Isolation and Characterization of a Novel Vibrio natriegens—Infecting Phage and Its Potential Therapeutic Application in Abalone Aquaculture
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Phage Isolation and Characterization
2.3. Phage Stability Characterization
2.4. Phage DNA Extraction, Genome Sequencing, and Genome Assembly
2.5. Phage Genomic and Phylogenetic Analyses
2.6. Phage Biocontrol Potential Bioassay
3. Results
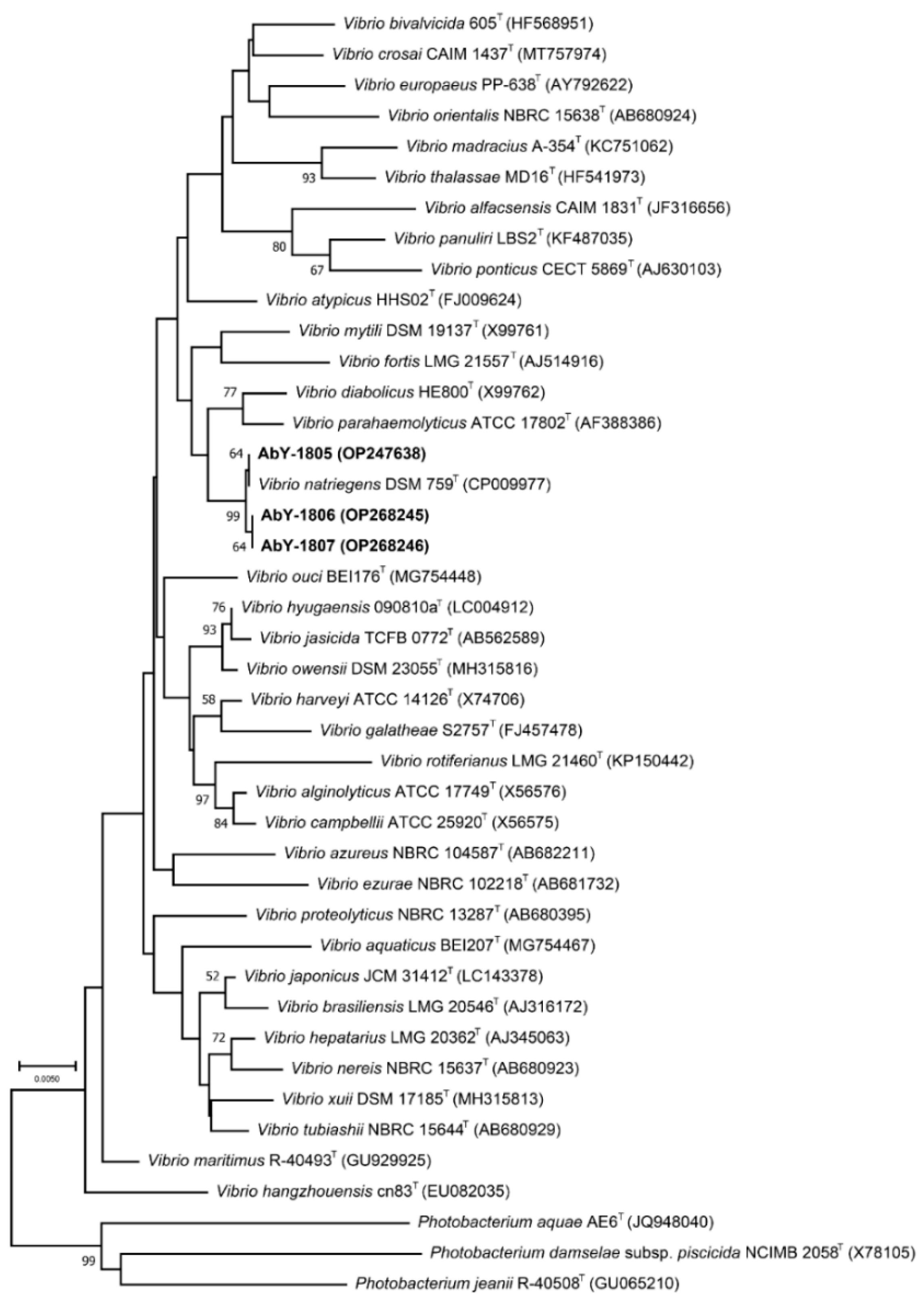
3.1. Isolation and Phylogenetic Characterization of the Pathogenetic Bacterial Strain AbY-1805
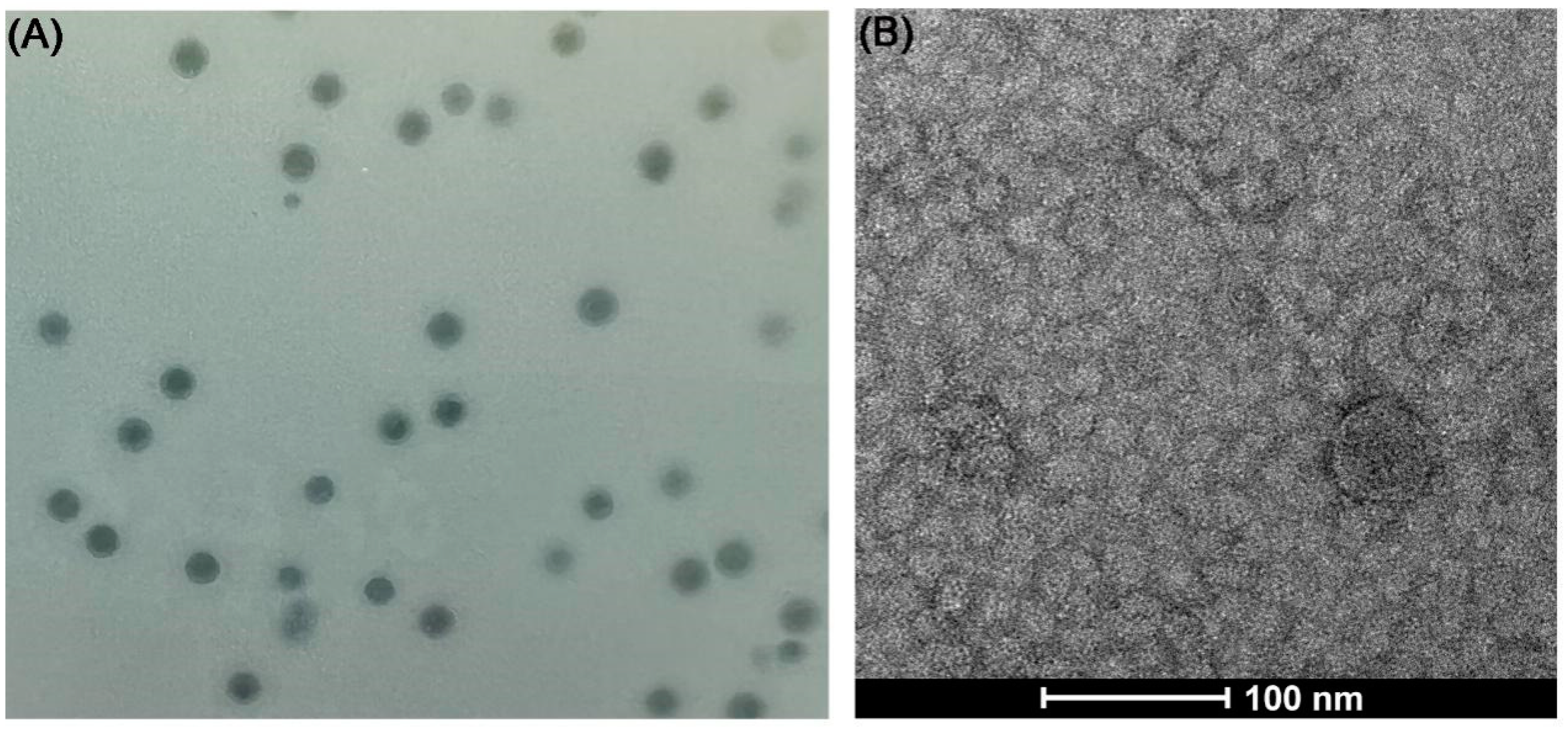
3.2. Isolation and Characterization of Phage vB_VnaS-L3
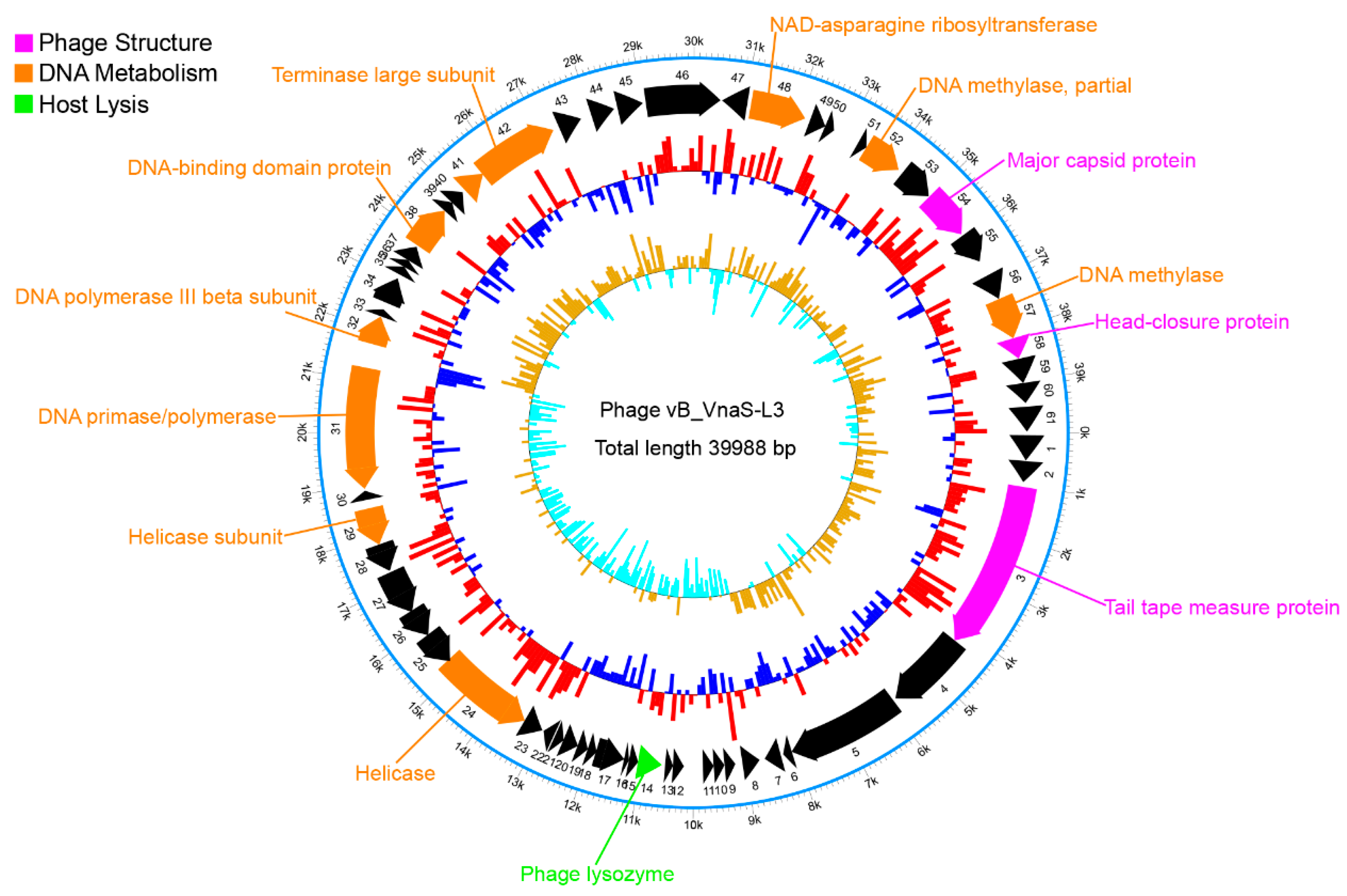
3.3. Genomic Characterization of Phage vB_VnaS-L3
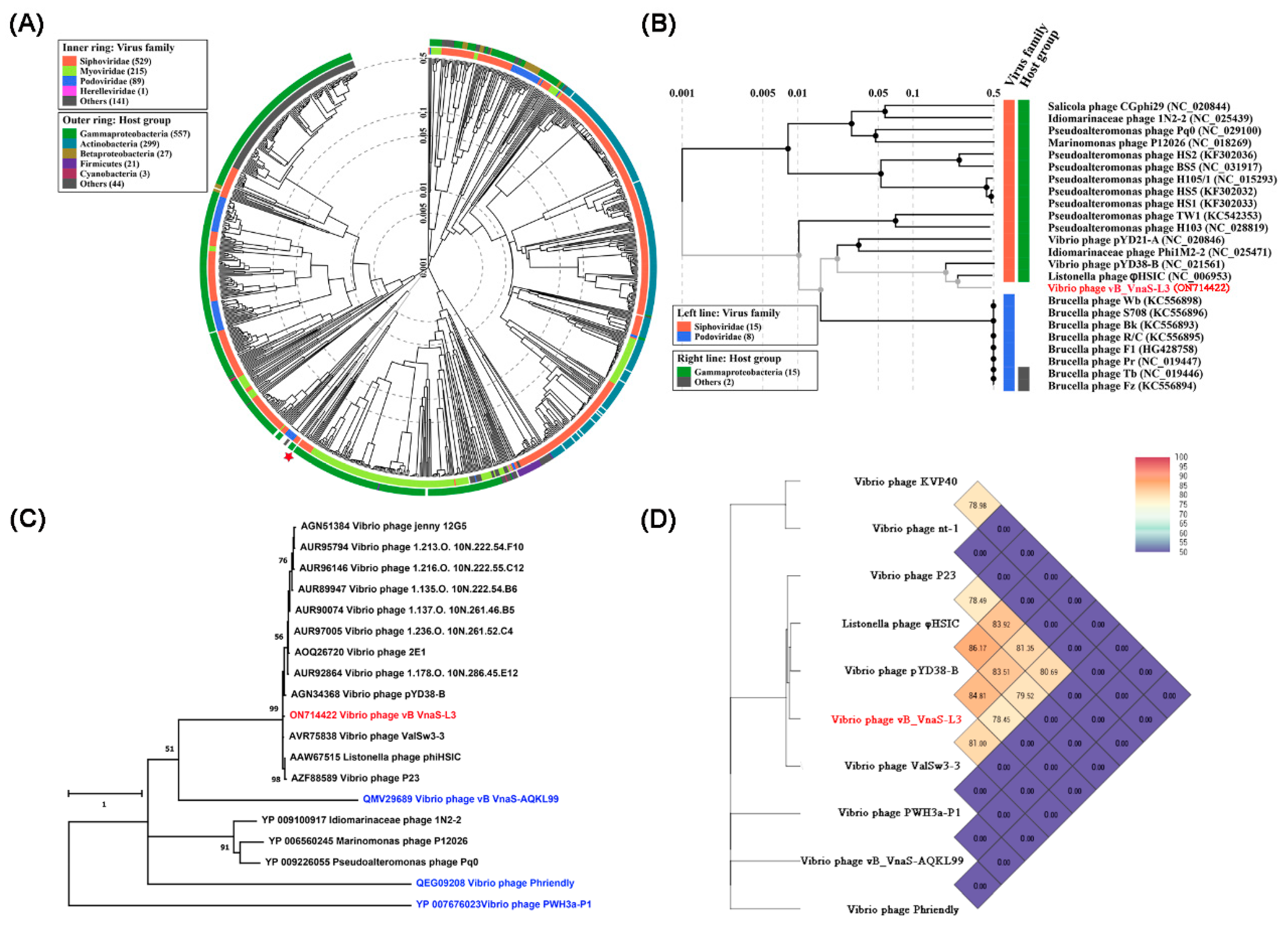
3.4. Phylogenetic and Comparative Genomic Analyses of Phage vB_VnaS-L3
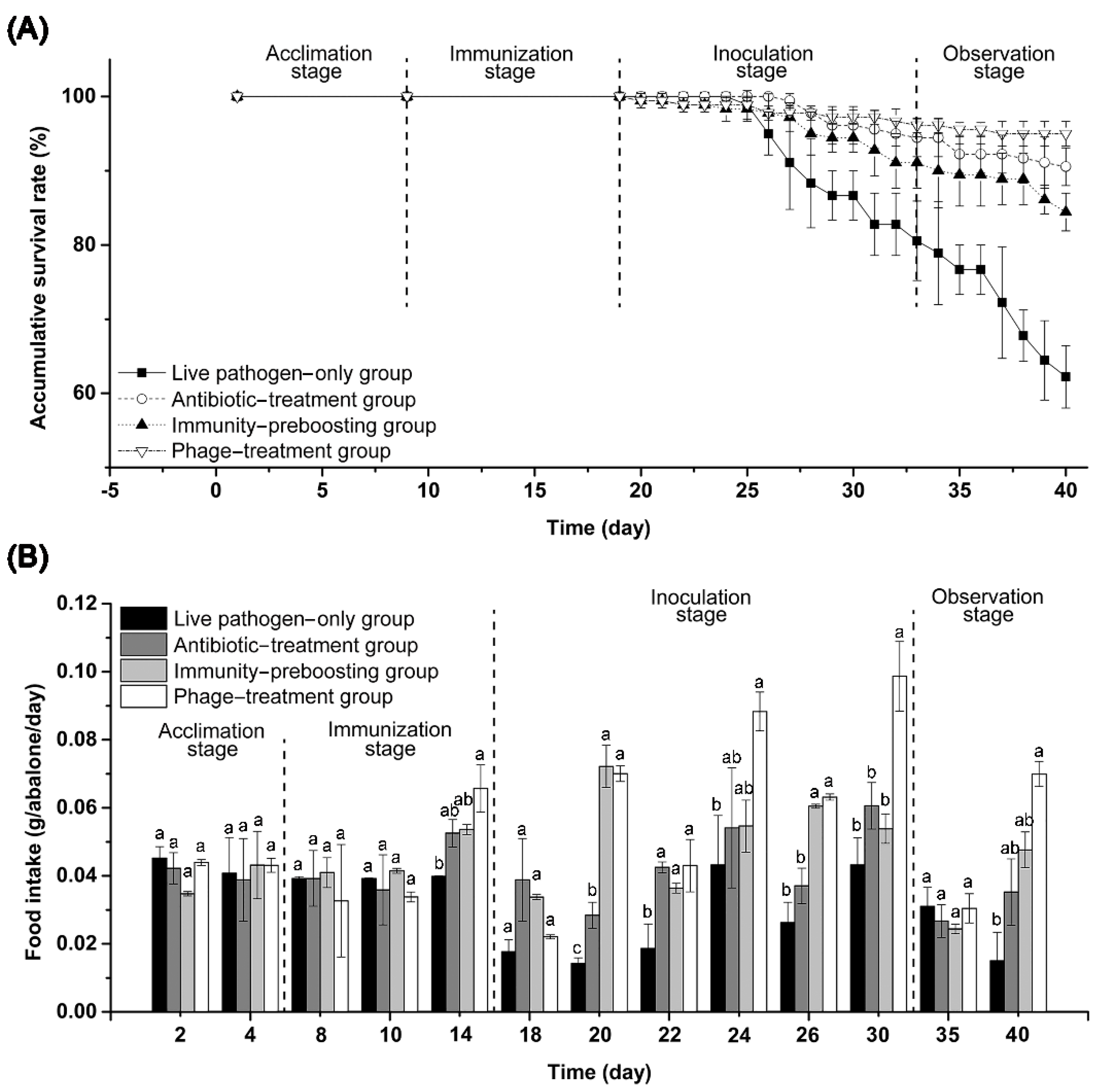
