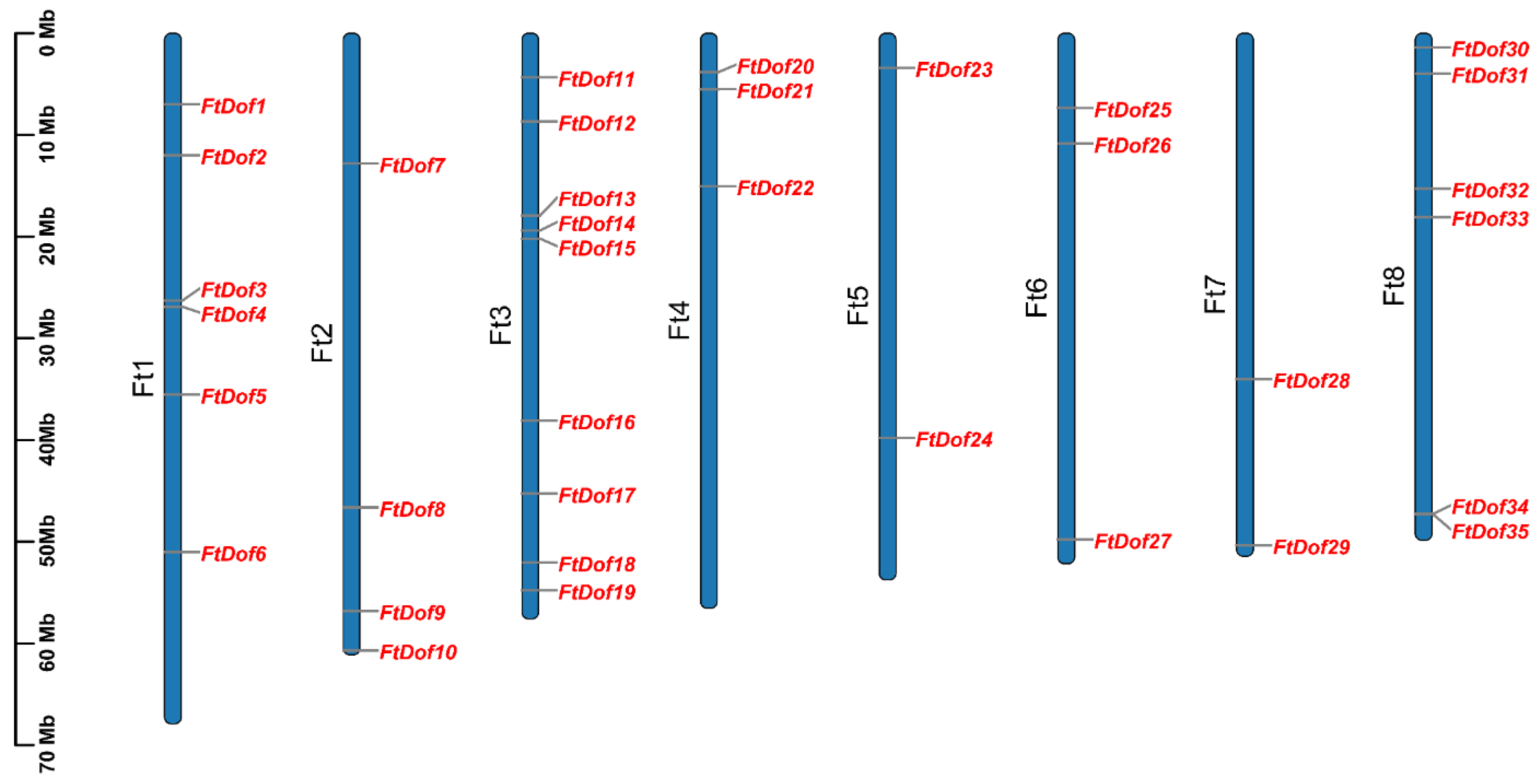
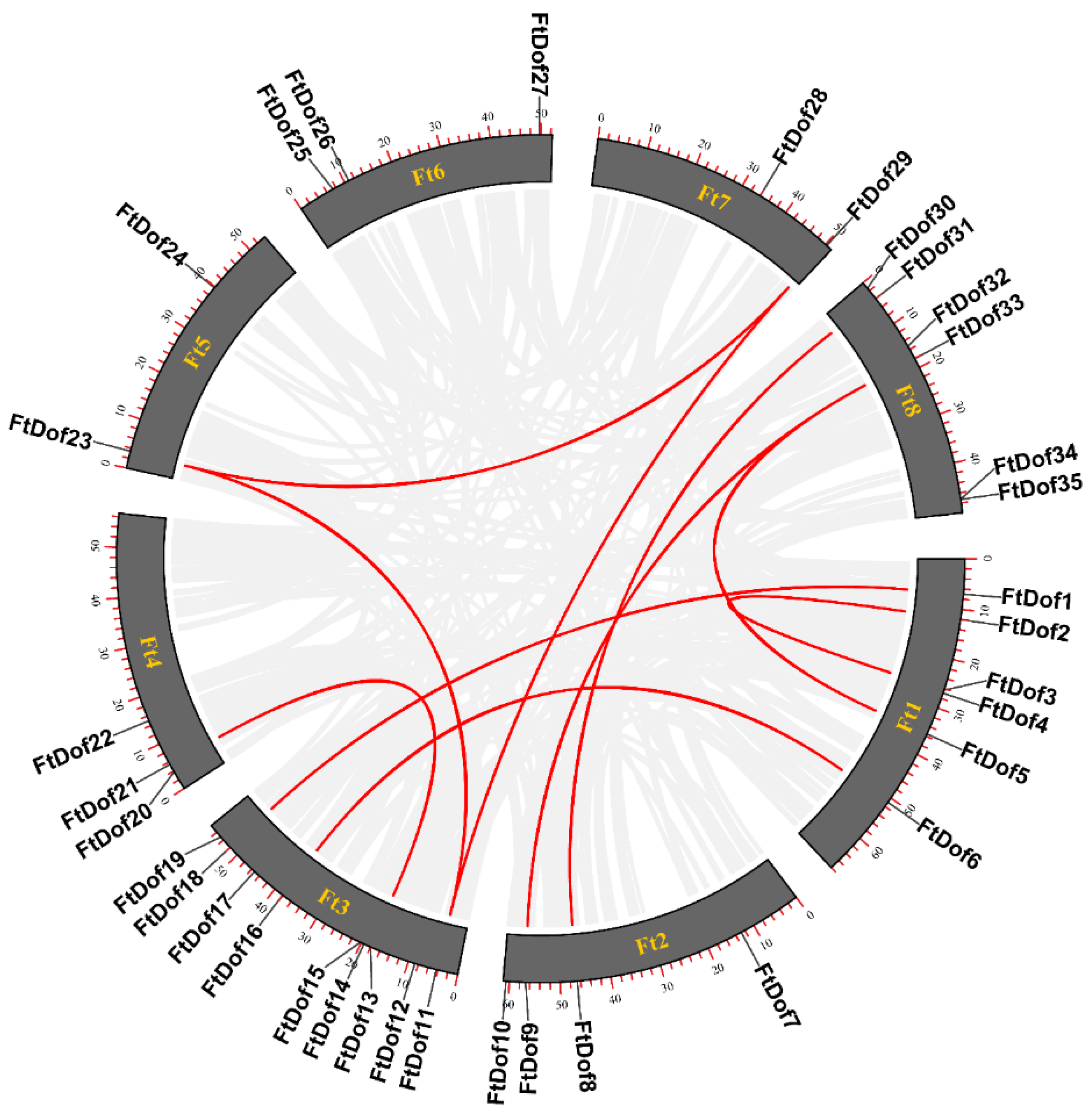
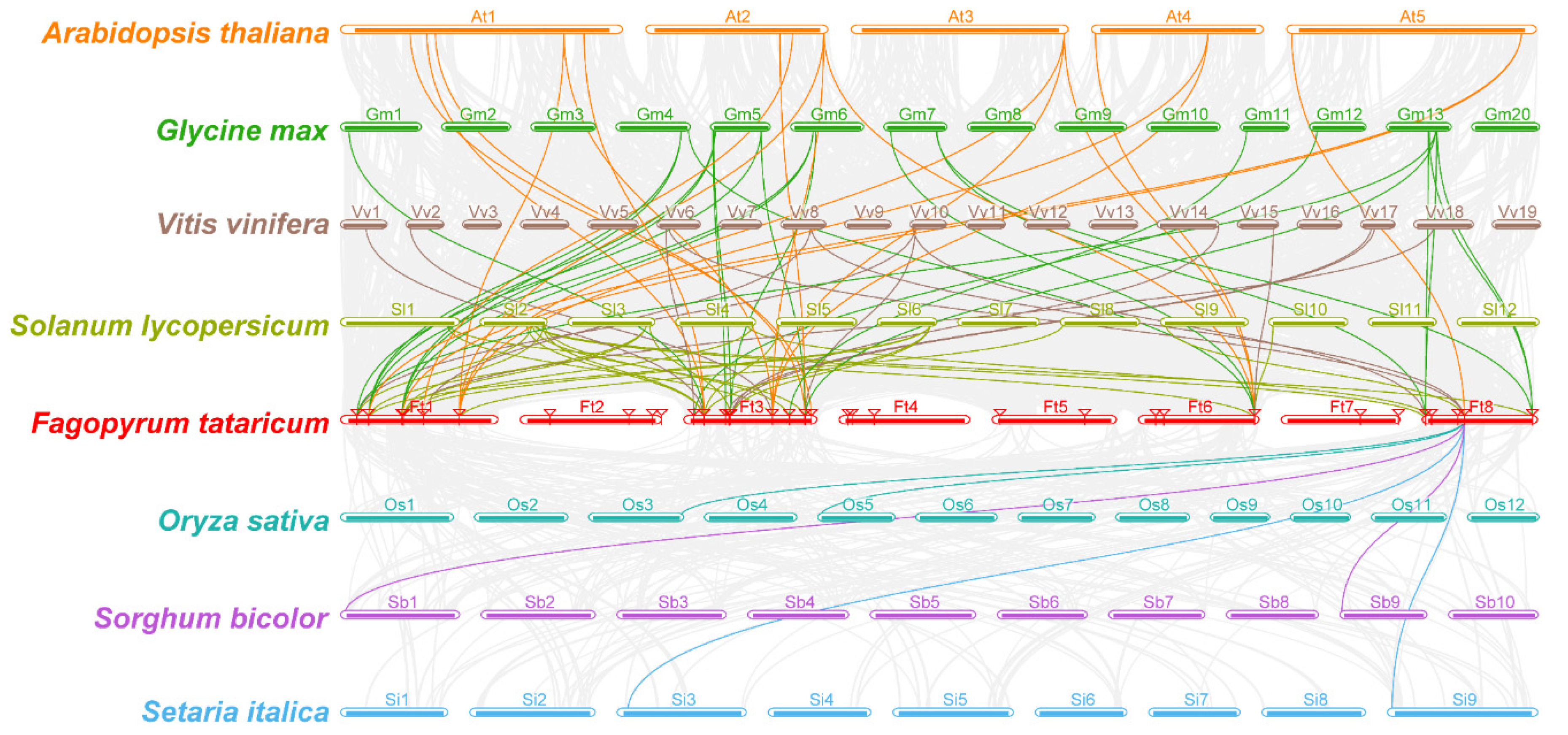
2.4. Chromosome Location, Gene Duplication and Synteny Analysis
2.5. Plant Materials and Treatments
2.6. Expression Analyses of FtDof Genes by qRT-PCR
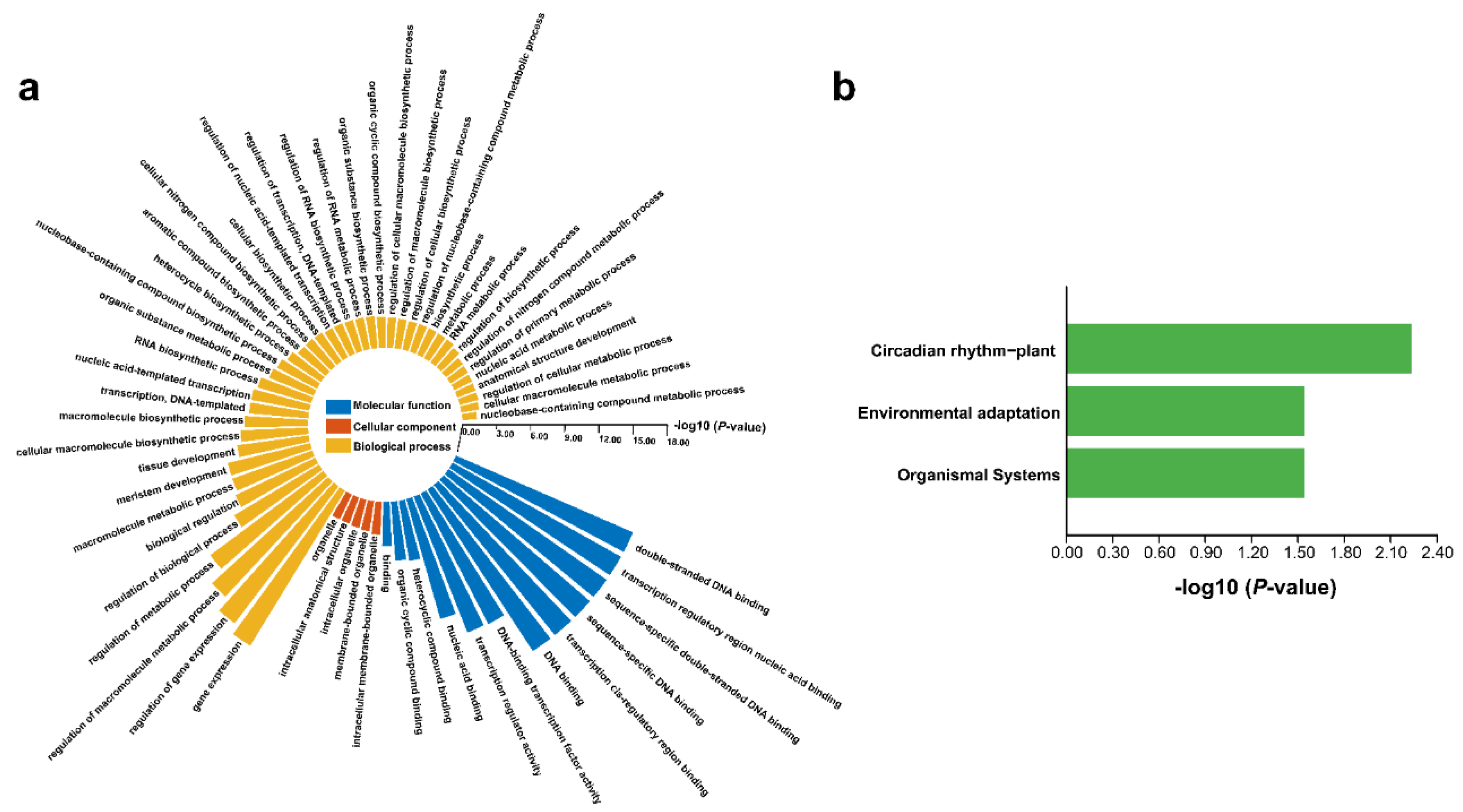
2.7. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis of FtDof Proteins
3. Results
3.1. Genome-Wide Identification of FtDofs in Tartary Buckwheat
3.2. Phylogenetic Analysis and Classification of FtDof Proteins
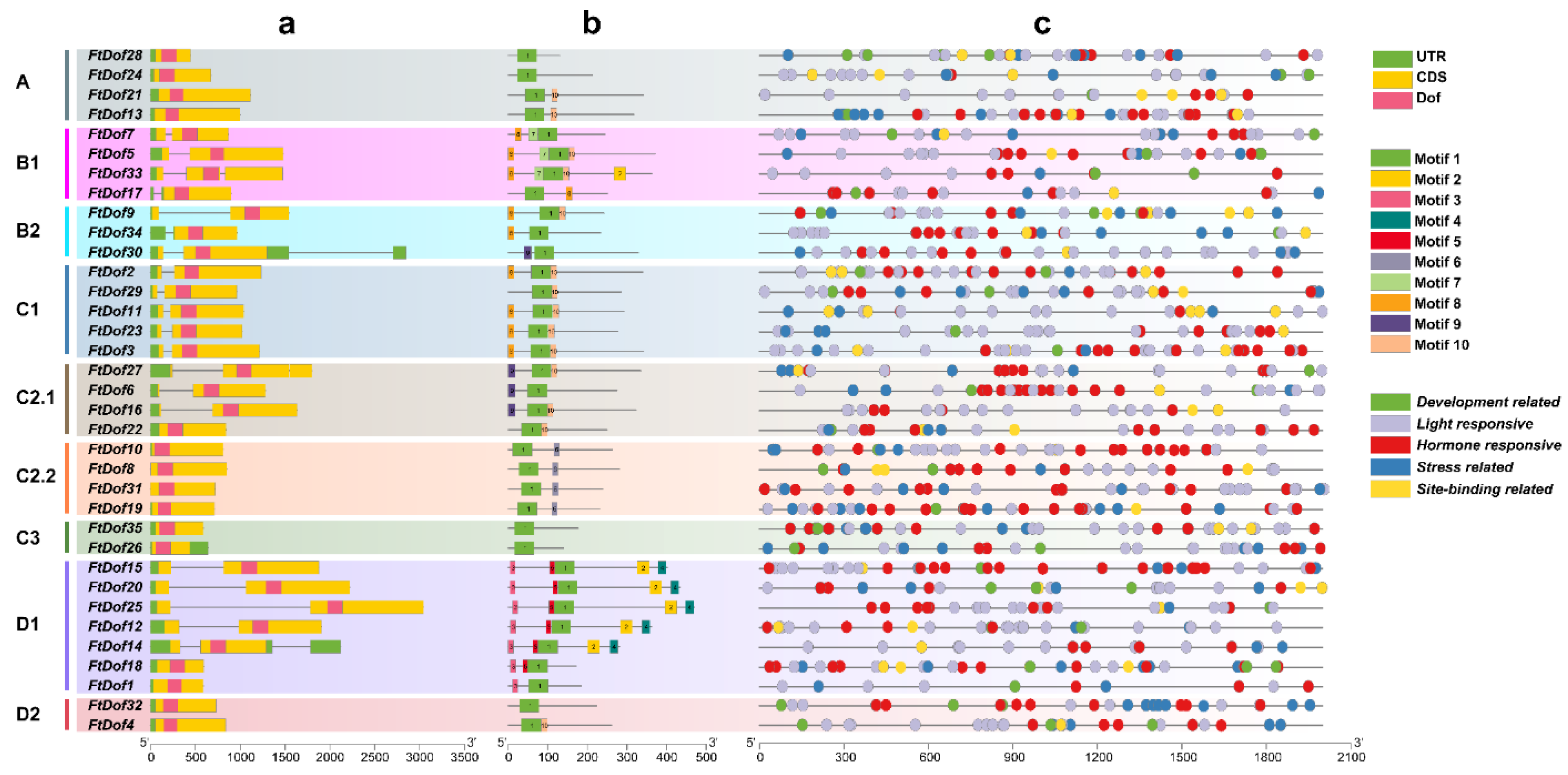
3.3. Gene Structure and Conserved Motif Analysis of FtDof Genes
3.4. Promoter cis-Acting Element Analysis of FtDof Genes
3.5. Gene Duplication and Synteny Analysis of FtDof Proteins
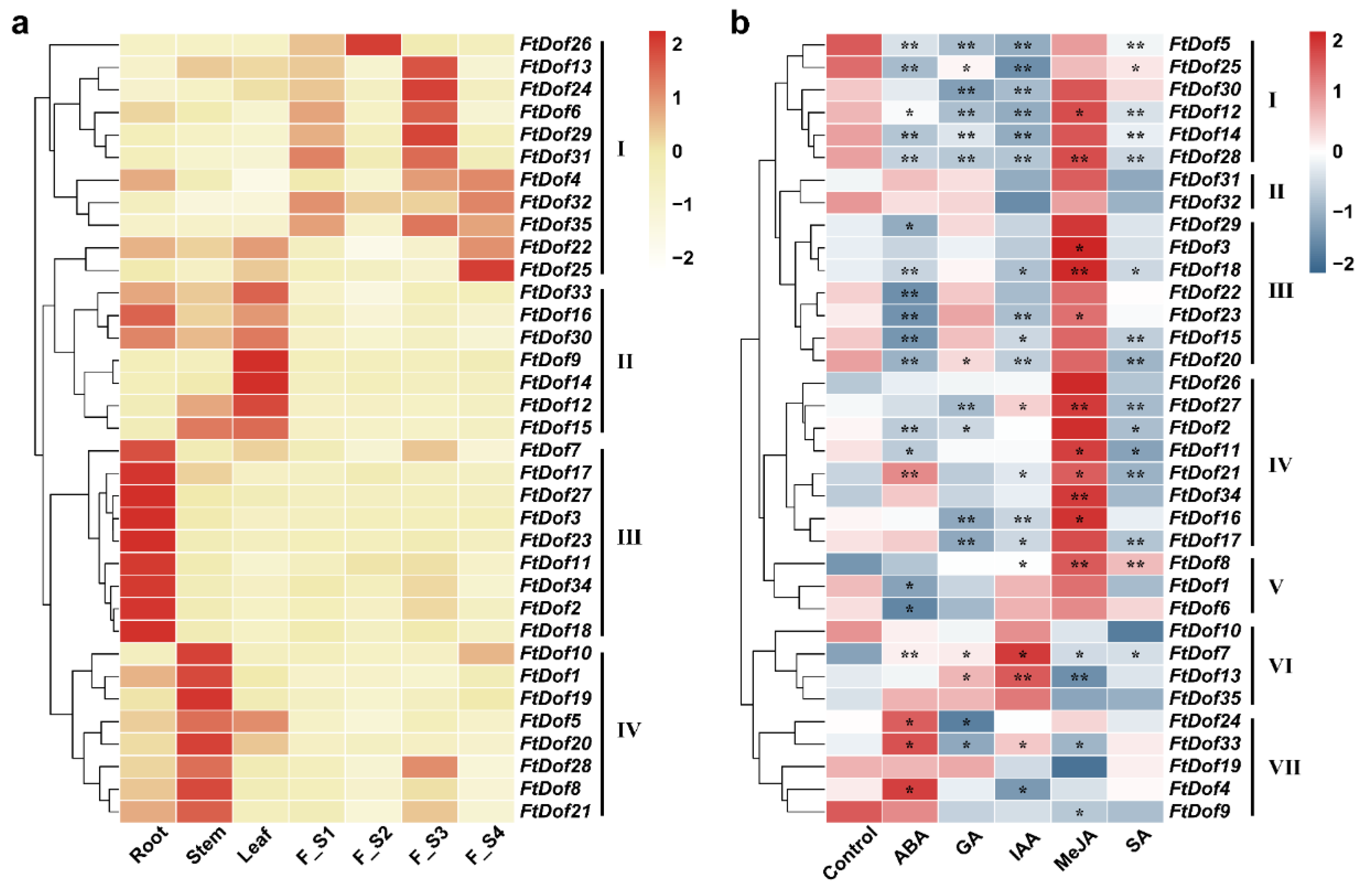
3.6. Expression Pattern of FtDof Genes in Different Tissues and in Different Fruit Developmental Stages of Tartary Buckwheat
3.7. Expression Pattern of FtDof Genes under Different Hormone Treatments
3.8. GO and KEGG Enrichment Analysis of FtDof Proteins
4. Discussion
4.1. Molecular Characterisation and Evolution of FtDof Proteinsin Tartary Buckwheat
4.2. Tissue-Specific Expression Characterisation of FtDof Genes in Tartary Buckwheat
4.3. Expression Pattern of FtDof Genes in Response to Hormones
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Todeschini, A.L.; Georges, A.; Veitia, R.A. Transcription factors: Specific DNA binding and specific gene regulation. Trends Genet. 2014, 30, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Malviya, N.; Kushwaha, H.; Nasim, J.; Bisht, N.C.; Singh, V.K.; Yadav, D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 2015, 241, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Sheen, J. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 1998, 10, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Umemura, Y.; Ishiduka, T.; Yamamoto, R.; Esaka, M. The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J. 2004, 37, 741–749. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999, 17, 209–214. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Izui, K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J. Biol. Chem. 1993, 268, 16028–16036. [Google Scholar] [CrossRef]
- Lijavetzky, D.; Carbonero, P.; Vicente-Carbajosa, J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003, 3, 17. [Google Scholar] [CrossRef]
- Moreno-Risueno, M.A.; Martinez, M.; Vicente-Carbajosa, J.; Carbonero, P. The family of DOF transcription factors: From green unicellular algae to vascular plants. Mol. Genet. Genom. 2007, 277, 379–390. [Google Scholar] [CrossRef]
