Stk10 Deficiency in Mice Promotes Tumor Growth by Dysregulating the Tumor Microenvironment
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Tumor Cell Lines and Mice
2.2. Bioinformatic Analysis
2.3. RNA Extraction and qRT-PCR Analyses
2.4. Label of RM-1 Cells with Luciferase
2.5. In Vivo Tumorigenicity Assay
2.6. In Vivo Bioluminescence Assay
2.7. Immunohistochemistry (IHC) and Immunofluorescence (IF) Staining
2.8. Flow Cytometry Analysis
2.9. Western Blot Analysis
2.10. Tube-Formation Assay
2.11. Statistical Analysis
3. Results
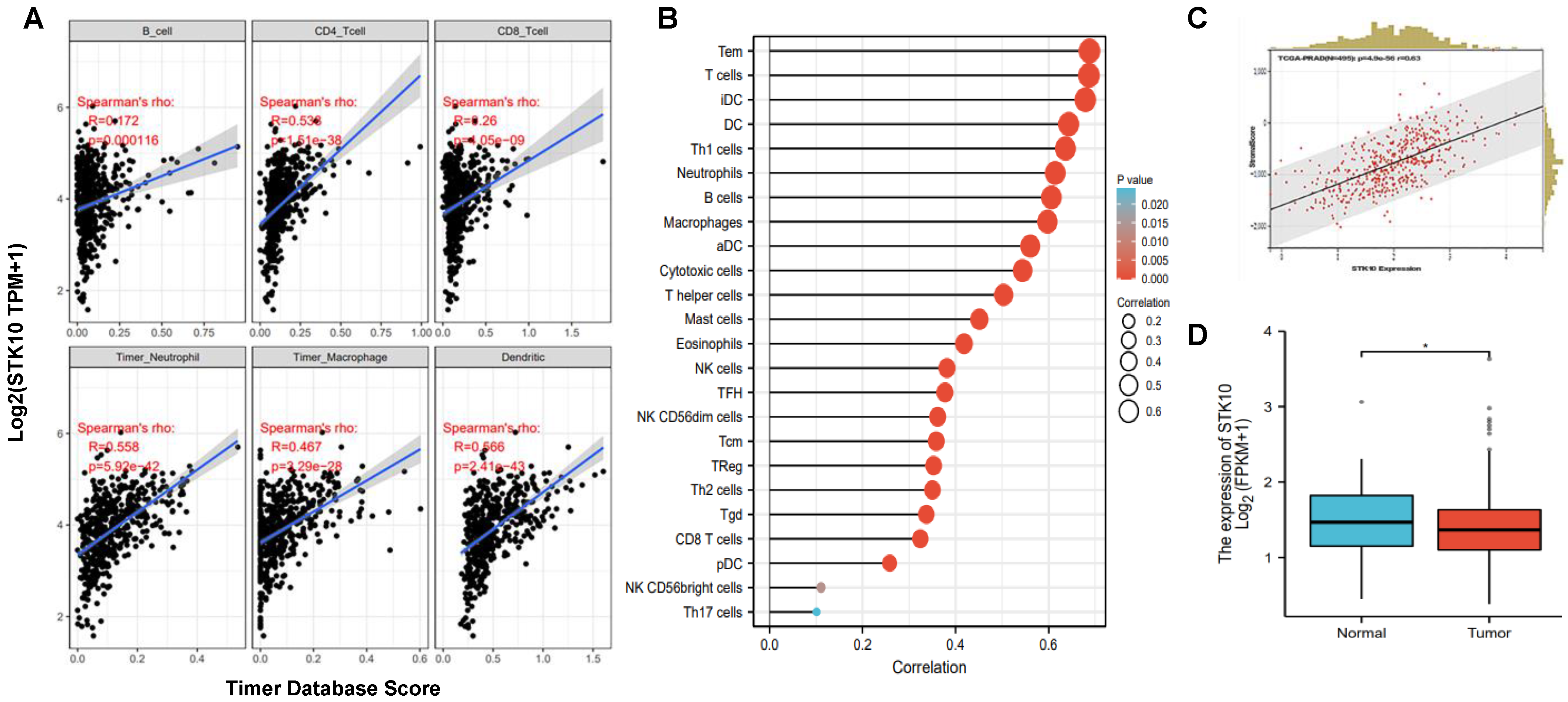
3.1. STK10 Was Associated with the Infiltrated Immune Cells in Prostate Cancer
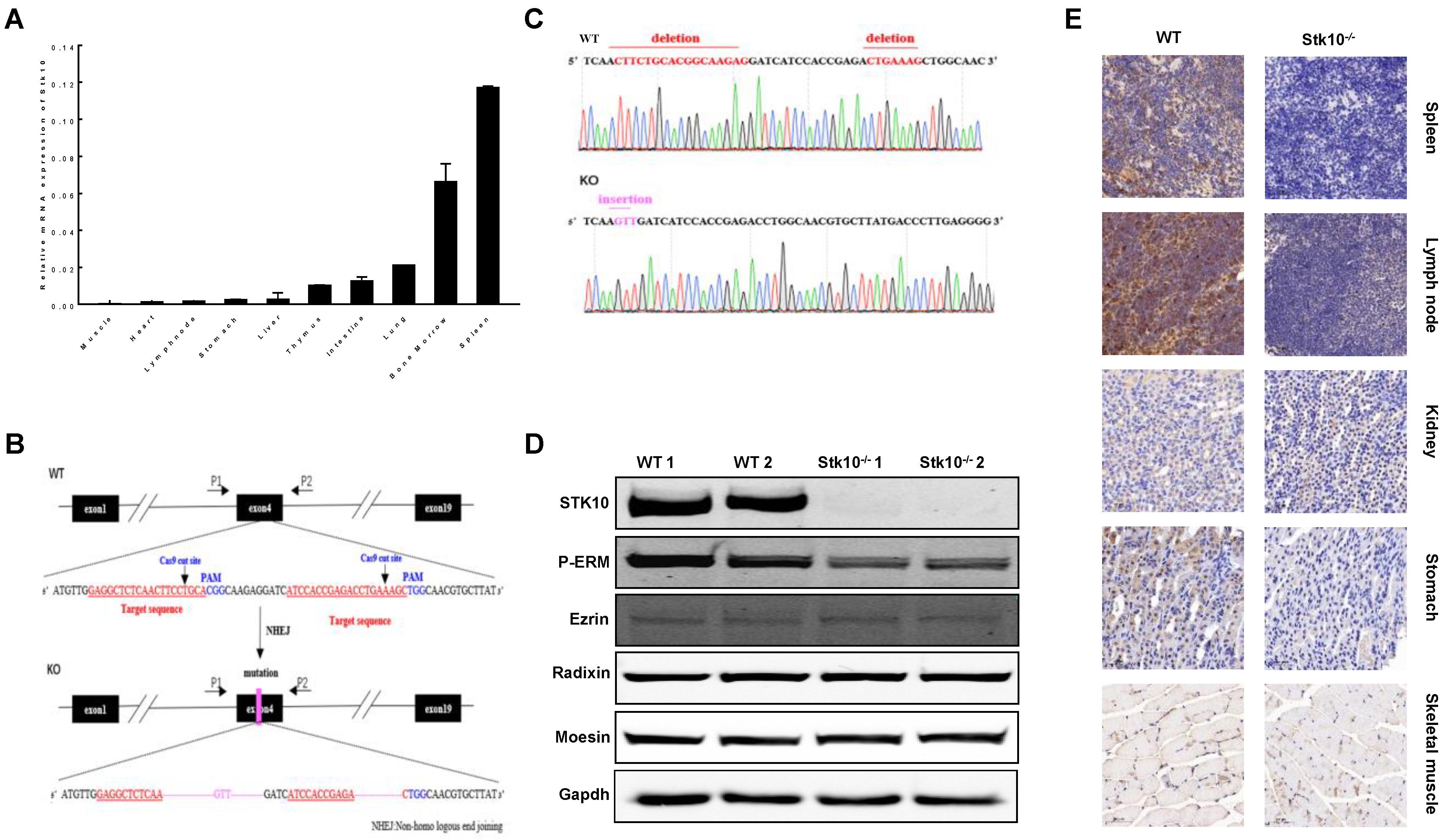
3.2. Generation of Stk10-Deficient Mice
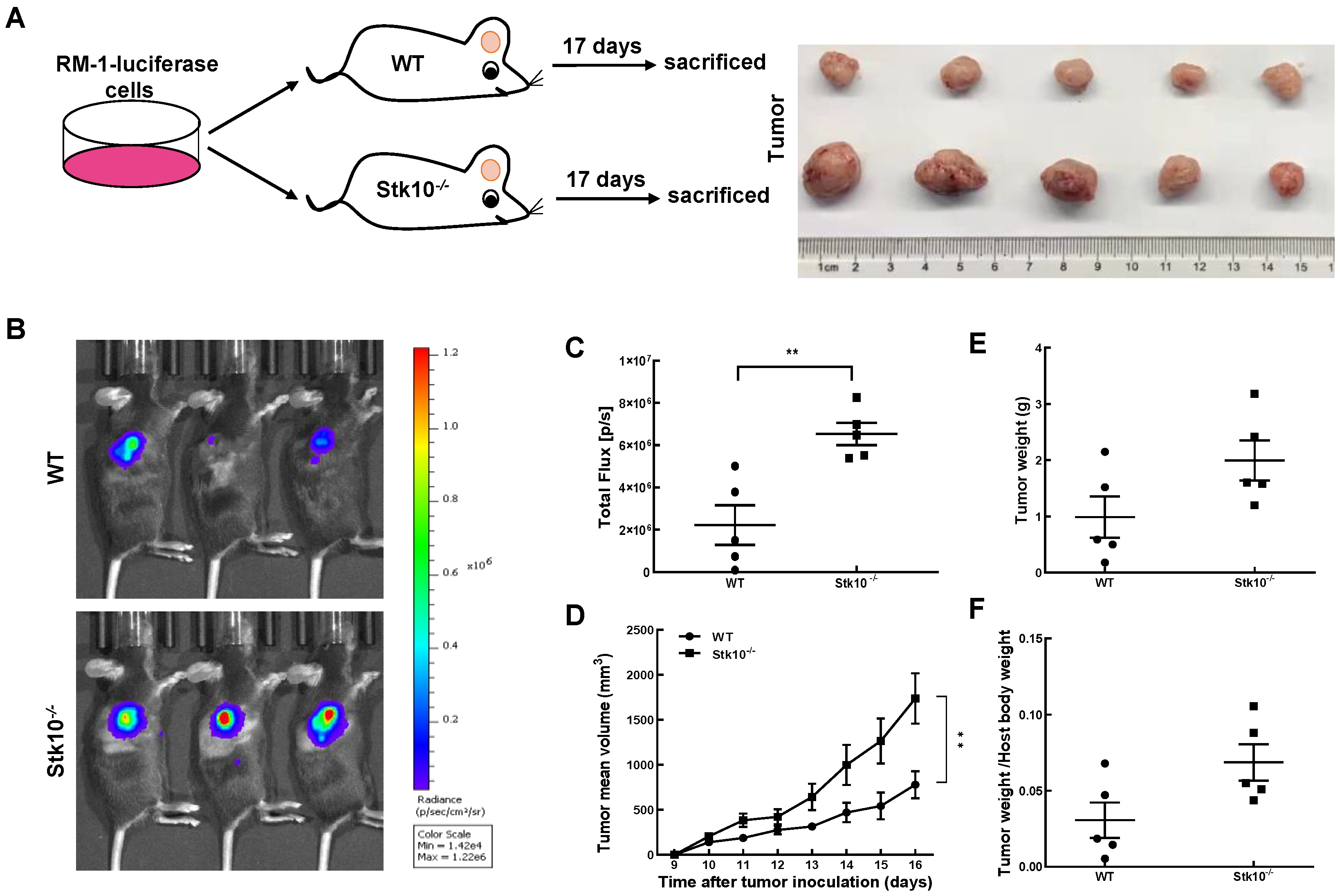
3.3. RM-1 Tumor Growth Was Promoted in Stk10 Knockout Mice
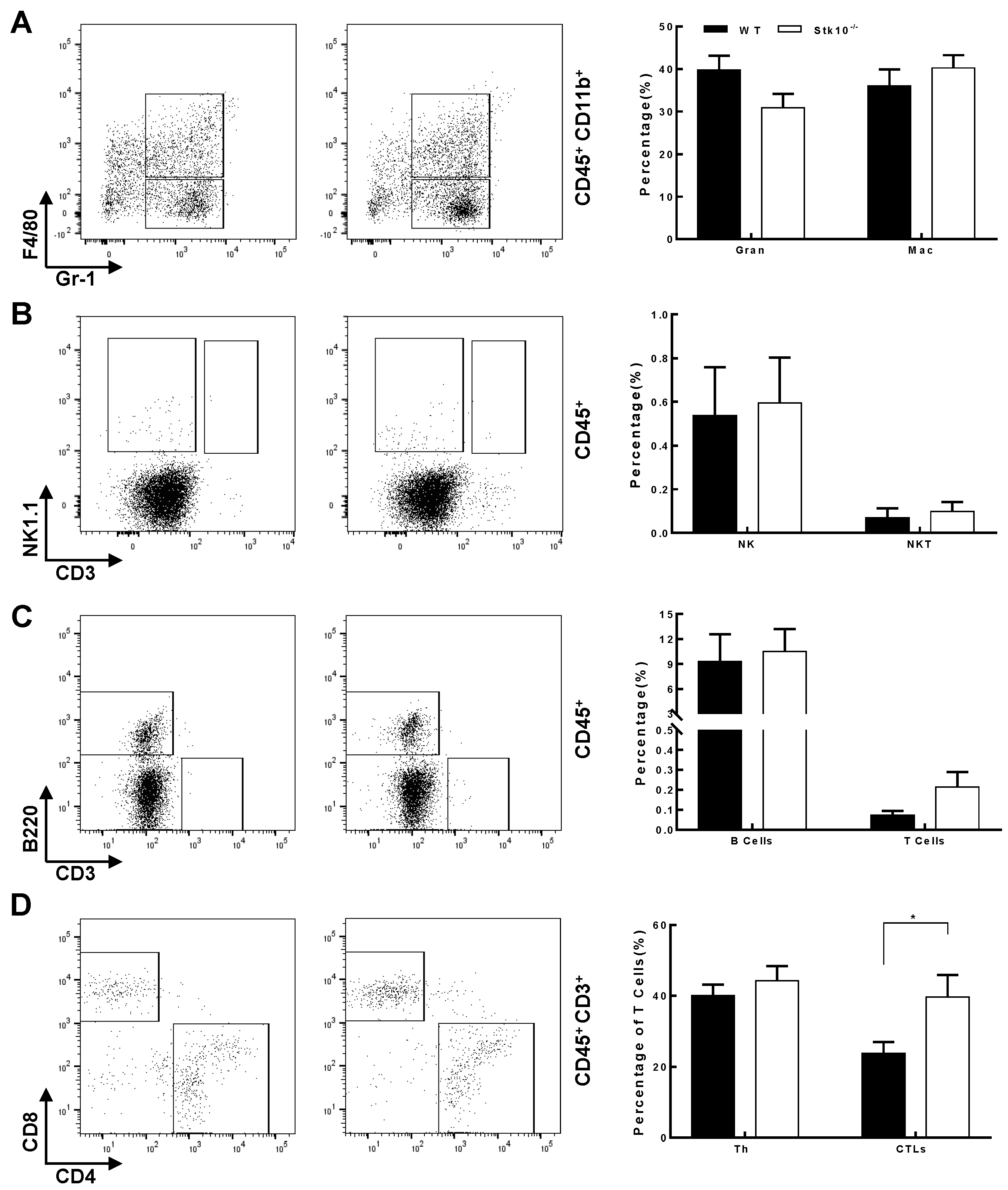
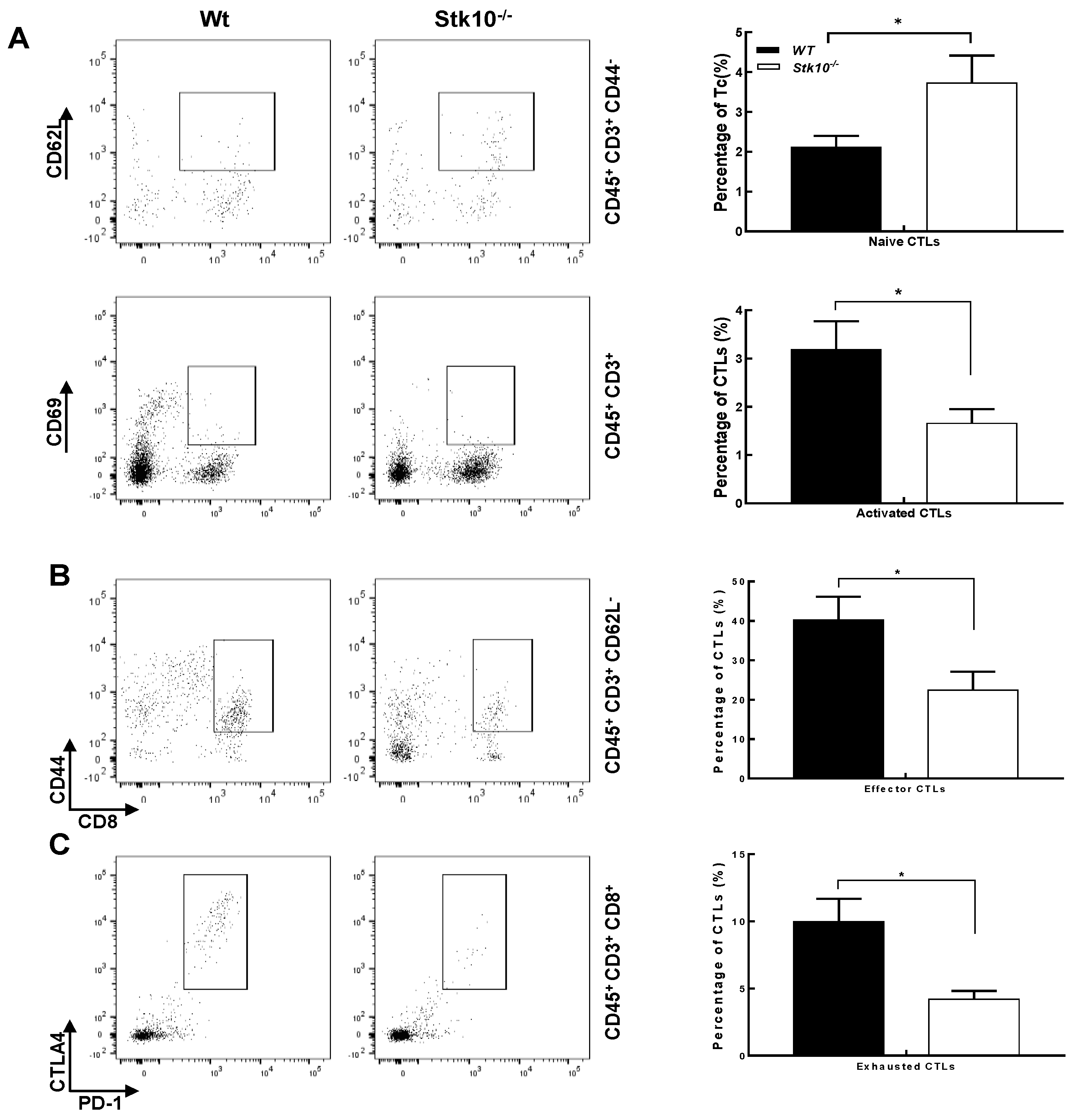
3.4. Stk10−/− Mice Show Tumors with Decreased Infiltration of Activated and Effector CTLs
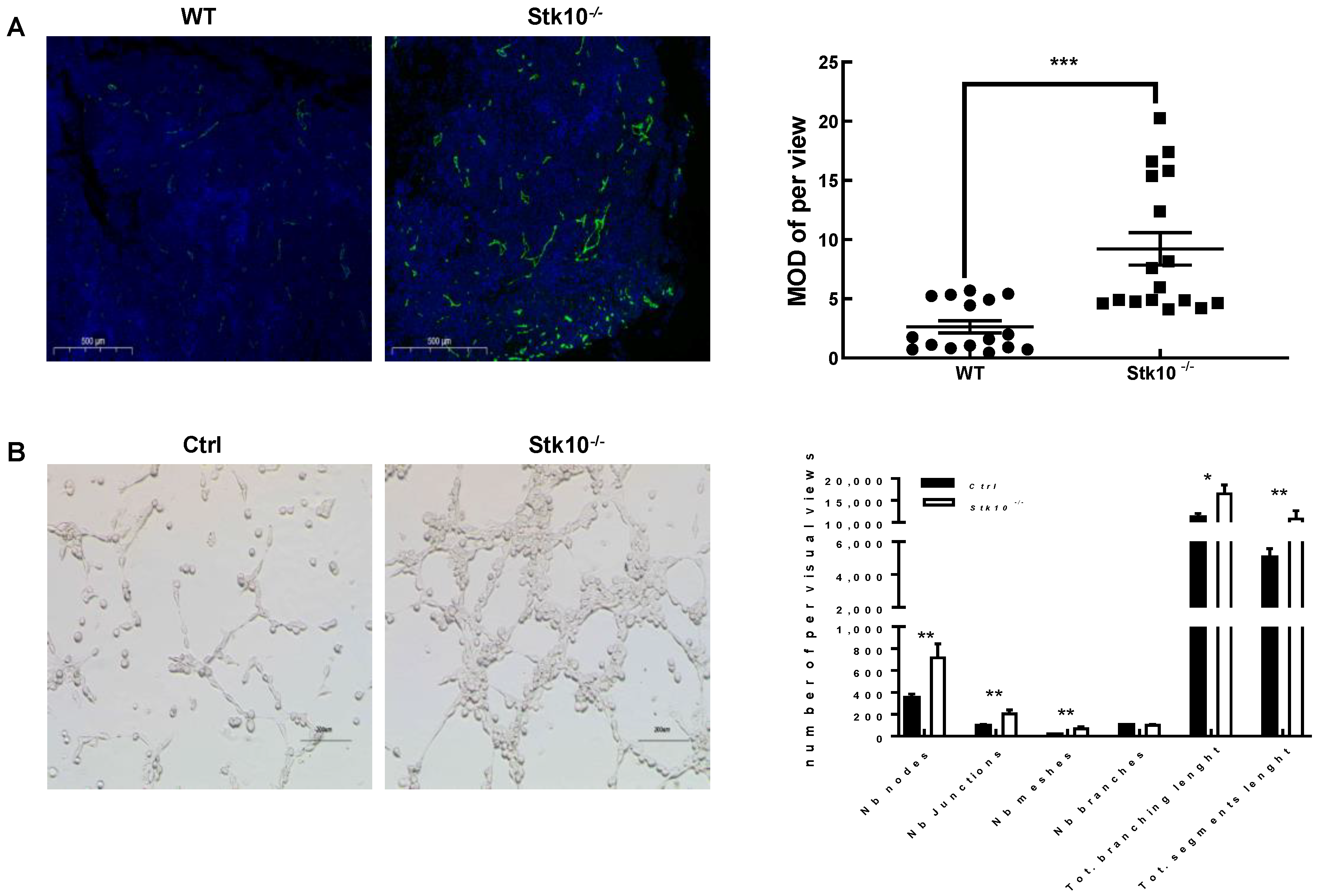
3.5. Stk10 Knockout Induces Angiogenesis in the Tumor Tissue