3.5. Bacterial Pathogen Biocontrol Potential of Phage vB_VnaS-L3
4. Discussion
4.1. vB_VnaS-L3 as a Novel V. natriegens Phage
4.2. Biological Properties of Phage vB_VnaS-L3
4.3. Potential Therapeutic Application of Phage vB_VnaS-L3 in Aquaculture
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grimes, D.J. The vibrios: Scavengers, symbionts, and pathogens from the sea. Microb. Ecol. 2020, 80, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Brumfield, K.D.; Usmani, M.; Chen, K.M.; Gangwar, M.; Jutla, A.S.; Huq, A.; Colwell, R.R. Environmental parameters associated with incidence and transmission of pathogenic Vibrio spp. Environ. Microbiol. 2021, 23, 7314–7340. [Google Scholar] [CrossRef] [PubMed]
- Orruño, M.; Kaberdin, V.; Arana, I. Survival strategies of Escherichia coli and Vibrio spp.: Contribution of the viable but nonculturable phenotype to their stress-resistance and persistence in adverse environments. World J. Microb. Biot. 2017, 33, 45. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Lovell, C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef] [Green Version]
- Sampaio, A.; Silva, V.; Poeta, P.; Aonofriesei, F. Vibrio spp.: Life strategies, ecology, and risks in a changing environment. Diversity 2022, 14, 97. [Google Scholar] [CrossRef]
- Destoumieux-Garzón, D.; Canesi, L.; Oyanedel, D.; Travers, M.A.; Charrière, G.M.; Pruzzo, C.; Vezzulli, L. Vibrio–bivalve interactions in health and disease. Environ. Microbiol. 2020, 22, 4323–4341. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Dölz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial use in aquaculture re-examined: Its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 2013, 15, 1917–1942. [Google Scholar] [CrossRef]
- Dang, H.; Song, L.; Chen, M.; Chang, Y. Concurrence of cat and tet genes in multiple antibiotic-resistant bacteria isolated from a sea cucumber and sea urchin mariculture farm in China. Microb. Ecol. 2006, 52, 634–643. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Ren, J.; Song, L.; Sun, S.; An, L. Diverse tetracycline resistant bacteria and resistance genes from coastal waters of Jiaozhou Bay. Microb. Ecol. 2008, 55, 237–246. [Google Scholar] [CrossRef]
- Zhao, J.; Dang, H. Coastal seawater bacteria harbor a large reservoir of plasmid-mediated quinolone resistance determinants in Jiaozhou Bay, China. Microb. Ecol. 2012, 64, 187–199. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Buschmann, A.H.; Dölz, H.J. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 2016, 16, e127–e133. [Google Scholar] [CrossRef]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. 2014. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 16 November 2021).
