A New Cell Line Derived from the Spleen of the Japanese Flounder (Paralichthys olivaceus) and Its Application in Viral Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Primary Cell Culture and Routine Maintenance
2.2. Cryopreservation and Thawing of Cells
2.3. Species Confirmation of JFSP Cells
2.4. Effects of Medium, Temperature, and FBS on Cell Growth
2.5. Chromosome Number/Analysis
2.6. Cell Transfection
2.7. Virus Susceptibility
2.8. Preparation of Sample for Transmission Electron Microscopy (TEM)
2.9. Detection of Viral Copies by qRT-PCR
2.10. qRT-PCR Analysis of Antiviral-Related Genes
3. Results
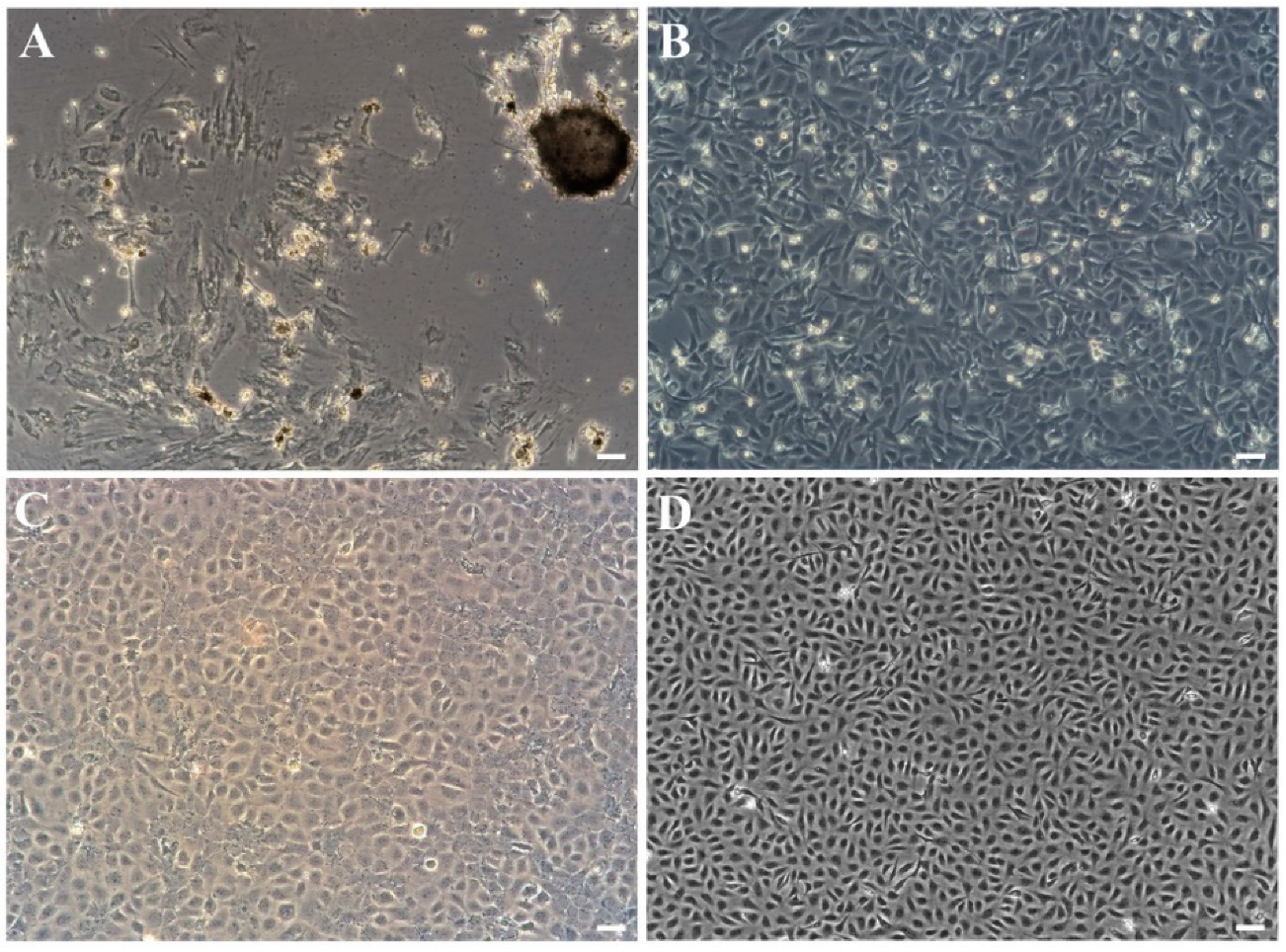
3.1. Primary Culture and Subculture
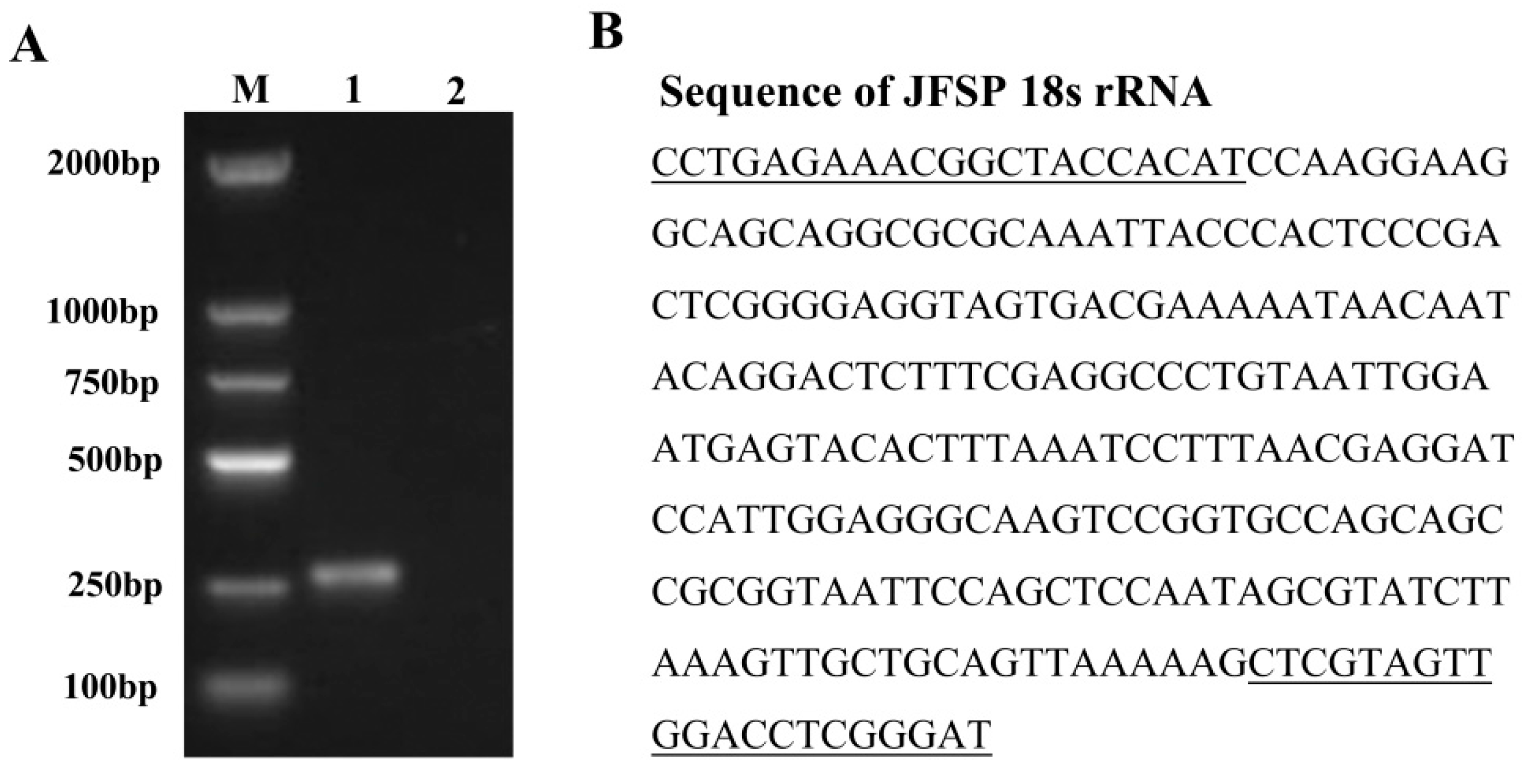
3.2. Species Confirmation of JFSP Cells
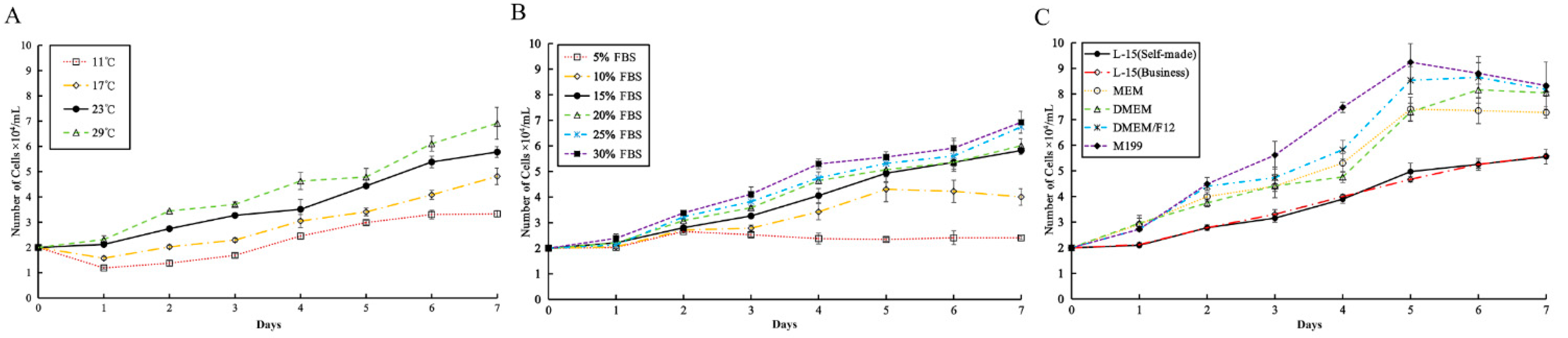
3.3. Effects of Temperature, FBS, and Medium, on Cell Growth
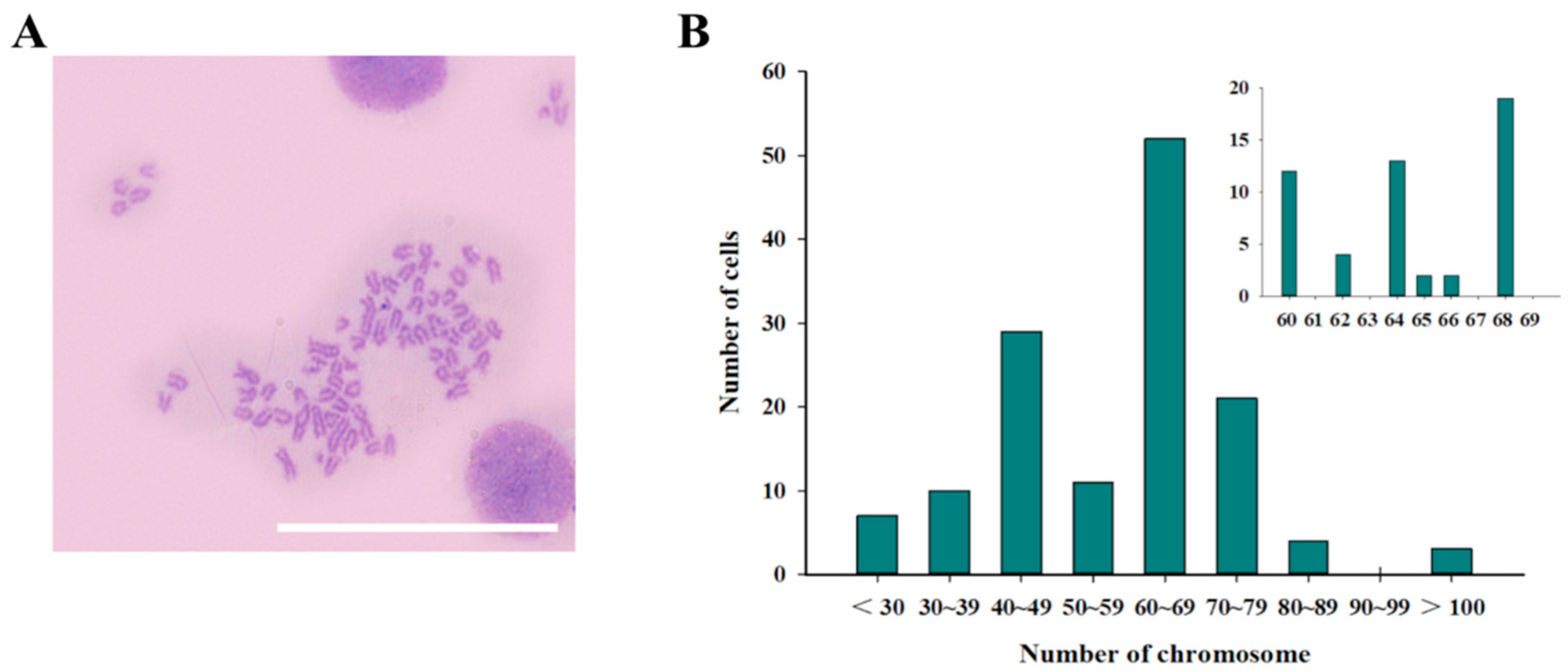
3.4. Chromosome Number
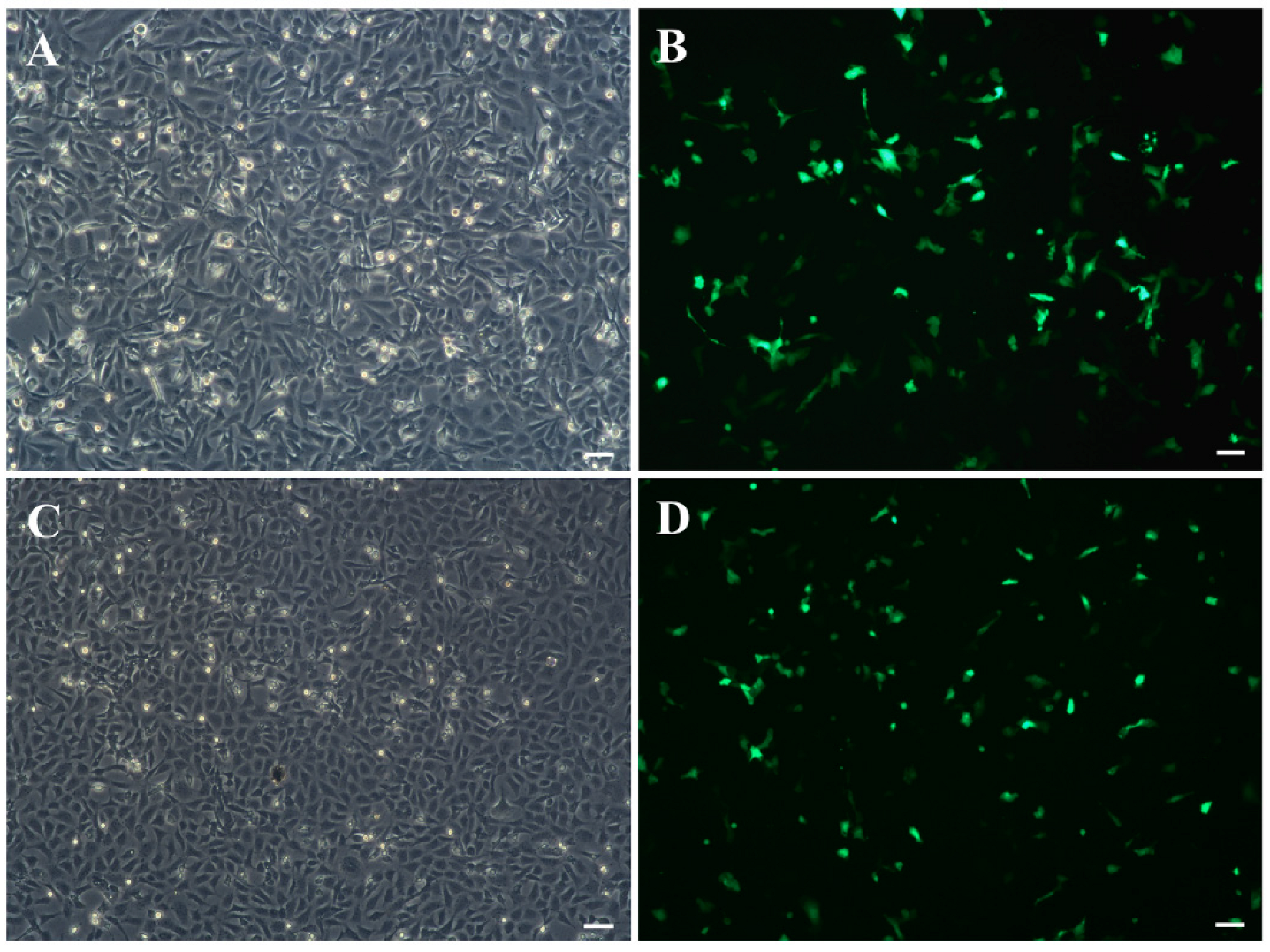
3.5. Cell Transfection
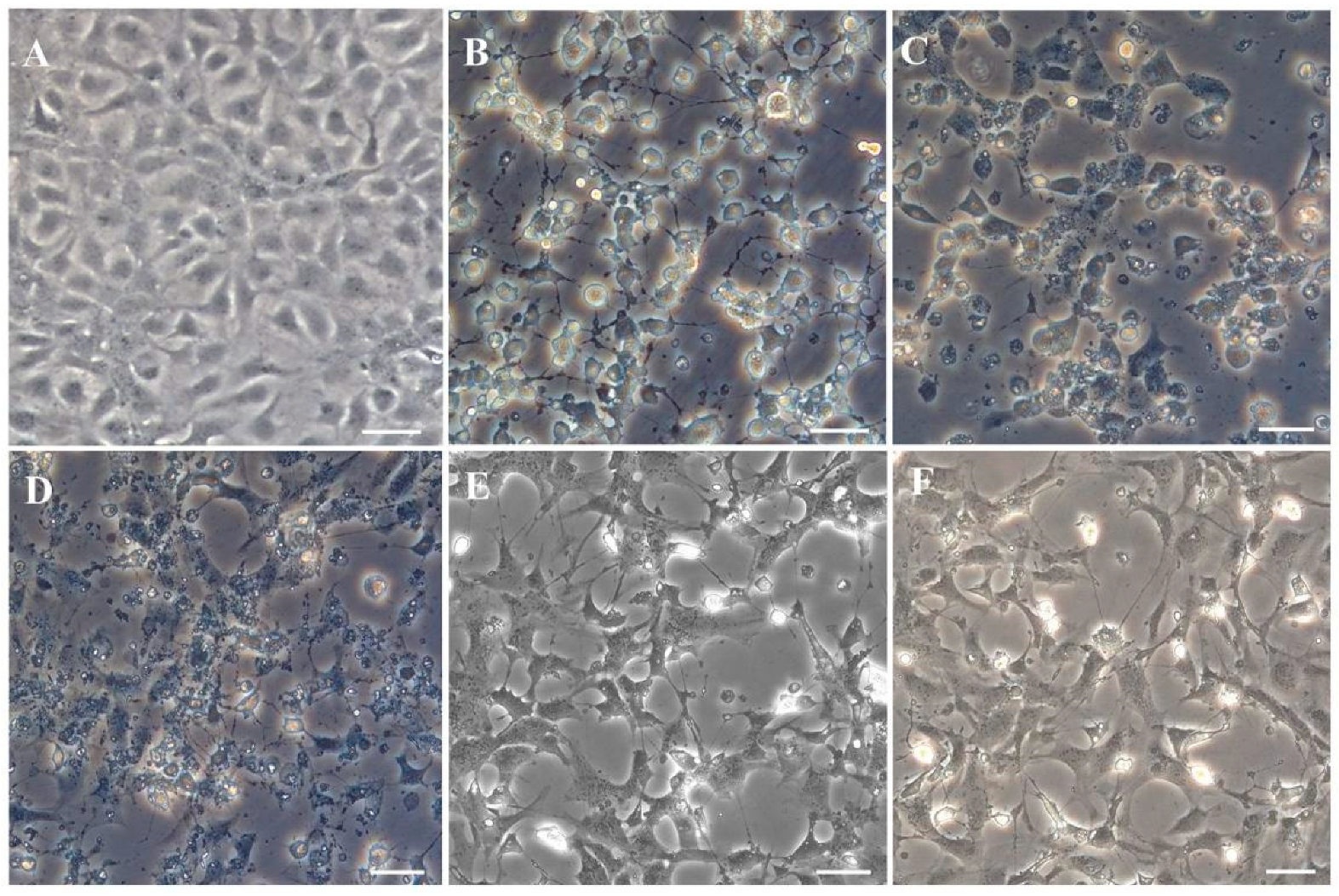
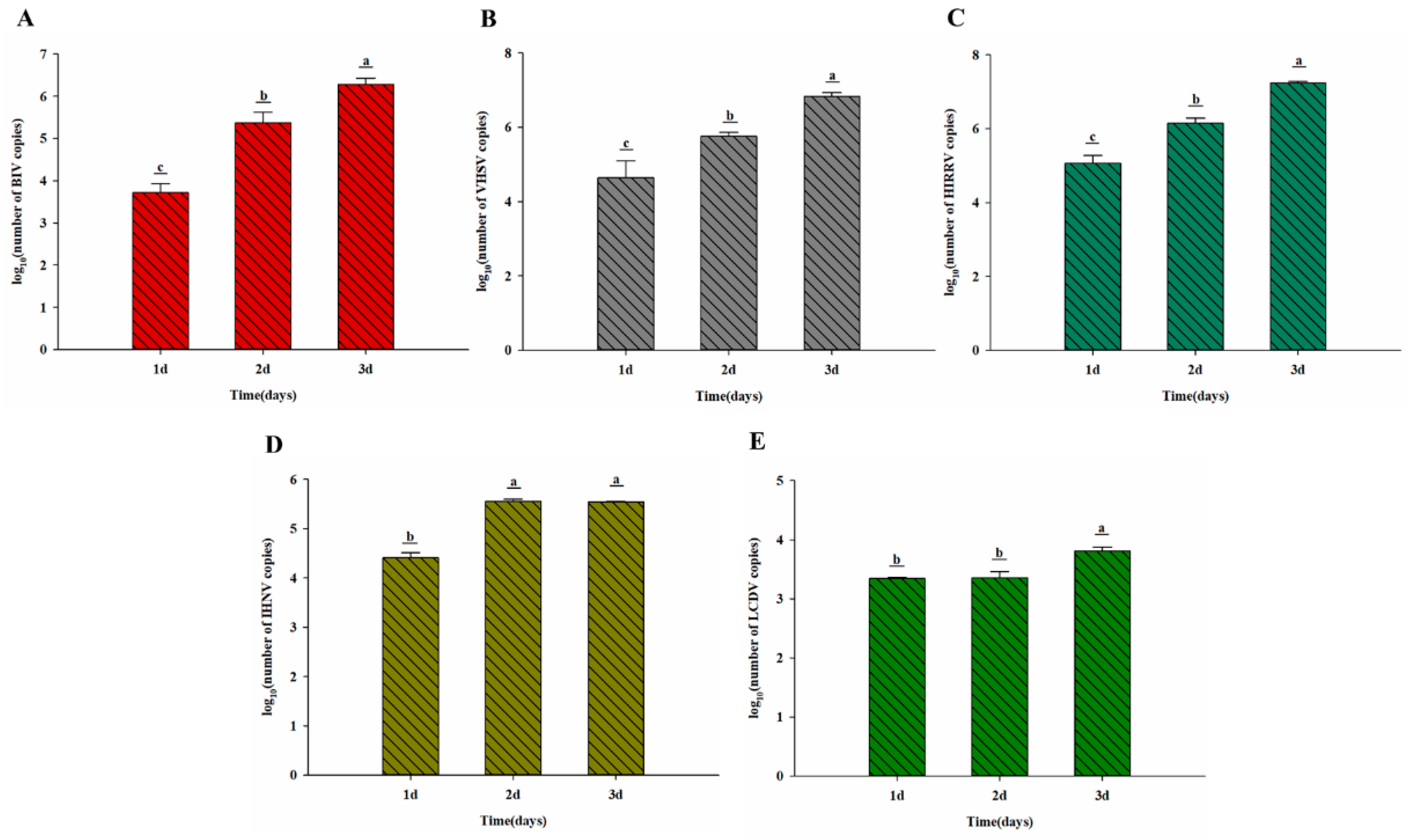
3.6. Virus Susceptibility
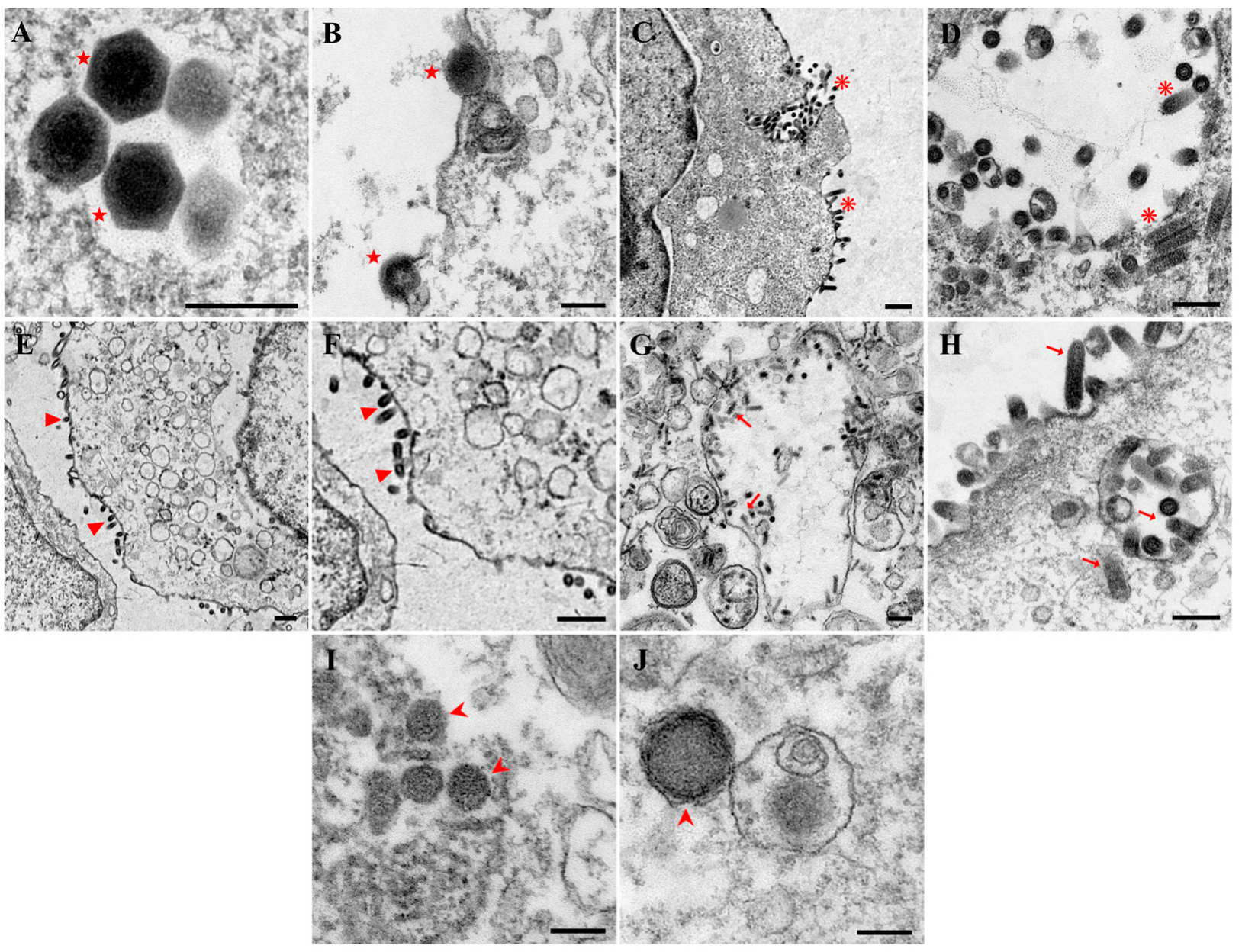
3.7. Virus Transmission Electron Microscopy Observation
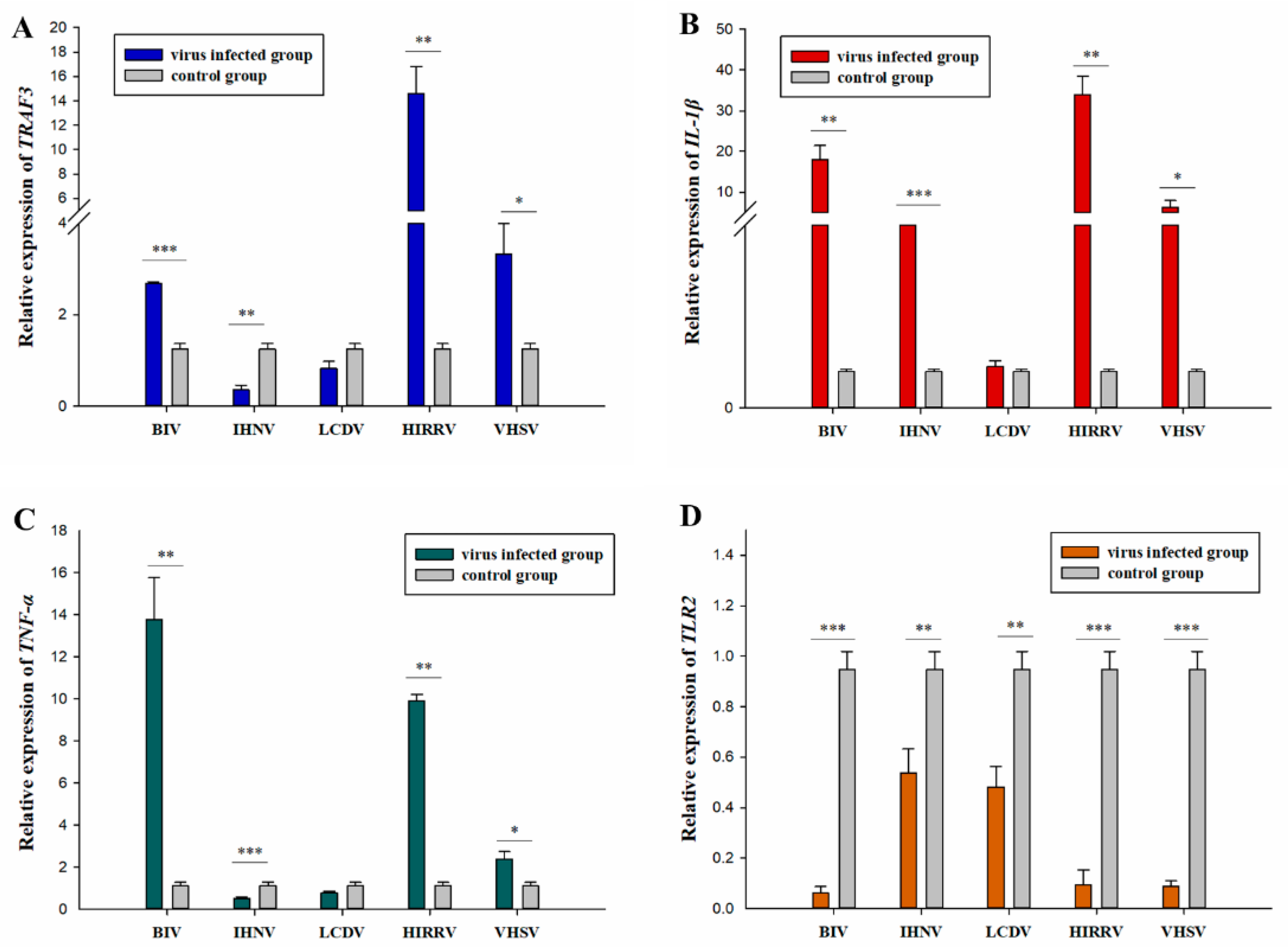
3.8. The Expression Profiles of Antiviral-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, Y.N.; Nan, X.Y.; Zhang, X.Y.; Wang, G.X.; Ren, Y.Q.; Wang, Y.F.; Fu, Y.S.; Hou, J.L. Molecular characterization and functional analysis of Japanese flounder (Paralichthys olivaceus) thbs2 in response to lymphocystis disease virus. Fish Shellfish Immunol. 