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuramochi, S.; Moriguchi, T.; Kuida, K.; Endo, J.; Semba, K.; Nishida, E.; Karasuyama, H. LOK Is a Novel Mouse STE20-like Protein Kinase that Is Expressed Predominantly in Lymphocytes. J. Biol. Chem. 1997, 272, 22679–22684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, Y. Purification and Cloning of a Protein Kinase that Phosphorylates and Activates the Polo-Like Kinase Plx1. Science 1998, 282, 1701–1704. [Google Scholar] [CrossRef] [PubMed]
- Belkina, N.V.; Liu, Y.; Hao, J.J.; Karasuyama, H.; Shaw, S. LOK is a major ERM kinase in resting lymphocytes and regulates cytoskeletal rearrangement through ERM phosphorylation. Proc. Natl. Acad. Sci. USA 2009, 106, 4707–4712. [Google Scholar] [CrossRef] [Green Version]
- Bignell, G.; Smith, R.; Hunter, C.; Stephens, P.; Davies, H.; Greenman, C.; Teague, J.; Butler, A.; Edkins, S.; Stevens, C.; et al. Sequence analysis of the protein kinase gene family in human testicular germ-cell tumors of adolescents and adults. Genes Chromosomes Cancer 2006, 45, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Fukumura, K.; Yamashita, Y.; Kawazu, M.; Sai, E.; Fujiwara, S.; Nakamura, N.; Takeuchi, K.; Ando, M.; Miyazono, K.; Ueno, T.; et al. STK10 missense mutations associated with anti-apoptotic function. Oncol. Rep. 2013, 30, 1542–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, L.; Jia, S.; Hu, W.; Su, X.; Chen, X.; Tang, H. Systematic analysis of prognostic significance, functional enrichment and immune implication of STK10 in acute myeloid leukemia. BMC Med. Genom. 2022, 15, 101. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, S.Y.; Guo, R.; Ma, J.X.; Tang, L.Y.; Wang, J.J.; Shen, C.L.; Lu, L.M.; Liu, J.; Wang, Z.G.; et al. STK10 knockout inhibits cell migration and promotes cell proliferation via modulating the activity of ERM and p38 MAPK in prostate cancer cells. Exp. Ther. Med. 2021, 22, 851. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, S.Y.; Guo, R.; Ma, J.X.; Tang, L.Y.; Shen, Y.; Shen, C.L.; Lu, L.M.; Wang, Z.G.; Liu, J.; et al. Knockout of STK10 promotes the migration and invasion of cervical cancer cells. Transl. Cancer Res. 2020, 9, 7079–7090. