Exploring Active Peptides with Antimicrobial Activity In Planta against Xylella fastidiosa
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Media
2.2. Peptide Synthesis
2.3. Antibiogram Assays
2.4. Effect of Selected AMPs on Xylella fastidiosa Growth
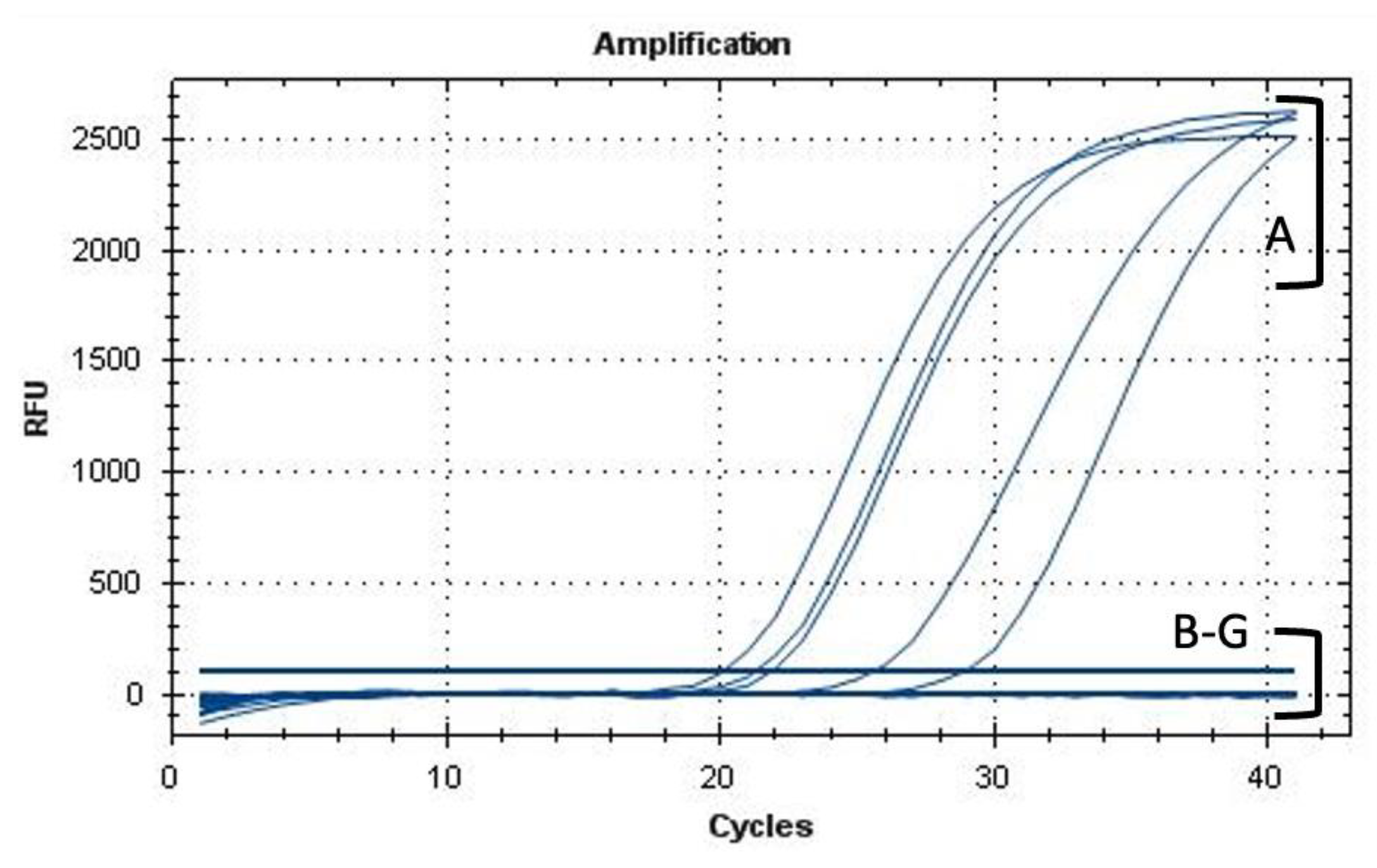
2.5. Viable-Quantitative PCR (v-qPCR) and Fluorescence Microscopy (FM)
2.6. Transmission Electron Microscopy (TEM) of AMPs against Xylella fastidiosa
2.7. Antibiofilm and Planktonic Cell Activity
2.8. Toxicity Evaluation of AMPs
2.9. In Planta Antagonistic Activity of AMPs against Xylella fastidiosa
3. Results
3.1. Screening of Potent AMP Activities against Xanthomonas albilineans and Xylella fastidiosa
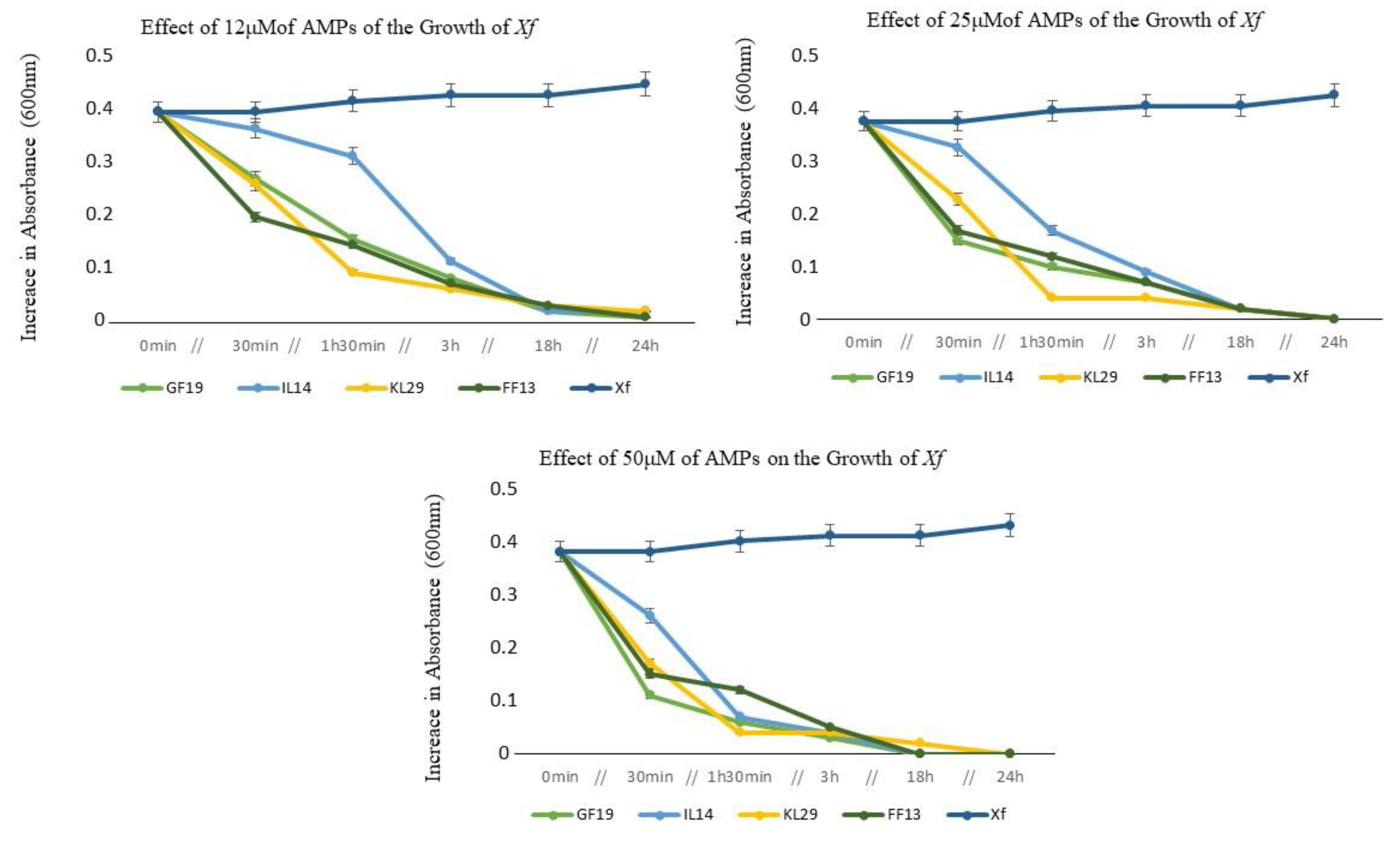
3.2. Optical Density of AMP-Treated Xylella fastidiosa Cells
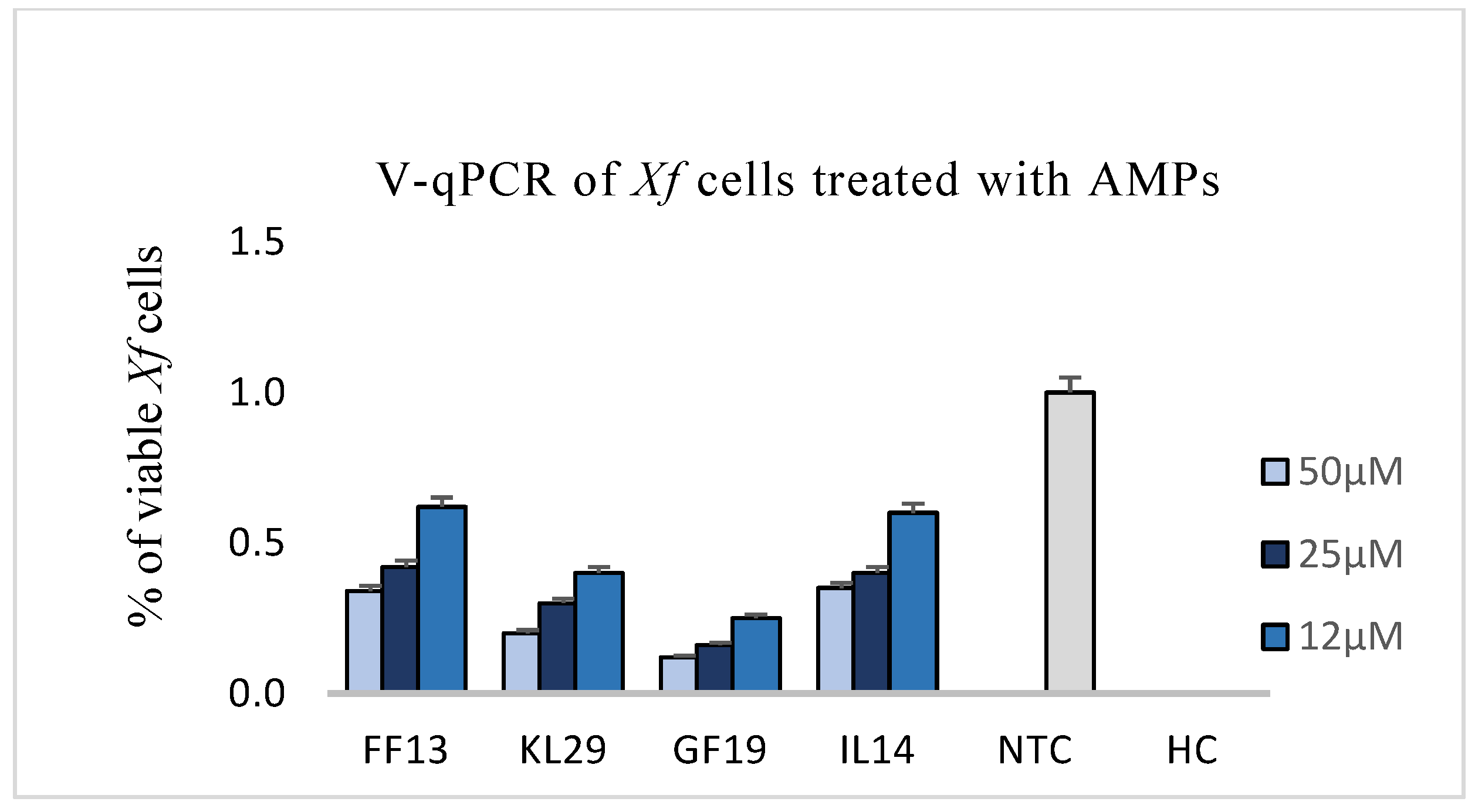
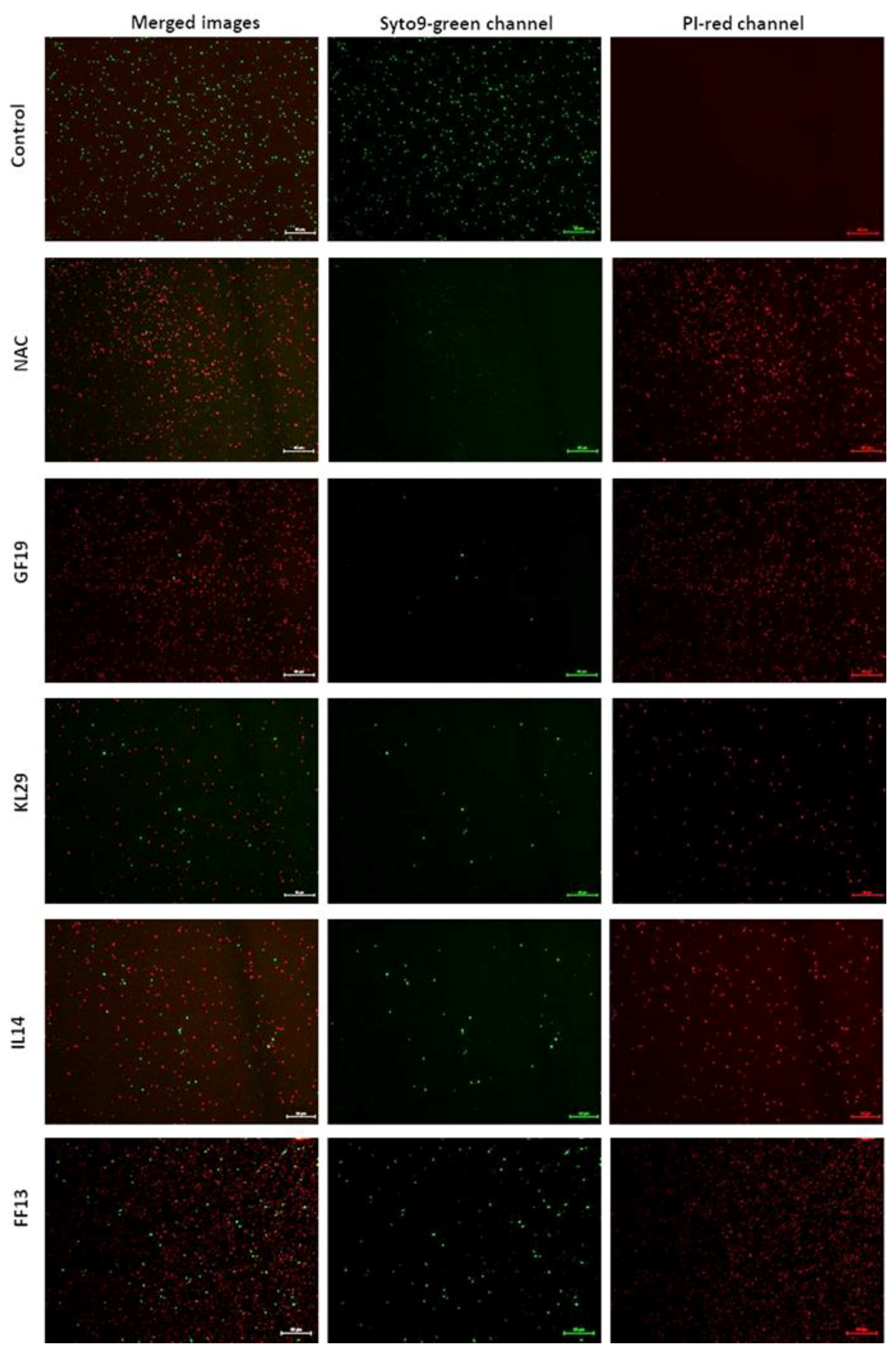
3.3. Effect of AMPs on Xylella fastidiosa: V-qPCR, Fluorescence, and Transmission Electron Microscopy
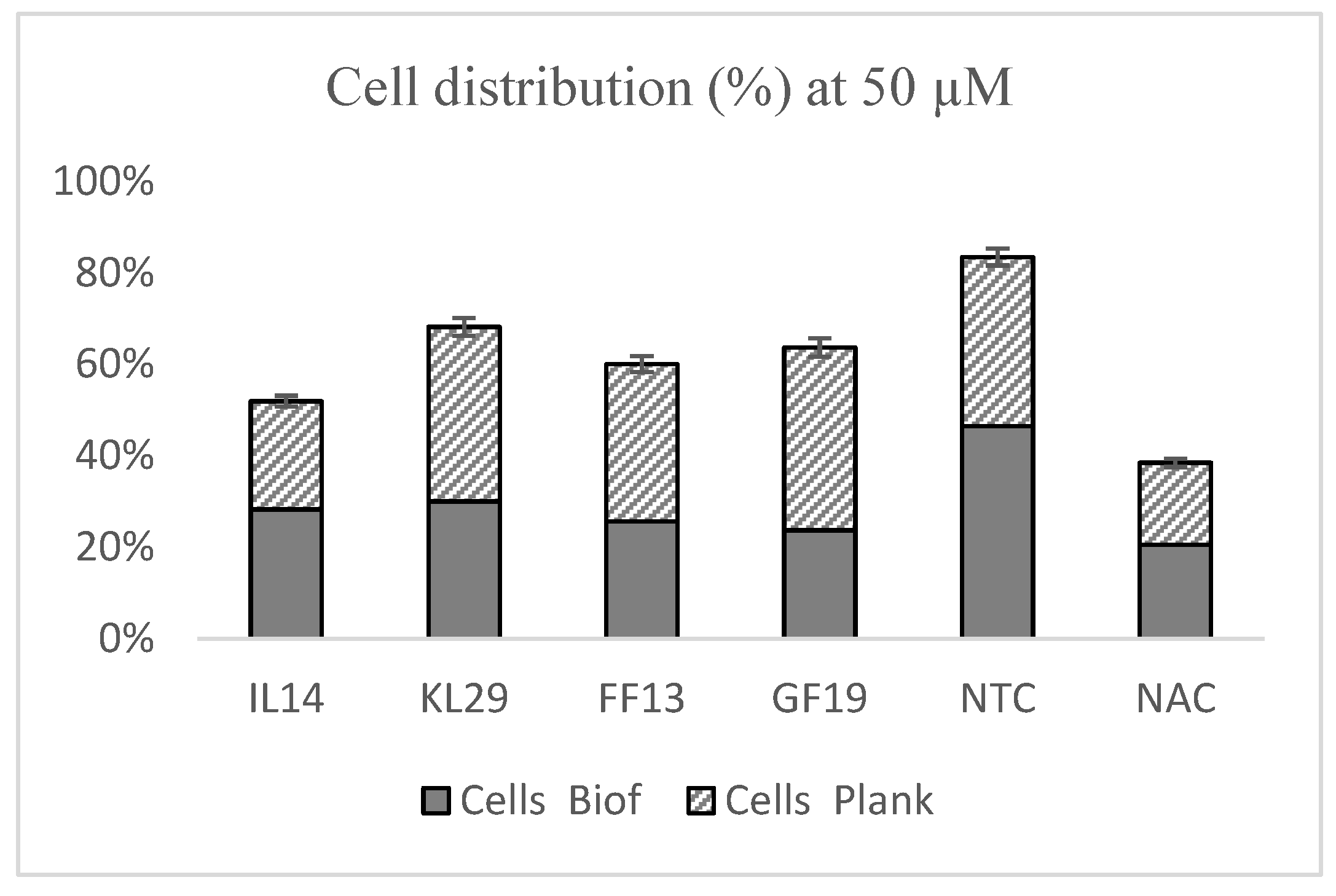
3.4. Effects of AMPs against Xylella fastidiosa Biofilm Formation
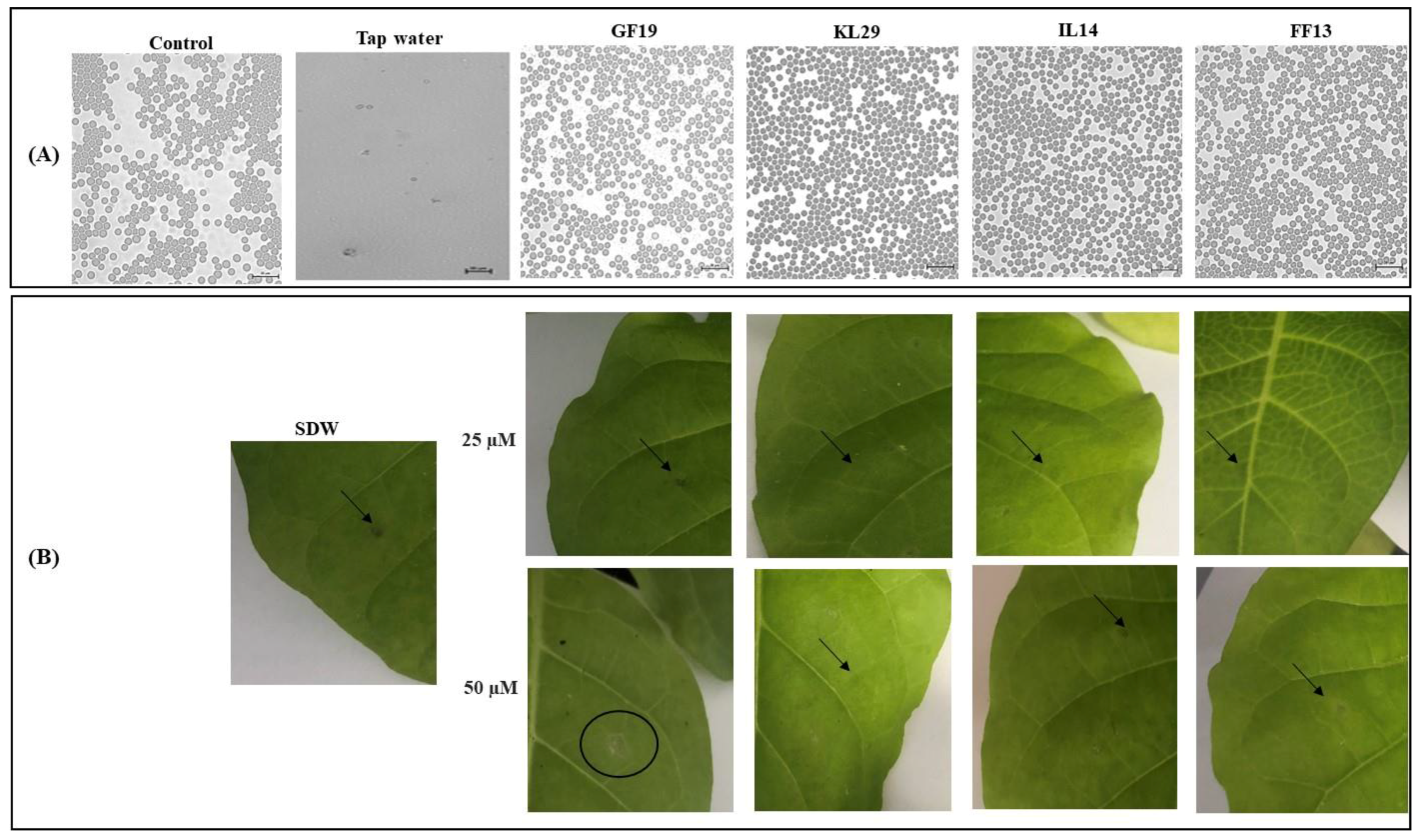
3.5. Evaluation of AMPs’ Toxicity on Eukaryotic Cells
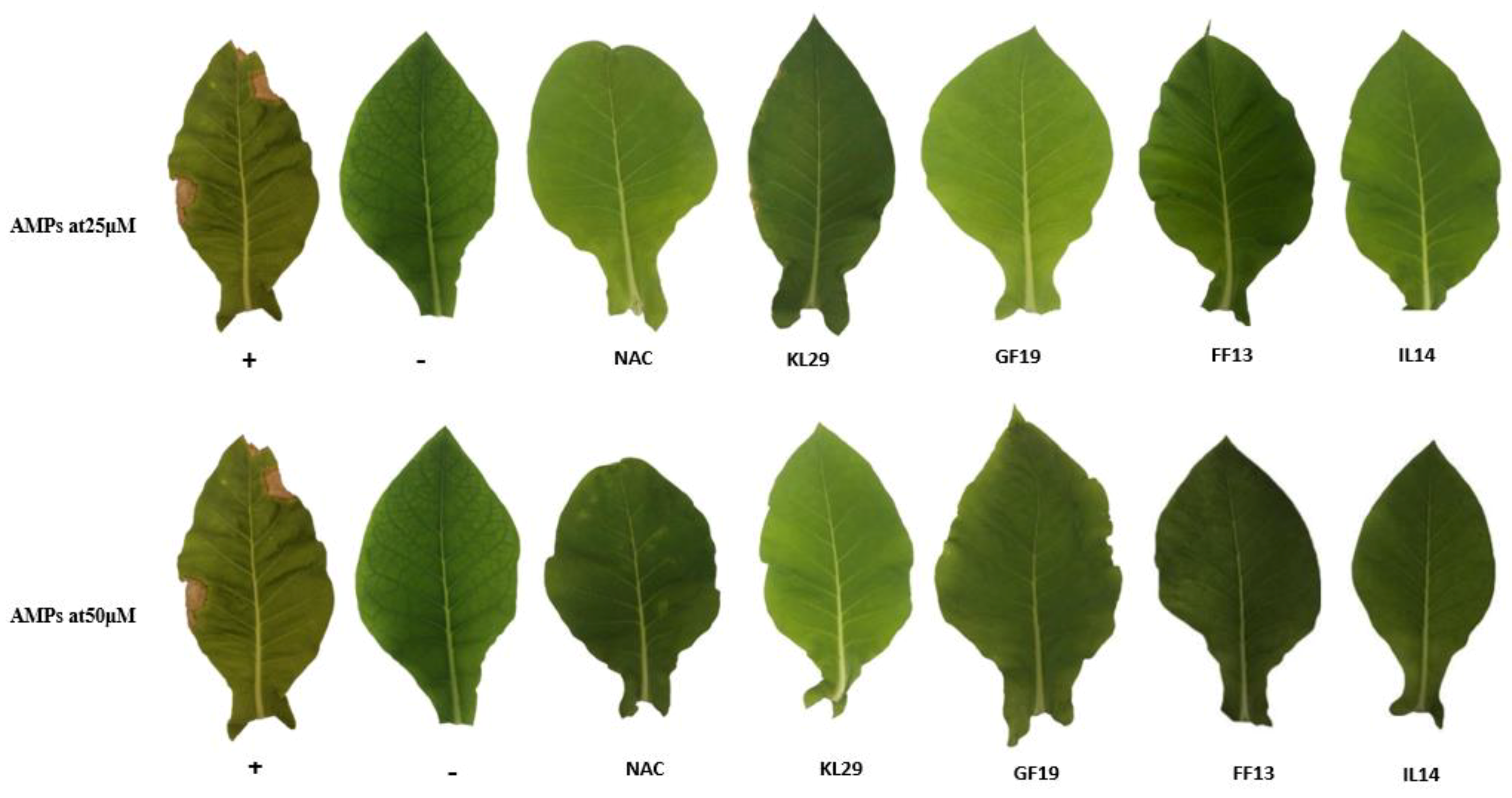
3.6. AMPs’ Efficiency in the Control of Xylella fastidiosa In Planta
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Food Safety Authority; Delbianco, A.; Gibin, D.; Pasinato, L.; Morelli, M. Update of the Xylella spp. host plant database–systematic literature searches up to 30 June 2022. EFSA J. 2022, 20, e07039. [Google Scholar] [PubMed]
- Burbank, L.P. Threat of Xylella fastidiosa and options for mitigation in infected plants. CABI Rev. 2022. [Google Scholar] [CrossRef]
- Bozzo, F.; Frem, M.; Fucilli, V.; Cardone, G.; Garofoli, P.F.; Geronimo, S.; Petrontino, A. Landscape and Vegetation Patterns Zoning Is a Methodological Tool for Management Costs Implications Due to Xylella fastidiosa Invasion. Land 2022, 11, 1105. [Google Scholar] [CrossRef]