2019, 93, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.L.; Tang, X.Q.; Sheng, X.Z.; Xing, J.; Zhan, W.B. Isolation and identification of a new strain of hirame rhabdovirus (HIRRV) from Japanese flounder Paralichthys olivaceus in China. Virol. J. 2017, 14, 73. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.W.; Li, Z.L.; Xiang, Y.; Jia, P.; Liu, W.; Yi, M.; Jia, K. Isolation and identification of a viral haemorrhagic septicaemia virus (VHSV) isolate from wild largemouth bass Micropterus salmoides in China. J. Fish Dis. 2019, 42, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.S.; Kim, S.R.; Kim, D.; Kim, J.O.; Park, M.A.; Kitamura, S.I.; Kim, H.Y.; Kim, D.H.; Han, H.J.; Jung, S.J.; et al. An outbreak of VHSV (viral hemorrhagic septicemia virus) infection in farmed olive flounder Paralichthys olivaceus in Korea. Aquaculture 2009, 296, 165–168. [Google Scholar] [CrossRef]
- Gong, C.G.; Zhang, Y.T.; Wang, G.X.; Liu, Y.F.; He, Z.W.; Ren, Y.Q.; Cao, W.; Zhao, H.T.; Xu, Y.H.; Wang, Y.F.; et al. The isolation and full-length transcriptome sequencing of a novel nidovirus and response of its infection in Japanese flounder (Paralichthys olivaceus). Viruses 2022, 14, 1216. [Google Scholar] [CrossRef]
- Zhang, J.L.; Tang, X.Q.; Sheng, X.Z.; Xing, J.; Zhan, W.B. The influence of temperature on viral replication and antiviral-related genes response in hirame rhabdovirus-infected flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2017, 68, 260–265. [Google Scholar] [CrossRef]
- Kimura, T.; Yoshimizu, M.; Gorie, S. A new rhabdovirus isolated in Japan from cultured hirame (Japanese flounder) Paralichthys olivaceus and ayu Plecoglossus altivelis. Dis. Aquat Organ. 1986, 1, 209–217. [Google Scholar] [CrossRef]