- Chen, Y.; Cao, J. Comparative analysis of Dof transcription factor family in maize. Plant Mol. Biol. Report. 2015, 33, 1245–1258. [Google Scholar] [CrossRef]
- Kushwaha, H.; Gupta, S.; Singh, V.K.; Rastogi, S.; Yadav, D. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol. Biol. Rep. 2011, 38, 5037–5053. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, Y.; Zhang, C.; Zhang, T.; Hu, T.; Ye, J.; Zhang, J.; Wang, T.; Li, H.; Ye, Z. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J. Integr. Plant Biol. 2013, 55, 552–566. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, N.; Deng, X.; Liu, D.; Li, M.; Cui, D.; Hu, Y.; Yan, Y. Genome-wide analysis of wheat DNA-binding with one finger (Dof) transcription factor genes: Evolutionary characteristics and diverse abiotic stress responses. BMC Genom. 2020, 21, 276. [Google Scholar] [CrossRef]
- Noguero, M.; Atif, R.M.; Ochatt, S.; Thompson, R.D. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 2013, 209, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana. Plant Physiol. Biochem. 2007, 45, 623–629. [Google Scholar] [CrossRef]
- Negi, J.; Moriwaki, K.; Konishi, M.; Yokoyama, R.; Nakano, T.; Kusumi, K.; Hashimoto-Sugimoto, M.; Schroeder, J.I.; Nishitani, K.; Yanagisawa, S.; et al. A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 2013, 23, 479–484. [Google Scholar] [CrossRef]
- Yang, J.; Yang, M.F.; Zhang, W.P.; Chen, F.; Shen, S.H. A putative flowering-time-related Dof transcription factor gene, JcDof3, is controlled by the circadian clock in Jatropha curcas. Plant Sci. 2011, 181, 667–674. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, X.; Yin, D.; Yuan, H.; Xie, Q.; Zhao, X.; Li, X.; Zhu, L.; Li, S.; Li, D. Constitutive expression of OsDof4, encoding a C2-C2 zinc finger transcription factor, confesses its distinct flowering effects under long- and short-day photoperiods in rice (Oryza sativa L.). BMC Plant Biol. 2017, 17, 166. [Google Scholar] [CrossRef]
- Chen, X.; Wang, D.; Liu, C.; Wang, M.; Wang, T.; Zhao, Q.; Yu, J. Maize transcription factor Zmdof1 involves in the regulation of Zm401 gene. Plant Growth Regul. 2012, 66, 271–284. [Google Scholar] [CrossRef]
- Gualberti, G.; Papi, M.; Bellucci, L.; Ricci, I.; Bouchez, D.; Camilleri, C.; Costantino, P.; Vittorioso, P. Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 2002, 14, 1253–1263. [Google Scholar] [CrossRef] [PubMed]
- Rueda-Romero, P.; Barrero-Sicilia, C.; Gomez-Cadenas, A.; Carbonero, P.; Onate-Sanchez, L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 2012, 63, 1937–1949. [Google Scholar] [CrossRef]
