Revised Annotation and Characterization of Novel Aedes albopictus miRNAs and Their Potential Functions in Dengue Virus Infection
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Virus
2.2. Virus Infection, RNA Extraction and Sequencing
2.3. Verification of DENV1 Infection in C6/36 Cells
2.4. miRNA Identification
2.5. Differential Expression of miRNA
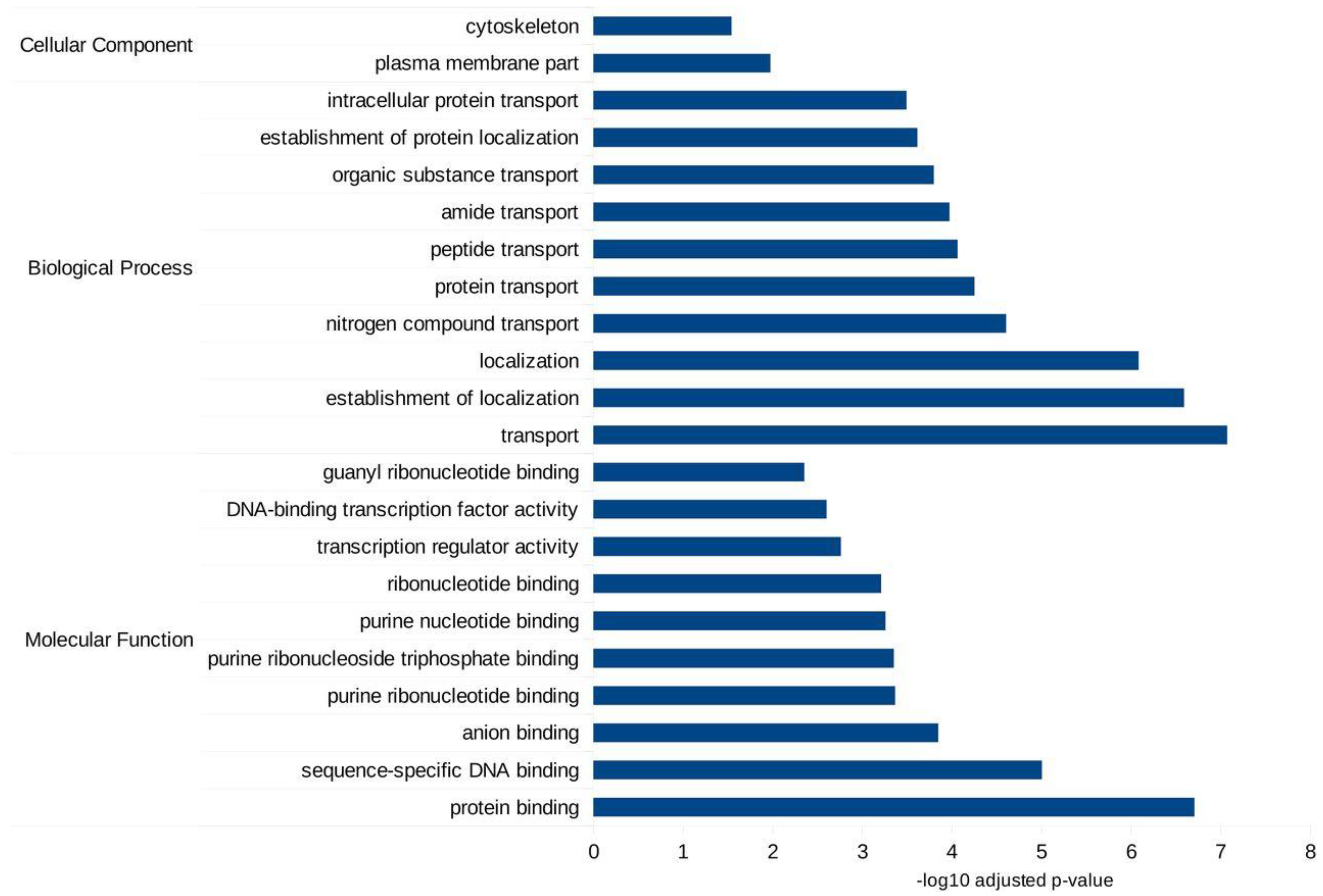
2.6. miRNA Target Prediction and Functional Analysis
3. Results and Discussion
3.1. RNA-Sequencing Libraries and Read Mapping
3.2. Identification of Novel miRNAs in Ae. albopictus
3.3. Expression of Novel miRNAs in Different Stages of Ae. albopictus Development