- Borrego, J.J.; Valverde, E.J.; Labella, A.M.; Castro, D. Lymphocystis disease virus: Its importance in aquaculture. Rev. Aquacult. 2017, 9, 179–193. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2011; pp. 1261–1291. [Google Scholar]
- Wolf, K.; Quimby, M.C. Established eurythermic line of fish cells in vitro. Science 1962, 135, 1065–1066. [Google Scholar] [CrossRef]
- Amos, B. The cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 2018, 29, 25–38. [Google Scholar]
- Swaminathan, T.R.; Lekshmi, N.; Preena, P.G.; Raja, S.A.; Arathi, D.; Raj, N.S. Comprehensive update on inventory of finfish cell lines developed during the last decade (2010–2020). Rev. Aquacult. 2021, 13, 2248–2288. [Google Scholar]
- Chen, S.L.; Ren, G.C.; Sha, Z.X.; Shi, C.Y. Establishment of a continuous embryonic cell line from Japanese flounder Paralichthys olivaceus for virus isolation. Dis. Aquat Organ. 2004, 60, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Puentes, C.F.; Novoa, B.A. Figueras, Initiation of a cell line from turbot (Scophthalmus Maximus L.). In Vitro Cell. Dev. Biol. Anim. 2007, 29, 899–900. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Peng, L.M.; You, F.; Zou, Y.X.; Zhang, P.J.; Chen, S.L. Establishment and characterization of a fish-cell line from the brain of Japanese flounder Paralichthys olivaceus. J. Fish Biol. 2015, 87, 115–122. [Google Scholar] [CrossRef]