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Endo, J.; Toyama-Sorimachi, N.; Taya, C.; Kuramochi-Miyagawa, S.; Nagata, K.; Kuida, K.; Takashi, T.; Yonekawa, H.; Yoshizawa, Y.; Miyasaka, N.; et al. Deficiency of a STE20/PAK family kinase LOK leads to the acceleration of LFA-1 clustering and cell adhesion of activated lymphocytes. FEBS Lett. 2000, 468, 234–238. [Google Scholar] [CrossRef]
- Lamplugh, Z.; Fan, Y. Vascular Microenvironment, Tumor Immunity and Immunotherapy. Front. Immunol. 2021, 12, 811485. [Google Scholar] [CrossRef] [PubMed]
- Li, M.O.; Wolf, N.; Raulet, D.H.; Akkari, L.; Pittet, M.J.; Rodriguez, P.C.; Kaplan, R.N.; Munitz, A.; Zhang, Z.; Cheng, S.; et al. Innate immune cells in the tumor microenvironment. Cancer Cell 2021, 39, 725–729. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef] [PubMed]
- Oura, K.; Morishita, A.; Tani, J.; Masaki, T. Tumor Immune Microenvironment and Immunosuppressive Therapy in Hepatocellular Carcinoma: A Review. Int. J. Mol. Sci. 2021, 22, 5801. [Google Scholar] [CrossRef]