- Marzabal, P.; Gas, E.; Fontanet, P.; Vicente-Carbajosa, J.; Torrent, M.; Ludevid, M.D. The maize Dof protein PBF activates transcription of gamma-zein during maize seed development. Plant Mol. Biol. 2008, 67, 441–454. [Google Scholar] [CrossRef]
- Santos, L.A.; de Souza, S.R.; Fernandes, M.S. OsDof25 expression alters carbon and nitrogen metabolism in Arabidopsis under high N-supply. Plant Biotechnol. Rep. 2012, 6, 327–337. [Google Scholar] [CrossRef]
- Venkatesh, J.; Park, S.W. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015, 94, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Boccaccini, A.; Lorrai, R.; Ruta, V.; Frey, A.; Mercey-Boutet, S.; Marion-Poll, A.; Tarkowska, D.; Strnad, M.; Costantino, P.; Vittorioso, P. The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels. BMC Plant Biol. 2016, 16, 198. [Google Scholar] [CrossRef]
- Lorrai, R.; Gandolfi, F.; Boccaccini, A.; Ruta, V.; Possenti, M.; Tramontano, A.; Costantino, P.; Lepore, R.; Vittorioso, P. Genome-wide RNA-seq analysis indicates that the DAG1 transcription factor promotes hypocotyl elongation acting on ABA, ethylene and auxin signaling. Sci. Rep. 2018, 8, 15895. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.G.; Foley, R.C.; Onate-Sanchez, L.; Lin, C.; Singh, K.B. Target genes for OBP3, a Dof transcription factor, include novel basic helix-loop-helix domain proteins inducible by salicylic acid. Plant J. 2003, 35, 362–372. [Google Scholar] [CrossRef]
- Su, Y.; Liang, W.; Liu, Z.; Wang, Y.; Zhao, Y.; Ijaz, B.; Hua, J. Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum. J. Plant Physiol. 2017, 218, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, D.; Deng, X.; Zhen, S.; Wang, Z.; Yan, Y. Effects of water deficit on breadmaking quality and storage protein compositions in bread wheat (Triticum aestivum L.). J. Sci. Food Agric. 2018, 98, 4357–4368. [Google Scholar] [CrossRef]
- Sasaki, N.; Matsumaru, M.; Odaira, S.; Nakata, A.; Nakata, K.; Nakayama, I.; Yamaguchi, K.; Nyunoya, H. Transient expression of tobacco BBF1-related Dof proteins, BBF2 and BBF3, upregulates genes involved in virus resistance and pathogen defense. Physiol. Mol. Plant Pathol. 2015, 89, 70–77. [Google Scholar] [CrossRef]
- Feng, S.; Li, J.; Qian, G.; Feng, B. Association between the yield and the main agronomic traits of Tartary buckwheat evaluated using the random forest model. Crop. Sci. 2020, 60, 2394–2407. [Google Scholar] [CrossRef]