- Saponari, M.; Giampetruzzi, A.; Loconsole, G.; Boscia, D.; Saldarelli, P. Xylella fastidiosa in olive in Apulia: Where we stand. Phytopathology 2019, 109, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Beretta, E.; Capasso, V.; Scacchi, S.; Brunetti, M.; Montagna, M. Prevention and control of OQDS (olive quick decline syndrome) outbreaks caused by Xylella fastidiosa. J. Theor. Biol. 2022, 542, 111118. [Google Scholar] [CrossRef] [PubMed]
- Sanna, F.; Mori, N.; Santoiemma, G.; D’Ascenzo, D.; Scotillo, M.A.; Marini, L. Ground cover management in olive groves reduces populations of Philaenus spumarius (Hemiptera: Aphrophoridae), vector of Xylella fastidiosa. J. Econ. Entomol. 2021, 114, 1716–1721. [Google Scholar] [CrossRef]
- Saponari, M.; Boscia, D.; Nigro, F.; Martelli, G.P. Identification of DNA sequences related to Xylella fastidiosa in oleander, almond and olive trees exhibiting leaf scorch symptoms in Apulia (Southern Italy). J. Plant Pathol. 2013, 95, 668. [Google Scholar]
- Giampetruzzi, A.; Chiumenti, M.; Saponari, M.; Donvito, G.; Italiano, A.; Loconsole, G.; Saldarelli, P. Draft genome sequence of the Xylella fastidiosa CoDiRO strain. Genome Announc. 2015, 3, e01538-14. [Google Scholar] [CrossRef] [Green Version]
- Anbumani, S.; da Silva, A.M.; Carvalho, I.G.B.; Fischer, E.R.; Silva, M.d.e.; von Zuben, A.A.G.; Carvalho, H.F.; de Souza, A.A.; Janissen, R.; Cotta, M.A. Controlled spatial organization of bacterial growth reveals key role of cell filamentation preceding Xylella fastidiosa biofilm formation. npj Biofilms Microbiomes 2021, 7, 86. [Google Scholar] [CrossRef]
- Roper, C.; Castro, C.; Ingel, B. Xylella fastidiosa: Bacterial parasitism with hallmarks of commensalism. Curr. Opin. Plant Biol. 2019, 50, 140–147. [Google Scholar] [CrossRef]