- Pérez-Sánchez, T.; Mora-Sánchez, B.; Balcázar, J.L. Biological approaches for disease control in aquaculture: Advantages, limitations and challenges. Trends Microbiol. 2018, 26, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Middelboe, M. Microbial disease in the sea: Effects of viruses on carbon and nutrient cycling. In Infectious Disease Ecology, 1st ed.; Richard, S.O., Felicia, K., Valerie, T.E., Eds.; Princeton University Press: Princeton, NJ, USA, 2008; Volume 11, pp. 242–259. [Google Scholar]
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage therapy for antibiotic-resistant bacterial infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lu, H.; Zhang, S.; Shi, Y.; Chen, Q. Phages against Pathogenic Bacterial Biofilms and Biofilm-Based Infections: A Review. Pharmaceutics 2022, 14, 427. [Google Scholar] [CrossRef] [PubMed]
- Nachimuthu, R.; Royam, M.M.; Manohar, P.; Leptihn, S. Application of bacteriophages and endolysins in aquaculture as a biocontrol measure. Biol. Control 2021, 160, 104678. [Google Scholar] [CrossRef]
- Rahimi-Midani, A.; Lee, S.-W.; Choi, T.-J. Potential Solutions Using Bacteriophages against Antimicrobial Resistant Bacteria. Antibiotics 2021, 10, 1496. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Q.; Fan, J.; Yan, T.; Zhang, H.; Yang, J.; Deng, D.; Liu, C.; Wei, T.; Ma, Y. Characterization and genomic analysis of ValSw3-3, a new Siphoviridae bacteriophage infecting Vibrio alginolyticus. J. Virol. 2020, 94, e00066-20. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhang, J.; Li, X.; Wang, X.; Cao, Z.; Wang, L.; Xu, Y. Efficiency of a bacteriophage in controlling vibrio infection in the juvenile sea cucumber Apostichopus japonicus. Aquaculture 2016, 451, 345–352. [Google Scholar] [CrossRef]
- Kim, S.G.; Jun, J.W.; Giri, S.S.; Yun, S.; Kim, H.J.; Kim, S.W.; Kang, J.W.; Han, S.J.; Jeong, D.; Park, S.C. Isolation and characterisation of pVa-21, a giant bacteriophage with anti-biofilm potential against Vibrio alginolyticus. Sci. Rep. 2019, 9, 6284. [Google Scholar] [CrossRef] [Green Version]
- Sieiro, C.; Areal-Hermida, L.; Pichardo-Gallardo, Á.; Almuiña-González, R.; De Miguel, T.; Sánchez, S.; Sánchez-Pérez, Á.; Villa, T.G. A hundred years of bacteriophages: Can phages replace antibiotics in agriculture and aquaculture? Antibiotics 2020, 9, 493. [Google Scholar] [CrossRef]
- Wittebole, X.; De Roock, S.; Opal, S.M. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Pajunen, M.; Kiljunen, S.; Skurnik, M. Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 2000, 182, 5114–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, L.; Cai, L.; Jiao, N.; Zhang, R. Isolation and characterization of the first phage infecting ecologically important marine bacteria Erythrobacter. Virol. J. 2017, 14, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 2001. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [Green Version]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [Green Version]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [Green Version]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA genes in genomic sequences. In Gene Prediction; Humana Press: New York, NY, USA, 2019; pp. 1–14. [Google Scholar]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.Y.; Mu, W.; Nikolic-Paterson, D.J.; Atkins, R.C. A novel, simple, reliable, and sensitive method for multiple immunoenzyme staining: Use of microwave oven heating to block antibody crossreactivity and retrieve antigens. J. Histochem. Cytochem. 1995, 43, 97–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, G.P.; Watson, M.A.; Madison, D.; Soffer, N.; Needleman, D.S.; Soroka, D.S.; Uknalis, J.; Baranzoni, G.M.; Church, K.M.; Polson, S.W. Bacteriophages against Vibrio coralliilyticus and Vibrio tubiashii: Isolation, characterization, and remediation of larval oyster mortalities. Appl. Environ. Microbiol. 2021, 87, e00008-21. [Google Scholar] [CrossRef]
- Jun, J.W.; Han, J.E.; Giri, S.S.; Tang, K.F.; Zhou, X.; Aranguren, L.F.; Kim, H.J.; Yun, S.; Chi, C.; Kim, S.G. Phage application for the protection from acute hepatopancreatic necrosis disease (AHPND) in Penaeus vannamei. Indian J. Microbiol. 2018, 58, 114–117. [Google Scholar] [CrossRef]
- Madison, D.; Richards, G.P.; Sulakvelidze, A.; Langdon, C. Bacteriophages improve survival and metamorphosis of larval Pacific oysters (Crassostrea gigas) exposed to Vibrio coralliilyticus strain RE98. Aquaculture 2022, 555, 738242. [Google Scholar] [CrossRef]
- Kim, H.J.; Jun, J.W.; Giri, S.S.; Chi, C.; Yun, S.; Kim, S.G.; Kim, S.W.; Kang, J.W.; Han, S.J.; Kwon, J. Application of the bacteriophage pVco-14 to prevent Vibrio coralliilyticus infection in Pacific oyster (Crassostrea gigas) larvae. J. Invertebr. Pathol. 2019, 167, 107244. [Google Scholar] [CrossRef]
- Wang, W.; Mai, K.; Zhang, W.; Ai, Q.; Yao, C.; Li, H.; Liufu, Z. Effects of dietary copper on survival, growth and immune response of juvenile abalone, Haliotis discus hannai Ino. Aquaculture 2009, 297, 122–127. [Google Scholar] [CrossRef]
- Naylor, R.L.; Hardy, R.W.; Buschmann, A.H.; Bush, S.R.; Cao, L.; Klinger, D.H.; Little, D.C.; Lubchenco, J.; Shumway, S.E.; Troell, M. A 20-year retrospective review of global aquaculture. Nature 2021, 591, 551–563. [Google Scholar] [CrossRef]