- Gao, C.; Song, H.; Wang, M.; Liu, X.; Zhao, J.; Wang, X.; Zhang, Q. Establishment and characterization of four long-term cultures of neural stem/progenitor cell lines from the Japanese flounder Paralichthys olivaceus. J. Ocean U China 2020, 19, 1153–1162. [Google Scholar] [CrossRef]
- Choi, H.J.; Lee, J.H.; Jung, H.J.; Lee, J.N.; Kim, K.I.; Kim, J.H.; Kang, Y.J. Establishment and characterization of a new cell line derived from the fin of olive flounder (Paralichthys olivaceus). Aquaculture 2020, 528, 735534. [Google Scholar] [CrossRef]
- Nie, M.; Wu, Z.; You, F. Derivation and characterization of a new embryonic cell line from the olive flounder Paralichthys olivaceus. Turk. J. Fish Aquat. Sci. 2021, 21, 159–167. [Google Scholar] [CrossRef]
- Tong, S.L.; Li, H.; Miao, H.Z. The establishment and partial characterization of a continuous fish cell line FG-9307 from the gill of flounder Paralichthys olivaceus. Aquaculture 1997, 156, 327–333. [Google Scholar] [CrossRef]
- Wang, R.Q.; Zhang, N.W.; Wang, R.K.; Wang, S.P.; Wang, N. Two skin cell lines from wild-type and albino Japanese flounder (Paralichthys olivaceus): Establishment, characterization, virus susceptibility, efficient transfection, and application to albinism study. Fish Physiol. Biochem. 2017, 43, 1477–1486. [Google Scholar] [CrossRef]
- Kang, M.S.; Oh, M.J.; Kim, Y.J.; Kawai, K.; Jung, S.J. Establishment and characterization of two new cell lines derived from flounder, Paralichthys olivaceus (Temminck Schlegel). J. Fish Dis. 2010, 26, 657–665. [Google Scholar] [CrossRef]
- Zhang, J.L.; Tang, X.Q.; Sheng, X.Z.; Xing, J.; Zhan, W.B. Primary cell culture of Japanese flounder (Paralichthys olivaceus) kidney and determination of their susceptibility to hirame rhabdovirus. Period. Ocean Univ. China 2018, 48, 19–24. [Google Scholar]