- Ge, Q.; Cobine, P.A.; De La Fuente, L. Copper supplementation in watering solution reaches the xylem but does not protect tobacco plants against Xylella fastidiosa infection. Plant Dis. 2020, 104, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Alves, D.; Olívia Pereira, M. Mini-review: Antimicrobial peptides and enzymes as promising candidates to functionalize biomaterial surfaces. Biofouling 2014, 30, 483–499. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.D.; Won, H.S.; Kim, J.H.; Mishig-Ochir, T.; Lee, B.J. Antimicrobial peptides for therapeutic applications: A review. Molecules 2012, 17, 12276–12286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Ahmed, T.A.; Hammami, R. Recent insights into structure–function relationships of antimicrobial peptides. J. Food Biochem. 2019, 43, e12546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Sani, M.A.; Separovic, F. Interaction of cationic antimicrobial peptides from Australian frogs with lipid membranes. Pept. Sci. 2018, 110, e24061. [Google Scholar] [CrossRef]
- Andersen, P.C.; Ishida, M.L.; Momol, E.A.; Brodbeck, B.V.; Leite, B.; Momol, M.T. Influence of Vitis xylem fluid and xylem fluid plus cecropin on growth of Xylella fastidiosa. Vitis 2004, 43, 19–26. [Google Scholar]
- Fogaça, A.C.; Zaini, P.A.; Wulff, N.A.; Da Silva, P.I.; Fázio, M.A.; Miranda, A.; Da Silva, A.M. Effects of the antimicrobial peptide gomesin on the global gene expression profile, virulence and biofilm formation of Xylella fastidiosa. FEMS Microbiol. Lett. 2014, 306, 152–159. [Google Scholar] [CrossRef] [Green Version]
- Muranaka, L.S.; Takita, M.A.; Olivato, J.C.; Kishi, L.T.; de Souza, A.A. Global expression profile of biofilm resistance to antimicrobial compounds in the plant-pathogenic bacterium Xylella fastidiosa reveals evidence of persister cells. J. Bacteriol. 2012, 194, 4561–4569. [Google Scholar] [CrossRef]
- Aldrich, T.J.; Rolshausen, P.E.; Roper, M.C.; Reader, J.M.; Steinhaus, M.J.; Rapicavoli, J.; Maloney, K.N. Radicinin from Cochliobolus sp. inhibits Xylella fastidiosa, the causal agent of Pierce’s Disease of grapevine. Phytochemistry 2015, 116, 130–137. [Google Scholar] [CrossRef]