- Paijens, S.T.; Vledder, A.; de Bruyn, M.; Nijman, H.W. Tumor-infiltrating lymphocytes in the immunotherapy era. Cell. Mol. Immunol. 2021, 18, 842–859. [Google Scholar] [CrossRef]
- Hui, L.; Chen, Y. Tumor microenvironment: Sanctuary of the devil. Cancer Lett. 2015, 368, 7–13. [Google Scholar] [CrossRef]
- Xu, L.; Zhu, S.; Lan, Y.; Yan, M.; Jiang, Z.; Zhu, J.; Liao, G.; Ping, Y.; Xu, J.; Pang, B.; et al. Revealing the contribution of somatic gene mutations to shaping tumor immune microenvironment. Brief. Bioinform. 2022, 23, bbac064. [Google Scholar] [CrossRef]
- Liu, K.; Cui, J.J.; Zhan, Y.; Ouyang, Q.Y.; Lu, Q.S.; Yang, D.H.; Li, X.P.; Yin, J.Y. Reprogramming the tumor microenvironment by genome editing for precision cancer therapy. Mol. Cancer 2022, 21, 98. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Frankel, T.; Lanfranca, M.P.; Zou, W. The Role of Tumor Microenvironment in Cancer Immunotherapy. Adv. Exp. Med. Biol. 2017, 1036, 51–64. [Google Scholar] [PubMed]