- Li, J.; Feng, S.; Qu, Y.; Gong, X.; Luo, Y.; Yang, Q.; Zhang, Y.; Dang, K.; Gao, X.; Feng, B. Identifying the primary meteorological factors affecting the growth and development of Tartary buckwheat and a comprehensive landrace evaluation using a multi-environment phenotypic investigation. J. Sci. Food Agric. 2021, 101, 6104–6116. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hossain, M.S.; Ma, H.; Yang, Q.; Gong, X.; Yang, P.; Feng, B. Comparative metabolomics reveals differences in flavonoid metabolites among different coloured buckwheat flowers. J. Food Compos. Anal. 2020, 85, 103335. [Google Scholar] [CrossRef]
- Li, J.; Yang, P.; Yang, Q.; Gong, X.; Ma, H.; Dang, K.; Chen, G.; Gao, X.; Feng, B. Analysis of Flavonoid Metabolites in Buckwheat Leaves Using UPLC-ESI-MS/MS. Molecules 2019, 24, 1310. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Wu, D.; Ren, G.; Hu, Y.; Peng, L.; Zhao, J.; Garcia-Perez, P.; Carpena, M.; Prieto, M.A.; Cao, H.; et al. Bioactive compounds, health benefits, and industrial applications of Tartary buckwheat (Fagopyrum tataricum). Crit. Rev. Food Sci. Nutr. 2021, 1–17. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Ma, B.; Gao, Q.; Du, H.; Han, Y.; Li, Y.; Cao, Y.; Qi, M.; Zhu, Y.; et al. The Tartary Buckwheat Genome Provides Insights into Rutin Biosynthesis and Abiotic Stress Tolerance. Mol. Plant 2017, 10, 1224–1237. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Z.; Wang, A.; Zheng, T.; Huang, L.; Sun, W.; Zhang, Y.; Jin, W.; Zhan, J.; Cai, Y.; et al. Genome-Wide Investigation of the Auxin Response Factor Gene Family in Tartary Buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 2018, 19, 3526. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Deng, R.; Zhao, H.; Xiao, Y.; Huang, Y.; Yao, P.; Lei, Y.; Li, C.; Chen, H.; Wu, Q. Cloning, Characterization, and Expression Analysis of Eight Stress-Related NAC Genes in Tartary Buckwheat. Crop. Sci. 2019, 59, 266–279. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Khaksar, G.; Sangchay, W.; Pinsorn, P.; Sangpong, L.; Sirikantaramas, S. Genome-wide analysis of the Dof gene family in durian reveals fruit ripening-associated and cultivar-dependent Dof transcription factors. Sci. Rep. 2019, 9, 12109. [Google Scholar] [CrossRef]
- Schneeberger, K.; Ossowski, S.; Ott, F.; Klein, J.D.; Wang, X.; Lanz, C.; Smith, L.M.; Cao, J.; Fitz, J.; Warthmann, N.; et al. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc. Natl. Acad. Sci. USA 2011, 108, 10249–10254. [Google Scholar] [CrossRef]
- Yu, J.; Hu, S.; Wang, J.; Li, S.; Wong, K.S.G.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice (Oryza sativa ssp. indica) genome. Chin. Sci. Bull. 2001, 46, 1937–1942. [Google Scholar] [CrossRef]