- Moll, L.; Badosa, E.; Planas, M.; Feliu, L.; Montesinos, E.; Bonaterra, A. Antimicrobial Peptides with Antibiofilm Activity Against Xylella fastidiosa. Front. Microbiol. 2021, 12, 753874. [Google Scholar] [CrossRef] [PubMed]
- Moll, L.; Baró, A.; Montesinos, L.; Badosa, E.; Bonaterra, A.; Montesinos, E. Induction of defense responses and protection of almond plants against Xylella fastidiosa by endotherapy with a bifunctional peptide. Phytopathology 2022, 112, 1907–1916. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.; Song, P.I.; Seo, C.H.; Cheong, H.; Park, Y. Colonization and infection of the skin by S. aureus: Immune system evasion and the response to cationic antimicrobial peptides. Int. J. Mol. Sci. 2014, 15, 8753–8772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Sun, T.L.; Huang, H.W. Mode of action of antimicrobial peptides on E. coli spheroplasts. Biophys. J. 2016, 111, 132–139. [Google Scholar] [CrossRef] [Green Version]
- Menousek, J.; Mishra, B.; Hanke, M.L.; Heim, C.E.; Kielian, T.; Wang, G. Database screening and in vivo efficacy of antimicrobial peptides against methicillin-resistant Staphylococcus aureus USA300. Int. J. Antimicrob. Agents 2012, 39, 402–406. [Google Scholar] [CrossRef] [Green Version]
- Clavijo-Coppens, F.; Ginet, N.; Cesbron, S.; Briand, M.; Jacques, M.A.; Ansaldi, M. Novel Virulent Bacteriophages Infecting Mediterranean Isolates of the Plant Pest Xylella fastidiosa and Xanthomonas albilineans. Viruses 2021, 13, 725. [Google Scholar] [CrossRef]
- D’Attoma, G.; Morelli, M.; De La Fuente, L.; Cobine, P.A.; Saponari, M.; de Souza, A.; Saldarelli, P. Phenotypic characterization and transformation attempts reveal peculiar traits of Xylella fastidiosa subspecies pauca strain De Donno. Microorganisms 2020, 8, 1832. [Google Scholar] [CrossRef]
- De La Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.M.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E.W. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef] [Green Version]