- Fiorella, K.J.; Okronipa, H.; Baker, K.; Heilpern, S. Contemporary aquaculture: Implications for human nutrition. Curr. Opin. Biotech. 2021, 70, 83–90. [Google Scholar] [CrossRef]
- Lulijwa, R.; Rupia, E.J.; Alfaro, A.C. Antibiotic use in aquaculture, policies and regulation, health and environmental risks: A review of the top 15 major producers. Rev. Aquacult. 2020, 12, 640–663. [Google Scholar] [CrossRef]
- Zhang, X.; Liao, S.; Li, Y.; Ji, W.; Xu, H. Studies on pathogenic bacteria (Vibrio natriegen) of Argopecten Irradins Lamarck. J. Ocean Univ. China 1998, 28, 426–432. [Google Scholar] [CrossRef]
- Li, G.; Yan, M.; Sun, J.; Lin, Z.; Ma, A.; Chang, W. Identification and biological characteristics of pathogen Vibrio natriegen from clam Meretrix meretrix. Prog. Fish. Sci. 2009, 30, 103–109. [Google Scholar] [CrossRef]
- Chen, L.; Fan, J.; Yan, T.; Liu, Q.; Yuan, S.; Zhang, H.; Yang, J.; Deng, D.; Huang, S.; Ma, Y. Isolation and characterization of specific phages to prepare a cocktail preventing Vibrio sp. Va-F3 infections in shrimp (Litopenaeus vannamei). Front. Microbiol. 2019, 10, 2337. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Bai, X.; Zhang, X.; Bi, K.; Qin, L.; Yan, B. Phenotypic and molecular characterization and the LAMP detection of pathogenic Vibrio natriegens isolated from diseased Penaeus japonicus. Haiyang Yu Huzhao 2012, 43, 1227–1232. [Google Scholar] [CrossRef]
- Bi, K.; Zhang, X.; Yan, B.; Gao, H.; Gao, X.; Sun, J. Isolation and molecular identification of Vibrio natriegens from diseased Portunus trituberculatus in China. J. World Aquacult. Soc. 2016, 47, 854–861. [Google Scholar] [CrossRef]
- Pratchett, M.S.; Caballes, C.F.; Rivera-Posada, J.A.; Sweatman, H.P.A. Limits to understanding and managing outbreaks of crown-of-thorns starfish (Acanthaster spp.). In Oceanography and Marine Biology: An Annual Review, 1st ed.; Hughes, R.N., Hughes, D.J., Smith, I.P., Eds.; CRC Press: Los Angeles, CA, USA, 2014; Volume 52, pp. 133–200. [Google Scholar]
- Hoff, J.; Daniel, B.; Stukenberg, D.; Thuronyi, B.W.; Waldminghaus, T.; Fritz, G. Vibrio natriegens: An ultrafast-growing marine bacterium as emerging synthetic biology chassis. Environ. Microbiol. 2020, 22, 4394–4408. [Google Scholar] [CrossRef]
- Eagon, R.G. Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes. J. Bacteriol. 1962, 83, 736–737. [Google Scholar] [CrossRef] [Green Version]
- Hoffart, E.; Grenz, S.; Lange, J.; Nitschel, R.; Müller, F.; Schwentner, A.; Feith, A.; Lenfers-Lücker, M.; Takors, R.; Blombach, B. High substrate uptake rates empower Vibrio natriegens as production host for industrial biotechnology. Appl. Environ. Microb. 2017, 83, e01614-17. [Google Scholar] [CrossRef] [Green Version]
- Thoma, F.; Blombach, B. Metabolic engineering of Vibrio natriegens. Essays Biochem. 2021, 65, 381–392. [Google Scholar] [CrossRef]
- Collins, J.H.; Young, E.M. Genetic engineering of host organisms for pharmaceutical synthesis. Curr. Opin. Biotech. 2018, 53, 191–200. [Google Scholar] [CrossRef]
- Ellis, G.A.; Tschirhart, T.; Spangler, J.; Walper, S.A.; Medintz, I.L.; Vora, G.J. Exploiting the feedstock flexibility of the emergent synthetic biology chassis Vibrio natriegens for engineered natural product production. Mar. Drugs 2019, 17, 679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Han, X.; Peng, Y.; Tan, C.; Wang, J.; Xue, H.; Xu, P.; Tao, F. Rapid production of L-DOPA by Vibrio natriegens, an emerging next-generation whole-cell catalysis chassis. Microb. Biotechnol. 2022, 15, 1610–1621. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tschirhart, T.; Schultzhaus, Z.; Kelly, E.E.; Chen, A.; Oh, E.; Nag, O.; Glaser, E.R.; Kim, E.; Lloyd, P.F. Melanin produced by the fast-growing marine bacterium Vibrio natriegens through heterologous biosynthesis: Characterization and application. Appl. Environ. Microb. 2020, 86, e02749-19. [Google Scholar] [CrossRef]
- Jana, B.; Keppel, K.; Salomon, D. Engineering a customizable antibacterial T6SS-based platform in Vibrio natriegens. EMBO Rep. 2021, 22, e53681. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Liu, D.; Zhang, Y.; Xiao, F.; Yuan, C.; Liang, F.; Liu, X.; Zhang, C. Dopamine-assisted sustainable antimicrobial peptide coating with antifouling and anticorrosion properties. Appl. Surf. Sci. 2022, 589, 153019. [Google Scholar] [CrossRef]
- Cheng, S.; Tian, J.; Chen, S.; Lei, Y.; Chang, X.; Liu, T.; Yin, Y. Microbially influenced corrosion of stainless steel by marine bacterium Vibrio natriegens: (I) Corrosion behavior. Mater. Sci. Eng. C 2009, 29, 751–755. [Google Scholar] [CrossRef]
- Cheng, S.; Lau, K.-T.; Chen, S.; Chang, X.; Liu, T.; Yin, Y. Microscopical observation of the marine bacterium Vibrio Natriegeus growth on metallic corrosion. Mater. Manuf. Process. 2010, 25, 293–297. [Google Scholar] [CrossRef]
- Conley, B.E.; Weinstock, M.T.; Bond, D.R.; Gralnick, J.A. A hybrid extracellular electron transfer pathway enhances the survival of Vibrio natriegens. Appl. Environ. Microbiol. 2020, 86, e01253-20. [Google Scholar] [CrossRef]