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cheng, Y.; Wan, C.; Li, J.; Yang, Y.; Chen, J. Genome-wide characterization and expression analysis of the Dof gene family related to abiotic stress in watermelon. PeerJ 2020, 8, e8358. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bahler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, S.; Gao, Y.; Yang, J. Characterization of Dof Transcription Factors and Their Responses to Osmotic Stress in Poplar (Populus trichocarpa). PLoS ONE 2017, 12, e0170210. [Google Scholar] [CrossRef]
- Li, H.; Dou, L.; Li, W.; Wang, P.; Zhao, Q.; Xi, R.; Pei, X.; Liu, Y.; Ren, Z. Genome-Wide Identification and Expression Analysis of the Dof Transcription Factor Gene Family in Gossypium hirsutum L. Agronomy 2018, 8, 186. [Google Scholar] [CrossRef]
- Liu, M.; Wang, X.; Sun, W.; Ma, Z.; Zheng, T.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; et al. Genome-wide investigation of the ZF-HD gene family in Tartary buckwheat (Fagopyrum tataricum). BMC Plant Biol. 2019, 19, 248. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification of the SPL gene family in Tartary Buckwheat (Fagopyrum tataricum) and expression analysis during fruit development stages. BMC Plant Biol. 2019, 19, 299. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 113. [Google Scholar] [CrossRef] [PubMed]
- Azam, S.M.; Liu, Y.; Rahman, Z.U.; Ali, H.; Yan, C.; Wang, L.; Priyadarshani, S.V.G.N.; Hu, B.; Huang, X.; Xiong, J.; et al. Identification, Characterization and Expression Profiles of Dof Transcription Factors in Pineapple (Ananas comosus L). Trop. Plant Biol. 2018, 11, 49–64. [Google Scholar] [CrossRef]
- Guo, Y.; Qin, G.; Gu, H.; Qu, L.J. Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 2009, 21, 3518–3534. [Google Scholar] [CrossRef] [PubMed]
- Derbyshire, P.; Drea, S.; Shaw, P.J.; Doonan, J.H.; Dolan, L. Proximal-distal patterns of transcription factor gene expression during Arabidopsis root development. J. Exp. Bot. 2008, 59, 235–245. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fornara, F.; Panigrahi, K.C.S.; Gissot, L.; Sauerbrunn, N.; Ruhl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF Transcription Factors Act Redundantly to Reduce CONSTANS Expression and Are Essential for a Photoperiodic Flowering Response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef]
- Sun, W.; Jin, X.; Ma, Z.; Chen, H.; Liu, M. Basic helix-loop-helix (bHLH) gene family in Tartary buckwheat (Fagopyrum tataricum): Genome-wide identification, phylogeny, evolutionary expansion and expression analyses. Int. J. Biol. Macromol. 2020, 155, 1478–1490. [Google Scholar] [CrossRef]