- Blander, J.M.; Sander, L.E. Beyond pattern recognition: Five immune checkpoints for scaling the microbial threat. Nat. Rev. Immunol. 2012, 12, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Meguro, Y.; Nakai, T.; Muroga, K.; Sorimachi, M. A cell line derived from the fin of Japanese flounder, Paralichthys olivaceus. Fish Pathol. 1991, 26, 69–75. [Google Scholar] [CrossRef][Green Version]
- Kim, J.W.; Bang, G.O.; Kim, J.; Kim, D.G.; Kong, H.J. Development and characterization of a new cell line from olive flounder Paralichthys olivaceus. Dev. Reprod. 2018, 22, 225–234. [Google Scholar] [CrossRef]
- Kim, J.W.; Cho, J.Y.; Kim, D.G.; Nam, B.H.; Kong, H.J. Establishment of conditions for long-term maintenance of primary embryonic cell cultures from olive flounder Paralichthys olivaceus. Dev. Reprod. 2020, 24, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Coupar, B.E.H.; Goldie, S.G.; Hyatt, A.D.; Pallister, J.A. Identification of a Bohle iridovirus thymidine kinase gene and demonstration of activity using vaccinia virus. Arch. Virol. 2005, 150, 1797–1812. [Google Scholar] [CrossRef]
- Maclaine, A.; Mashkour, N.; Scott, J.; Ariel, E. Susceptibility of eastern water dragons Intellagama lesueurii lesueurii to Bohle iridovirus. Dis. Aquat. Organ. 2018, 127, 97–105. [Google Scholar] [CrossRef]
- Subramaniam, K.; Waltzek, T.B.; Gregory, C.V. Genomic sequence of a Bohle iridovirus strain isolated from a diseased boreal toad (Anaxyrus boreas boreas) in a North American aquarium. Arch. Virol. 2019, 164, 1923–1926. [Google Scholar] [CrossRef]