- Datta, M.; Coussens, L.M.; Nishikawa, H.; Hodi, F.S.; Jain, R.K. Reprogramming the Tumor Microenvironment to Improve Immunotherapy: Emerging Strategies and Combination Therapies. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Kaymak, I.; Williams, K.S.; Cantor, J.R.; Jones, R.G. Immunometabolic Interplay in the Tumor Microenvironment. Cancer Cell 2021, 39, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Kuramochi, S.; Matsuda, Y.; Okamoto, M.; Kitamura, F.; Yonekawa, H.; Karasuyama, H. Molecular cloning of the human gene STK10 encoding lymphocyte-oriented kinase, and comparative chromosomal mapping of the human, mouse, and rat homologues. Immunogenetics 1999, 49, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Forster, J.C.; Harriss-Phillips, W.M.; Douglass, M.J.; Bezak, E. A review of the development of tumor vasculature and its effects on the tumor microenvironment. Hypoxia 2017, 5, 21–32. [Google Scholar] [CrossRef] [Green Version]
- De Palma, M.; Biziato, D.; Petrova, T.V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 2017, 17, 457–474. [Google Scholar] [CrossRef]
- Ghosh, S.; Di Bartolo, V.; Tubul, L.; Shimoni, E.; Kartvelishvily, E.; Dadosh, T.; Feigelson, S.W.; Alon, R.; Alcover, A.; Haran, G. ERM-Dependent Assembly of T Cell Receptor Signaling and Co-stimulatory Molecules on Microvilli prior to Activation. Cell Rep. 2020, 30, 3434–3447.e6. [Google Scholar] [CrossRef] [Green Version]
- Esen, E.; Sergin, I.; Jesudason, R.; Himmels, P.; Webster, J.D.; Zhang, H.; Xu, M.; Piskol, R.; McNamara, E.; Gould, S.; et al. MAP4K4 negatively regulates CD8 T cell-mediated antitumor and antiviral immunity. Sci. Immunol. 2020, 5, eaay2245. [Google Scholar] [CrossRef]
- Wang, X.; Li, N.; Han, A.; Wang, Y.; Lin, Z.; Yang, Y. Ezrin promotes hepatocellular carcinoma progression by modulating glycolytic reprogramming. Cancer Sci. 2020, 111, 4061–4074. [Google Scholar] [CrossRef]
- Chen, Q.; Fan, K.; Chen, X.; Xie, X.; Huang, L.; Song, G.; Qi, W. Ezrin regulates synovial angiogenesis in rheumatoid arthritis through YAP and Akt signalling. J. Cell. Mol. Med. 2021, 25, 9378–9389. [Google Scholar] [CrossRef]