- Zhong, R.; Richardson, E.A.; Ye, Z.H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Kim, S.J.; Abbasi, N.; Bressan, R.A.; Yun, D.J.; Yoo, S.D.; Kwon, S.Y.; Choi, S.B. The DOF transcription factor Dof5.1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis. Plant J. 2010, 64, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Skirycz, A.; Reichelt, M.; Burow, M.; Birkemeyer, C.; Rolcik, J.; Kopka, J.; Zanor, M.I.; Gershenzon, J.; Strnad, M.; Szopa, J.; et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006, 47, 10–24. [Google Scholar] [CrossRef]
- Kang, H.G.; Singh, K.B. Characterization of salicylic acid-responsive, Arabidopsis Dof domain proteins: Overexpression of OBP3 leads to growth defects. Plant J. 2000, 21, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.F.; Zhang, Y.Q.; Wei, W.; Chen, H.W.; Song, Q.X.; Liu, Y.F.; Zhao, M.Y.; Wang, F.; Zhang, B.C.; Lin, Q.; et al. The transcription factor AtDOF4.2 regulates shoot branching and seed coat formation in Arabidopsis. Biochem. J. 2013, 449, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Siepel, A.; Arbiza, L. Cis-regulatory elements and human evolution. Curr. Opin. Genet. Dev. 2014, 29, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.H.; Bian, L.; Wan, Y.T.; Jiao, Z.L.; Yu, K.K.; Zhang, G.H.; Guo, D.L. Grape (Vitis vinifera) VvDOF3 functions as a transcription activator and enhances powdery mildew resistance. Plant Physiol. Biochem. 2019, 143, 183–189. [Google Scholar] [CrossRef]
- Wang, C.M.; Zeng, Z.X.; Su, X.G.; Lakshmanan, P.; Shan, W.; Kuang, J.F.; Lu, W.J.; Chen, J.Y.; Zhao, Y.T. A transcriptional repressor BrDof2.4 regulates protease genes involved in postharvest leaf senescence in Chinese flowering cabbage. Postharvest Biol. Tec. 2021, 181, 111680. [Google Scholar] [CrossRef]
- Jin, Z.; Chandrasekaran, U.; Liu, A. Genome-wide analysis of the Dof transcription factors in castor bean (Ricinus communis L.). Genes Genom. 2014, 36, 527–537. [Google Scholar] [CrossRef]
- Song, A.; Gao, T.; Li, P.; Chen, S.; Guan, Z.; Wu, D.; Xin, J.; Fan, Q.; Zhao, K.; Chen, F. Transcriptome-Wide Identification and Expression Profiling of the DOF Transcription Factor Gene Family in Chrysanthemum morifolium. Front. Plant Sci. 2016, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Weirauch, M.T.; Hughes, T.R. Conserved expression without conserved regulatory sequence: The more things change, the more they stay the same. Trends Genet. 2010, 26, 66–74. [Google Scholar] [CrossRef]
- Yang, C.; Wang, D.; Zhang, C.; Ye, M.; Kong, N.; Ma, H.; Chen, Q. Comprehensive Analysis and Expression Profiling of PIN, AUX/LAX, and ABCB Auxin Transporter Gene Families in Solanum tuberosum under Phytohormone Stimuli and Abiotic Stresses. Biology 2021, 10, 127. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Gene ID | Chr Location | CDS Length (bp) | Protein Length (aa) | Mw (kDa) | pI | Subcellular Location |