- Moody, N.J.G.; Owens, L. Experimental demonstration of the pathogenicity of a frog virus, Bohle iridovirus, for a fish species, barramundi Lates calcarifer. Dis. Aquat. Organ. 1994, 18, 95–102. [Google Scholar] [CrossRef]
- McGrogan, D.G.; Ostland, V.E.; Byrne, P.J.; Ferguson, H.W. Systemic disease involving an iridovirus-like agent in cultured tilapia, Oreochromis niloticus L. a case report. J. Fish Dis. 1998, 21, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Evans, J.E.; Rock, K.L. Molecular identification of a danger signal that alerts the immune system to dying cells. Nature 2003, 425, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Peddie, S.; McLauchlan, P.E.; Ellis, A.E.; Secombes, C.J. Effect of intraperitoneally administered IL-1beta-derived peptides on resistance to viral haemorrhagic septicaemia in rainbow trout Oncorhynchus mykiss. Dis. Aquat. Organ. 2003, 56, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Emmadi, D.; Iwahori, A.; Hirono, I.; Aoki, T. cDNA microarray analysis of interleukin-1β-induced Japanese flounder Paralichthys olivaceus kidney cells. Fish. Sci. 2010, 71, 519–530. [Google Scholar] [CrossRef]
- Peñaranda, M.M.; Purcell, M.K.; Kurath, G. Differential virulence mechanisms of infectious hematopoietic necrosis virus in rainbow trout (Oncorhynchus mykiss) include host entry and virus replication kinetics. J. Gen Virol. 2009, 90, 2172. [Google Scholar] [CrossRef]
- Wei, X.X.; Li, X.Z.; Zheng, X.C.; Jia, P.; Wang, J.J.; Yang, X.L.; Yu, L.; Shi, X.J.; Tong, G.X.; Liu, H. Toll-like receptors and interferon associated immune factors responses to spring viraemia of carp virus infection in common carp (Cyprinus carpio). Fish Shellfish Immunol. 2016, 55, 568–576. [Google Scholar] [CrossRef]