- Badosa, E.; Moiset, G.; Montesinos, L.; Talleda, M.; Bardají, E.; Feliu, L.; Planas, M.; Montesinos, E. Derivatives of the antimicrobial peptide BP100 for expression in plant systems. PLoS ONE 2013, 8, e85515. [Google Scholar] [CrossRef] [Green Version]
- Baró, A.; Badosa, E.; Montesinos, L.; Feliu, L.; Planas, M.; Montesinos, E.; Bonaterra, A. Screening and identification of BP100 peptide conjugates active against Xylella fastidiosa using a viability-qPCR method. BMC Microbiol. 2020, 20, 229. [Google Scholar] [CrossRef]
- Vobruba, S.; Kadlcik, S.; Janata, J.; Kamenik, Z. TldD/TldE peptidases and N-deacetylases: A structurally unique yet ubiquitous protein family in the microbial metabolism. Microbiol. Res. 2022, 265, 127186. [Google Scholar] [CrossRef] [PubMed]
- Kuzina, L.V.; Miller, T.A.; Cooksey, D.A. In vitro activities of antibiotics and antimicrobial peptides against the plant pathogenic bacterium Xylella fastidiosa. Lett. Appl. Microbiol. 2006, 42, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Badosa Romañó, E.; Planas i Grabuleda, M.; Feliu Soley, L.; Montesinos Barreda, L.; Bonaterra i Carreras, A.; Montesinos Seguí, E. Synthetic Peptides against Plant Pathogenic Bacteria. Microorganisms 2022, 10, 1784. [Google Scholar] [CrossRef] [PubMed]
- Praveena, R.; Srekha, K.; Revathy, R.; Srinivasan, V.; Sarathambal, C.; George, P.; Dinesh, R. New rhizobacteria strains with effective antimycotic compounds against rhizome rot pathogens and identification of genes encoding antimicrobial peptides. Rhizosphere 2022, 22, 100515. [Google Scholar] [CrossRef]
- Wang, Y.; Chang, R.Y.K.; Britton, W.J.; Chan, H.K. Advances in the development of antimicrobial peptides and proteins for inhaled therapy. Adv. Drug Deliv. Rev. 2022, 180, 114066. [Google Scholar] [CrossRef]