|---|---|---|---|---|---|---|---|
| FtDof1 | FtPinG0002490200.01.T01 | Ft1:6984896-6985480 | 555 | 184 | 20.91976 | 9.23 | Nuclear |
| FtDof2 | FtPinG0005819100.01.T01 | Ft1:11995949-11997181 | 1023 | 340 | 37.16502 | 6.88 | Nuclear |
| FtDof3 | FtPinG0006052400.01.T01 | Ft1:26283542-26284756 | 1029 | 342 | 37.13006 | 7.13 | Nuclear |
| FtDof4 | FtPinG0000038100.01.T01 | Ft1:26894633-26895469 | 786 | 261 | 26.95576 | 6.29 | Nuclear |
| FtDof5 | FtPinG0006014000.01.T01 | Ft1:35519701-35521178 | 1116 | 371 | 39.08230 | 9.15 | Nuclear |
| FtDof6 | FtPinG0000383700.01.T01 | Ft1:51008719-51009996 | 828 | 275 | 30.39486 | 8.88 | Nuclear |
| FtDof7 | FtPinG0007854200.01.T01 | Ft2:12819999-12820866 | 735 | 244 | 26.69874 | 9.84 | Nuclear |
| FtDof8 | FtPinG0009668700.01.T01 | Ft2:46641998-46642846 | 846 | 281 | 31.58071 | 4.46 | Nuclear |
| FtDof9 | FtPinG0003838400.01.T01 | Ft2:56803328-56804871 | 729 | 242 | 26.43628 | 9.23 | Nuclear |
| FtDof10 | FtPinG0000870700.01.T01 | Ft2:60683203-60684009 | 792 | 263 | 29.30853 | 7.13 | Nuclear |
| FtDof11 | FtPinG0003987600.01.T01 | Ft3:4327917-4328955 | 879 | 292 | 32.09061 | 8.60 | Nuclear |
| FtDof12 | FtPinG0006042300.01.T01 | Ft3:8679735-8681642 | 1089 | 362 | 39.37413 | 6.55 | Nuclear |
| FtDof13 | FtPinG0001985100.01.T01 | Ft3:17940022-17941017 | 954 | 317 | 34.10505 | 8.84 | Nuclear |
| FtDof14 | FtPinG0009592200.01.T01 | Ft3:19407541-19409658 | 846 | 281 | 31.00104 | 9.56 | Nuclear |
| FtDof15 | FtPinG0006943800.01.T01 | Ft3:20185865-20187740 | 1209 | 402 | 44.58969 | 7.58 | Nuclear |
| FtDof16 | FtPinG0001746600.01.T01 | Ft3:38071212-38072846 | 969 | 322 | 35.98977 | 8.57 | Nuclear |
| FtDof17 | FtPinG0006517000.01.T01 | Ft3:45271193-45272090 | 756 | 251 | 27.20933 | 9.32 | Nuclear |
| FtDof18 | FtPinG0001221400.01.T01 | Ft3:52038929-52039519 | 519 | 172 | 19.48991 | 9.08 | Nuclear |
| FtDof19 | FtPinG0002126900.01.T01 | Ft3:54786097-54786807 | 696 | 231 | 26.33179 | 4.83 | Nuclear |
| FtDof20 | FtPinG0006352100.01.T01 | Ft4:3820883-3823102 | 1305 | 434 | 47.49093 | 6.05 | Nuclear |
| FtDof21 | FtPinG0000044800.01.T01 | Ft4:5503745-5504860 | 1029 | 342 | 37.27505 | 6.00 | Nuclear |
| FtDof22 | FtPinG0005157600.01.T01 | Ft4:15029533-15030375 | 750 | 249 | 27.95802 | 9.15 | Nuclear |
| FtDof23 | FtPinG0000644200.01.T01 | Ft5:3393885-3394904 | 831 | 276 | 31.16242 | 6.13 | Nuclear |
| FtDof24 | FtPinG0007868100.01.T01 | Ft5:39770127-39770799 | 639 | 212 | 23.72748 | 8.61 | Nuclear |