- Avunje, S.; Kim, W.S.; Park, C.S.; Jung, S.J. Toll-like receptors and interferon associated immune factors in viral haemorrhagic septicaemia virus-infected olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2011, 31, 407–414. [Google Scholar] [CrossRef]
- Wu, R.; Sheng, X.; Tang, X.; Xing, J.; Zhan, W. Transcriptome analysis of flounder (Paralichthys olivaceus) gill in response to lymphocystis disease virus (LCDV) infection: Novel insights into fish defense mechanisms. Int. J. Mol. Sci. 2018, 19, 160. [Google Scholar] [CrossRef]
| Primer | Sequences (5′-3′) | Taa | Source or Reference |
|---|---|---|---|
| Genetic properties analysis | |||
| 18s rRNA-F | CCTGAGAAACGGCTACCACAT | 60 | self-designed |
| 18s rRNA-R | ATCCCGAGGTCCAACTACGAG | ||
| qRT-PCR analysis of viral replication | |||
| Q-LCDV-F | TTGACAGCAGGCGATTTAGAC | 55 | self-designed |
| Q-LCDV-R | AACGACGTTCCTCATTGGTTA | ||
| Q-HIRRV-F | CGACCTGACATTCTACTATACGAC | 55 | self-designed |
| Q-HIRRV-R | TGGAGCACTTCCCTTCAATAA | ||
| Q-IHNV-F | GACCCTTTGGGGATGAGTGG | 60 | self-designed |
| Q-IHNV-R | ATGCTCGTCTTGTACTGGGC | ||
| Q-VHSV-F | GTCAAGGCAATTGTGGCTGG | 60 | self-designed |
| Q-VHSV-R | TGGAGTCAGTTTCCTCGTGC | ||
| Q-BIV-F | AAAGTACATACTCTACCAGCTCCTC | 55 | self-designed |
| Q-BIV-R | GCTTCCAGTCTTTACGGTCAG | ||
| qRT-PCR analysis of immune-related genes | |||
| TNF-α-F | AAACACCTCACGTCCATCA | 60 | [23] |
| TNF-α-R | GCGTCCTCCTGACTCTTCT | ||
| IL-1β-F | AAAGAAGCATCACCACTGTCT | 60 | [23] |