- Lebeaux, D.; Ghigo, J.-M.; Beloin, C. Biofilm-related infections: Bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef]
- Wang, K.-L.; Dou, Z.-R.; Gong, G.-F.; Li, H.-F.; Jiang, B.; Xu, Y. Anti-Larval and Anti-Algal Natural Products from Marine Microorganisms as Sources of Anti-Biofilm Agents. Mar. Drugs 2022, 20, 90. [Google Scholar] [CrossRef] [PubMed]
- Zachary, A. Isolation of bacteriophages of the marine bacterium Beneckea natriegens from coastal salt marshes. Appl. Microbiol. 1974, 27, 980–982. [Google Scholar] [CrossRef] [PubMed]
- Zachary, A. An ecological study of bacteriophages of Vibrio natriegens. Can. J. Microbiol. 1978, 24, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.; Awah, A.; Moreland, R.; Liu, M.; Gill, J.J.; Ramsey, J. Complete genome sequence of Vibrio natriegens phage phriendly. Microbiol. Resour. Announce. 2019, 8, e01096-19. [Google Scholar] [CrossRef] [Green Version]
- Yonas, N.; Boleman, P.; Nguyen, Y.; Kerr, M.; Malki, K.; Greco, A.M.; Breitbart, M. Genome Sequence of Vibrio natriegens Phage vB_VnaS-AQKL99. Microbiol. Resour. Announc. 2020, 9, e00967-20. [Google Scholar] [CrossRef]
- Cook, P.A. The worldwide abalone industry. Mod. Econ. 2014, 5, 1181. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhang, G. Pacific abalone farming in China: Recent innovations and challenges. J. Shellfish Res. 2016, 35, 703–710. [Google Scholar] [CrossRef]
- Sawabe, T.; Inoue, S.; Fukui, Y.; Yoshie, K.; Nishihara, Y.; Miura, H. Mass mortality of Japanese abalone Haliotis discus hannai caused by Vibrio harveyi infection. Microbes Environ. 2007, 22, 300–308. [Google Scholar] [CrossRef] [Green Version]
- Kamaishi, T.; Miwa, S.; Goto, E.; Matsuyama, T.; Oseko, N. Mass mortality of giant abalone Haliotis gigantea caused by a Francisella sp. bacterium. Dis. Aquat. Org. 2010, 89, 145–154. [Google Scholar] [CrossRef]
- Cai, J.; Wang, Z.; Cai, C.; Zhou, Y. Characterization and identification of virulent Klebsiella oxytoca isolated from abalone (Haliotis diversicolor supertexta) postlarvae with mass mortality in Fujian, China. J. Invertebr. Pathol. 2008, 97, 70–75. [Google Scholar] [CrossRef]
- Zachary, A. Physiology and ecology of bacteriophages of the marine bacterium Beneckea natriegens: Salinity. Appl. Environ. Microbiol. 1976, 31, 415–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suttle, C.A.; Chen, F. Mechanisms and rates of decay of marine viruses in seawater. Appl. Environ. Microbiol. 1992, 58, 3721–3729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, J.H.; Williamson, S.J.; Long, A.; Authement, R.N.; John, D.; Segall, A.M.; Rohwer, F.L.; Androlewicz, M.; Patterson, S. Complete genome sequence of φHSIC, a pseudotemperate marine phage of Listonella pelagia. Appl. Environ. Microbiol. 2005, 71, 3311–3320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhao, L.; Wang, M.; Wang, Q.; Zhang, X.; Han, Y.; Wang, M.; Jiang, T.; Shao, H.; Jiang, Y. Complete genomic sequence of bacteriophage P23: A novel Vibrio phage isolated from the Yellow Sea, China. Virus Genes 2019, 55, 834–842. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A roadmap for genome-based phage taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef] [PubMed]
- Dy, R.L.; Rigano, L.A.; Fineran, P.C. Phage-based biocontrol strategies and their application in agriculture and aquaculture. Biochem. Soc. Trans. 2018, 46, 1605–1613. [Google Scholar] [CrossRef]
- Liu, R.; Han, G.; Li, Z.; Cun, S.; Hao, B.; Zhang, J.; Liu, X. Bacteriophage therapy in aquaculture: Current status and future challenges. Folia Microbiol. 2022, 67, 573–590. [Google Scholar] [CrossRef]
- Ye, M.; Sun, M.; Huang, D.; Zhang, Z.; Zhang, H.; Zhang, S.; Hu, F.; Jiang, X.; Jiao, W. A review of bacteriophage therapy for pathogenic bacteria inactivation in the soil environment. Environ. Int. 2019, 129, 488–496. [Google Scholar] [CrossRef]
- Watts, G. Phage therapy: Revival of the bygone antimicrobial. Lancet 2017, 390, 2539–2540. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, Z.; Zhou, Y.; Bao, H.; Wang, R.; Li, T.; Pang, M.; Sun, L.; Zhou, X. Application of a phage in decontaminating Vibrio parahaemolyticus in oysters. Int. J. Food Microbiol. 2018, 275, 24–31. [Google Scholar] [CrossRef]
- Mateus, L.; Costa, L.; Silva, Y.; Pereira, C.; Cunha, A.; Almeida, A. Efficiency of phage cocktails in the inactivation of Vibrio in aquaculture. Aquaculture 2014, 424, 167–173. [Google Scholar] [CrossRef]
- Kalatzis, P.G.; Bastías, R.; Kokkari, C.; Katharios, P. Isolation and characterization of two lytic bacteriophages, φSt2 and φGrn1; phage therapy application for biological control of Vibrio alginolyticus in aquaculture live feeds. PLoS ONE 2016, 11, e0151101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duerkop, B.A.; Huo, W.; Bhardwaj, P.; Palmer, K.L.; Hooper, L.V. Molecular basis for lytic bacteriophage resistance in Enterococci. mBio 2016, 7, e01304-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Yuan, S.; Liu, Q.; Mai, G.; Yang, J.; Deng, D.; Zhang, B.; Liu, C.; Ma, Y. In vitro design and evaluation of phage cocktails against Aeromonas salmonicida. Front. Microbiol. 2018, 9, 1476. [Google Scholar] [CrossRef] [Green Version]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [Green Version]