| FtDof25 | FtPinG0007057700.01.T01 | Ft6:7340538-7343583 | 1416 | 471 | 50.79071 | 6.20 | Nuclear |
| FtDof26 | FtPinG0006338000.01.T01 | Ft6:10848783-10849422 | 423 | 140 | 15.48469 | 9.21 | Nuclear |
| FtDof27 | FtPinG0000330300.01.T01 | Ft6:49780770-49782567 | 1008 | 335 | 36.87670 | 9.15 | Nuclear |
| FtDof28 | FtPinG0002571600.01.T01 | Ft7:33993961-33994407 | 393 | 130 | 14.84745 | 9.32 | Nuclear |
| FtDof29 | FtPinG0009543500.01.T01 | Ft7:50345602-50346563 | 858 | 285 | 31.47978 | 8.16 | Nuclear |
| FtDof30 | FtPinG0002293400.01.T01 | Ft8:1391182-1394031 | 990 | 329 | 36.68763 | 7.62 | Nuclear |
| FtDof31 | FtPinG0002167800.01.T01 | Ft8:3967827-3968546 | 720 | 239 | 26.66127 | 5.13 | Nuclear |
| FtDof32 | FtPinG0008252900.01.T01 | Ft8:15269916-15270644 | 675 | 224 | 23.02970 | 8.57 | Nuclear |
| FtDof33 | FtPinG0004209600.01.T01 | Ft8:18068180-18069652 | 1092 | 363 | 37.55589 | 9.03 | Nuclear |
| FtDof34 | FtPinG0005937800.01.T01 | Ft8:47262582-47263547 | 702 | 233 | 26.2462 | 6.44 | Nuclear |
| FtDof35 | FtPinG0000702500.01.T01 | Ft8:47316870-47317455 | 531 | 176 | 19.33684 | 9.39 | Nuclear |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Zhang, Y.; Xu, L.; Wang, C.; Luo, Y.; Feng, S.; Yuan, Y.; Yang, Q.; Feng, B. Genome-Wide Identification of DNA Binding with One Finger (Dof) Gene Family in Tartary Buckwheat (Fagopyrum tataricum) and Analysis of Its Expression Pattern after Exogenous Hormone Stimulation. Biology 2022, 11, 173. https://doi.org/10.3390/biology11020173
Li J, Zhang Y, Xu L, Wang C, Luo Y, Feng S, Yuan Y, Yang Q, Feng B. Genome-Wide Identification of DNA Binding with One Finger (Dof) Gene Family in Tartary Buckwheat (Fagopyrum tataricum) and Analysis of Its Expression Pattern after Exogenous Hormone Stimulation. Biology. 2022; 11(2):173. https://doi.org/10.3390/biology11020173
Chicago/Turabian StyleLi, Jing, Yuchuan Zhang, Lei Xu, Chenyang Wang, Yan Luo, Shan Feng, Yuhao Yuan, Qinghua Yang, and Baili Feng. 2022. "Genome-Wide Identification of DNA Binding with One Finger (Dof) Gene Family in Tartary Buckwheat (Fagopyrum tataricum) and Analysis of Its Expression Pattern after Exogenous Hormone Stimulation" Biology 11, no. 2: 173. https://doi.org/10.3390/biology11020173
APA StyleLi, J., Zhang, Y., Xu, L., Wang, C., Luo, Y., Feng, S., Yuan, Y., Yang, Q., & Feng, B. (2022). Genome-Wide Identification of DNA Binding with One Finger (Dof) Gene Family in Tartary Buckwheat (Fagopyrum tataricum) and Analysis of Its Expression Pattern after Exogenous Hormone Stimulation. Biology, 11(2), 173. https://doi.org/10.3390/biology11020173