| IL-1β-R | TGGTAGCACCGGGCATTCT | ||
| TLR2-F | GCTACATCTGCGACTCTCCT | 58 | self-designed |
| TLR2-R | CACAGGGACACGAACAAATC | ||
| TRAF3-F | CACATCATTCCGCTCCTCTTA | 60 | self-designed |
| TRAF3-R | GCGTTCATTCACGACTTTACC | ||
| β-actin-F | GGAAATCGTGCGTGACATTAAG | 60 | self-designed |
| β-actin-R | CCTCTGGACAACGGAACCTCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Ren, Y.; Zhang, Y.; Wang, G.; He, Z.; Liu, Y.; Cao, W.; Wang, Y.; Chen, S.; Fu, Y.; et al. A New Cell Line Derived from the Spleen of the Japanese Flounder (Paralichthys olivaceus) and Its Application in Viral Study. Biology 2022, 11, 1697. https://doi.org/10.3390/biology11121697
Yang Y, Ren Y, Zhang Y, Wang G, He Z, Liu Y, Cao W, Wang Y, Chen S, Fu Y, et al. A New Cell Line Derived from the Spleen of the Japanese Flounder (Paralichthys olivaceus) and Its Application in Viral Study. Biology. 2022; 11(12):1697. https://doi.org/10.3390/biology11121697
Chicago/Turabian StyleYang, Yucong, Yuqin Ren, Yitong Zhang, Guixing Wang, Zhongwei He, Yufeng Liu, Wei Cao, Yufen Wang, Songlin Chen, Yuanshuai Fu, and et al. 2022. "A New Cell Line Derived from the Spleen of the Japanese Flounder (Paralichthys olivaceus) and Its Application in Viral Study" Biology 11, no. 12: 1697. https://doi.org/10.3390/biology11121697
APA StyleYang, Y., Ren, Y., Zhang, Y., Wang, G., He, Z., Liu, Y., Cao, W., Wang, Y., Chen, S., Fu, Y., & Hou, J. (2022). A New Cell Line Derived from the Spleen of the Japanese Flounder (Paralichthys olivaceus) and Its Application in Viral Study. Biology, 11(12), 1697. https://doi.org/10.3390/biology11121697