- Silva, A.R.P.; Guimarães, M.S.; Rabelo, J.; Belén, L.H.; Perecin, C.J.; Farías, J.G.; Santos, J.H.P.M.; Rangel-Yagui, C.O. Recent advances in the design of antimicrobial peptide conjugates. J. Mater. Chem. B 2022, 10, 3587–3600. [Google Scholar] [CrossRef]
- Wei, D.; Zhang, X. Biosynthesis, bioactivity, biotoxicity and applications of antimicrobial peptides for human health. Biosaf. Health 2022, 4, 118–134. [Google Scholar] [CrossRef]
- Mendes, R.J.; Sario, S.; Luz, J.P.; Tassi, N.; Teixeira, C.; Gomes, P.; Tavares, F.; Santos, C. Evaluation of Three Antimicrobial Peptides Mixtures to Control the Phytopathogen Responsible for Fire Blight Disease. Plants 2021, 10, 2637. [Google Scholar] [CrossRef]
- Scudiero, O.; Brancaccio, M.; Mennitti, C.; Laneri, S.; Lombardo, B.; De Biasi, M.G.; De Gregorio, E.; Pagliuca, C.; Colicchio, R.; Salvatore, P.; et al. Human defensins: A novel approach in the fight against skin colonizing staphylococcus aureus. Antibiotics 2020, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Maddox, C.E.; Laur, L.M.; Tian, L. Antibacterial activity of phenolic compounds against the phytopathogen Xylella fastidiosa. Curr. Microbiol. 2010, 60, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Luo, Y. Recent advances of polysaccharide-based nanoparticles for oral insulin delivery. Int. J. Biol. Macromol. 2018, 120, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.; Li, M.; Ge, S.; Yang, J.; Sun, Q.; Xiong, L. Glucose-responsive biopolymer nanoparticles prepared by co-assembly of concanavalin A and amylopectin for insulin delivery. Ind. Crops Prod. 2018, 112, 98–104. [Google Scholar] [CrossRef]
- Xu, Y.; Zheng, Y.; Wu, L.; Zhu, X.; Zhang, Z.; Huang, Y. Novel solid lipid nanoparticle with endosomal escape function for oral delivery of insulin. ACS Appl. Mater. Interfaces 2018, 10, 9315–9324. [Google Scholar] [CrossRef] [PubMed]
- Gaballa, H.; Theato, P. Glucose-responsive polymeric micelles via boronic acid–diol complexation for insulin delivery at neutral pH. Biomacromolecules 2019, 20, 871–881. [Google Scholar] [CrossRef]