- Torres-Barceló, C.; Hochberg, M.E. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol. 2016, 24, 249–256. [Google Scholar] [CrossRef]
- Droubogiannis, S.; Katharios, P. Genomic and biological profile of a novel bacteriophage, Vibrio phage Virtus, which improves survival of Sparus aurata larvae challenged with Vibrio harveyi. Pathogens 2022, 11, 630. [Google Scholar] [CrossRef]
- Kim, H.J.; Jun, J.W.; Giri, S.S.; Kim, S.G.; Kim, S.W.; Kwon, J.; Lee, S.B.; Chi, C.; Park, S.C. Bacteriophage cocktail for the prevention of multiple-antibiotic-resistant and mono-phage-resistant Vibrio coralliilyticus infection in Pacific oyster (Crassostrea gigas) larvae. Pathogens 2020, 9, 831. [Google Scholar] [CrossRef]
- Wang, Y.; Barton, M.; Elliott, L.; Li, X.; Abraham, S.; O’Dea, M.; Munro, J. Bacteriophage therapy for the control of Vibrio harveyi in greenlip abalone (Haliotis laevigata). Aquaculture 2017, 473, 251–258. [Google Scholar] [CrossRef] [Green Version]
- Silva, Y.J.; Costa, L.; Pereira, C.; Mateus, C.; Cunha, A.; Calado, R.; Gomes, N.C.; Pardo, M.A.; Hernandez, I.; Almeida, A. Phage therapy as an approach to prevent Vibrio anguillarum infections in fish larvae production. PLoS ONE 2014, 9, e114197. [Google Scholar] [CrossRef]
- Dubey, S.; Singh, A.; Kumar, B.; Singh, N.K.; Tyagi, A. Isolation and characterization of bacteriophages from inland saline aquaculture environments to control Vibrio parahaemolyticus contamination in shrimp. Indian J. Microbiol. 2021, 61, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Kemp, I.K.; Coyne, V.E. Identification and characterisation of the Mpeg1 homologue in the South African abalone, Haliotis midae. Fish Shellfish Immunol. 2011, 31, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Tran, N.T.; Zhu, C.H.; Yao, D.F.; Aweya, J.J.; Gong, Y.; Ma, H.Y.; Zhang, Y.L.; Li, G.L.; Li, S.K. Immune priming in shellfish: A review and an updating mechanistic insight focused on cellular and humoral responses. Aquaculture 2021, 530, 735831. [Google Scholar] [CrossRef]
- Carroll-Portillo, A.; Lin, H.C. Bacteriophage and the innate immune system: Access and signaling. Microorganisms 2019, 7, 625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miernikiewicz, P.; Kłopot, A.; Soluch, R.; Szkuta, P.; Kęska, W.; Hodyra-Stefaniak, K.; Konopka, A.; Nowak, M.; Lecion, D.; Kaźmierczak, Z. T4 phage tail adhesin gp12 counteracts LPS-induced inflammation in vivo. Front. Microbiol. 2016, 7, 1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Przerwa, A.; Zimecki, M.; Świtała-Jeleń, K.; Dąbrowska, K.; Krawczyk, E.; Łuczak, M.; Weber-Dąbrowska, B.; Syper, D.; Międzybrodzki, R.; Górski, A. Effects of bacteriophages on free radical production and phagocytic functions. Med. Microbiol. Immun. 2006, 195, 143–150. [Google Scholar] [CrossRef]
- Bauer, A. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin pathol. 1966, 45, 149–158. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. In Approved Standard, 2nd ed.; M45-A2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- National Committee for Clinical Laboratory Standards (NCCLS). Performance Standards for Antimicrobial Disk Susceptibility Tests. Approved Standard, 8th ed.; M2-A8; National Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2003. [Google Scholar]



| Groups | Acclimation Stage | Immunization Stage | Inoculation Stage | Observation Stage | |||
|---|---|---|---|---|---|---|---|
| Day 1 to Day 6 | Day 7 to Day 16 | Day 17 to Day 30 | Day 31 to Day 40 | ||||
| No Treatment | Heat-Inactivated Strain AbY-1805 | Phage vB_VnaS-L3 | Strain AbY-1805 | Phage vB_VnaS-L3 | Oxytetracycline | No Treatment | |
| Live pathogen-only group | − | − | + | − | − | ||
| Immunity-preboosting group | + | − | + | − | − | ||
| Antibiotic-treatment group | − | − | + | − | + | ||
| Phage-treatment group | − | + | + | + | − | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Liang, Y.; Wang, Z.; Yao, Y.; Chen, X.; Shao, A.; Lu, L.; Dang, H. Isolation and Characterization of a Novel Vibrio natriegens—Infecting Phage and Its Potential Therapeutic Application in Abalone Aquaculture. Biology 2022, 11, 1670. https://doi.org/10.3390/biology11111670
Li X, Liang Y, Wang Z, Yao Y, Chen X, Shao A, Lu L, Dang H. Isolation and Characterization of a Novel Vibrio natriegens—Infecting Phage and Its Potential Therapeutic Application in Abalone Aquaculture. Biology. 2022; 11(11):1670. https://doi.org/10.3390/biology11111670
Chicago/Turabian StyleLi, Xuejing, Yantao Liang, Zhenhua Wang, Yanyan Yao, Xiaoli Chen, Anran Shao, Longfei Lu, and Hongyue Dang. 2022. "Isolation and Characterization of a Novel Vibrio natriegens—Infecting Phage and Its Potential Therapeutic Application in Abalone Aquaculture" Biology 11, no. 11: 1670. https://doi.org/10.3390/biology11111670
APA StyleLi, X., Liang, Y., Wang, Z., Yao, Y., Chen, X., Shao, A., Lu, L., & Dang, H. (2022). Isolation and Characterization of a Novel Vibrio natriegens—Infecting Phage and Its Potential Therapeutic Application in Abalone Aquaculture. Biology, 11(11), 1670. https://doi.org/10.3390/biology11111670