- Degryse, B.; Britto, M.; Shan, C.X.; Wallace, R.G.; Rochfort, K.D.; Cummins, P.M.; Meade, G.; Murphy, R.P. Moesin and merlin regulate urokinase receptor-dependent endothelial cell migration, adhesion and angiogenesis. Int. J. Biochem. Cell Biol. 2017, 88, 14–22. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.-X.; Xu, D.-D.; Lu, S.-Y.; Wang, Q.-L.; Zhang, L.; Guo, R.; Tang, L.-Y.; Shen, Y.; Shen, C.-L.; Wang, J.-J.; et al. Stk10 Deficiency in Mice Promotes Tumor Growth by Dysregulating the Tumor Microenvironment. Biology 2022, 11, 1668. https://doi.org/10.3390/biology11111668
Ma J-X, Xu D-D, Lu S-Y, Wang Q-L, Zhang L, Guo R, Tang L-Y, Shen Y, Shen C-L, Wang J-J, et al. Stk10 Deficiency in Mice Promotes Tumor Growth by Dysregulating the Tumor Microenvironment. Biology. 2022; 11(11):1668. https://doi.org/10.3390/biology11111668
Chicago/Turabian StyleMa, Jin-Xia, Dan-Dan Xu, Shun-Yuan Lu, Qian-Lan Wang, Lu Zhang, Rui Guo, Ling-Yun Tang, Yan Shen, Chun-Ling Shen, Jin-Jin Wang, and et al. 2022. "Stk10 Deficiency in Mice Promotes Tumor Growth by Dysregulating the Tumor Microenvironment" Biology 11, no. 11: 1668. https://doi.org/10.3390/biology11111668