- Wong, C.Y.; Al-Salami, H.; Dass, C.R. Recent advancements in oral administration of insulin-loaded liposomal drug delivery systems for diabetes mellitus. Int. J. Pharm. 2018, 549, 201–217. [Google Scholar] [CrossRef]
- Shrestha, N.; Araújo, F.; Shahbazi, M.A.; Mäkilä, E.; Gomes, M.J.; Herranz-Blanco, B.; Lindgren, R.; Granroth, S.; Kukk, E.; Salonen, J.; et al. Thiolation and cell-penetrating peptide surface functionalization of porous silicon nanoparticles for oral delivery of insulin. Adv. Funct. Mater. 2016, 26, 3405–3416. [Google Scholar] [CrossRef]
- Giuliani, A.; Rinaldi, A.C. Beyond natural antimicrobial peptides: Multimeric peptides and other peptidomimetic approaches. Cell. Mol. Life Sci. 2011, 68, 2255–2266. [Google Scholar] [CrossRef]
- Hao, G.; Stover, E.; Gupta, G. Overexpression of a modified plant thionin enhances disease resistance to citrus canker and huanglongbing (HLB). Front. Plant Sci. 2016, 7, 1078. [Google Scholar] [CrossRef]
- Hao, G.; Zhang, S.; Stover, E. Transgenic expression of antimicrobial peptide D2A21 confers resistance to diseases incited by Pseudomonas syringae pv. tabaci and Xanthomonas citri, but not Candidatus Liberibacter asiaticus. PLoS ONE 2017, 12, e0186810. [Google Scholar]


| Peptide | Code | Sequence | Source | MIC (μM) | Reference | ||
|---|---|---|---|---|---|---|---|
| S. aureus | E. coli | X. fastidiosa | |||||
| Ascaphin-8 | GF19 | GFKDLLKGAAKALVKTVLF-NH2 | Frog | 3.1 | 12.5 | - | [25] |
| DASamP1 | FF13 | FFGKVLKLIRKIF-NH2 | Synthetic | 3.1 | >100 | - | [25] |
| DASamP2 | IL14 | IKWKKLLRAAKRIL-NH2 | Synthetic | 6.2 | 3.1 | - | [25] |
| Lycotoxin I | IL25 | IWLTALKFLGKHAAKHLAKQQLSKL | Spider | 3.1 | 25 | - | [25] |
| Maculatin 1.3 | GF21 | GLLGLLGSVVSHVVPAIVGHF-NH2 | Frog | 6.2 | >100 | - | [25] |
| Piscidin 1 | FG22 | FFHHIFRGIVHVGKTIHRLVTG | Fish | 3.1 | 12.5 | - | [25] |
| 1036 | VK13 | VQFRIRVRIVIRK-NH2 | Synthetic | - | - | 12.5 | [28] |
| RIJK2 | RV12 | RIVWVRIRRWFV-NH2 | Synthetic | - | - | 12.5 | [21] |
| BP178 | KL29 | KKLFKKILKYL-AGPA-GIGKFLHSAK-KDEL-OH | Synthetic | - | - | 12.5 | [29] |
| AMP | Code | 1 mM | 150 μM | 100 μM | 50 μM | 25 μM | 12 μM | 6 μM |
|---|---|---|---|---|---|---|---|---|
| Ascaphin-8 | GF19 | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
| DASamP1 | FF13 | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
| DASamP2 | IL14 | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
| Lycotoxin I | IL25 | +++ | +++ | +++ | +++ | ++ | - | - |
| Maculatin 1.3 | GF21 | +++ | +++ | +++ | +++ | ++ | - | - |
| Piscidin 1 | FG22 | +++ | +++ | +++ | +++ | ++ | - | - |
| 1036 | VK13 | +++ | +++ | +++ | +++ | +++ | ++ | + |
| RIJK2 | RV12 | +++ | +++ | +++ | +++ | +++ | ++ | + |
| BP178 | KL29 | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Handi, K.; Sabri, M.; Valentini, F.; De Stradis, A.; Achbani, E.H.; Hafidi, M.; El Moujabber, M.; Elbeaino, T. Exploring Active Peptides with Antimicrobial Activity In Planta against Xylella fastidiosa. Biology 2022, 11, 1685. https://doi.org/10.3390/biology11111685
El Handi K, Sabri M, Valentini F, De Stradis A, Achbani EH, Hafidi M, El Moujabber M, Elbeaino T. Exploring Active Peptides with Antimicrobial Activity In Planta against Xylella fastidiosa. Biology. 2022; 11(11):1685. https://doi.org/10.3390/biology11111685
Chicago/Turabian StyleEl Handi, Kaoutar, Miloud Sabri, Franco Valentini, Angelo De Stradis, El Hassan Achbani, Majida Hafidi, Maroun El Moujabber, and Toufic Elbeaino. 2022. "Exploring Active Peptides with Antimicrobial Activity In Planta against Xylella fastidiosa" Biology 11, no. 11: 1685. https://doi.org/10.3390/biology11111685
APA StyleEl Handi, K., Sabri, M., Valentini, F., De Stradis, A., Achbani, E. H., Hafidi, M., El Moujabber, M., & Elbeaino, T. (2022). Exploring Active Peptides with Antimicrobial Activity In Planta against Xylella fastidiosa. Biology, 11(11), 1685. https://doi.org/10.3390/biology11111685