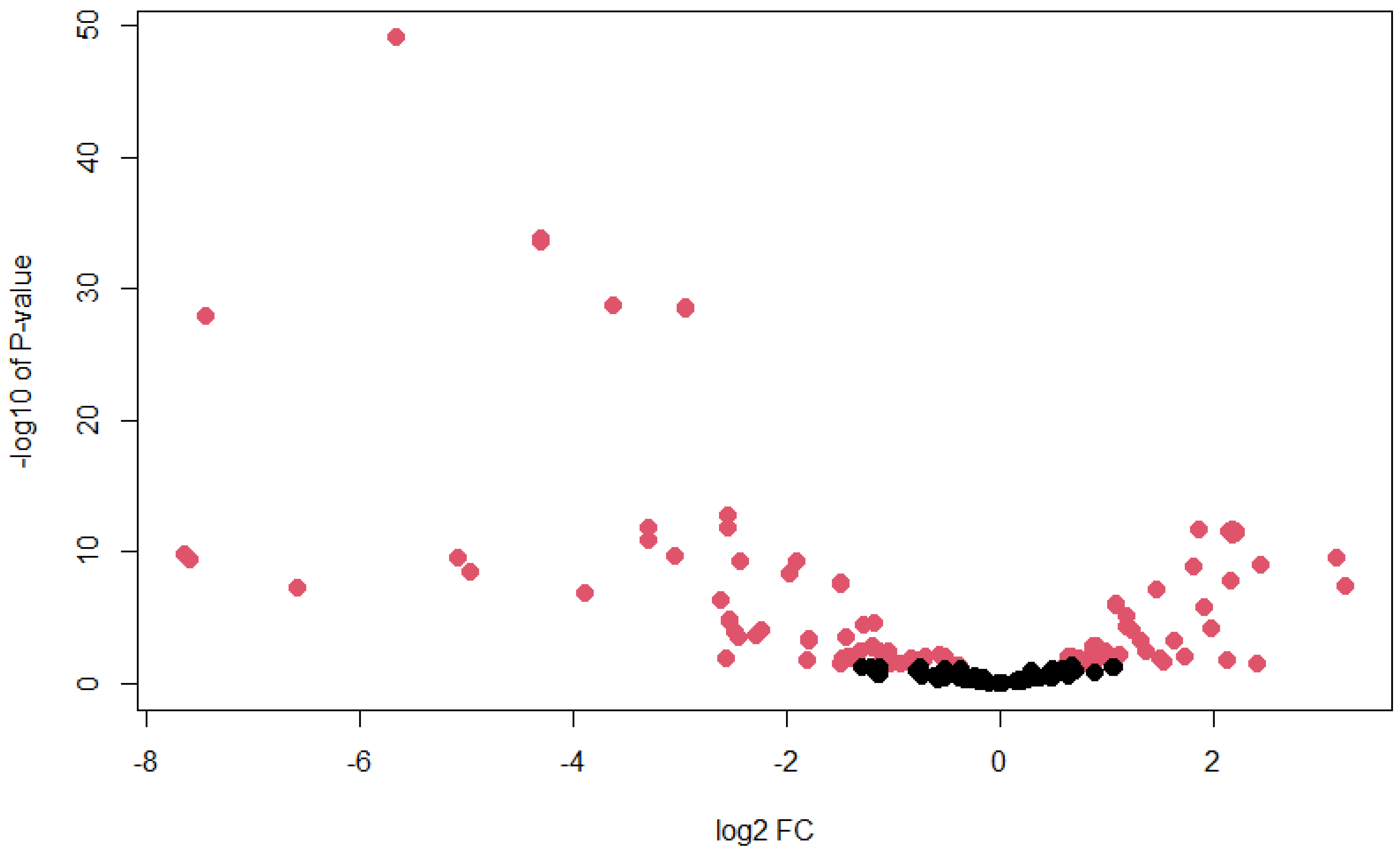
3.4. Ae. albopictus Novel miRNAs Were Differentially Expressed upon DENV Infection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gloria-Soria, A.; Payne, A.F.; Bialosuknia, S.M.; Stout, J.; Mathias, N.; Eastwood, G.; Ciota, A.T.; Kramer, L.D.; Armstrong, P.M. Vector Competence of Aedes albopictus Populations from the Northeastern United States for Chikungunya, Dengue, and Zika Viruses. Am. J. Trop. Med. Hyg. 2021, 104, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.J.; Carlson, C.J.; Mordecai, E.A.; Johnson, L.R. Global expansion and redistribution of Aedes-borne virus transmission risk with climate change. PLoS Negl. Trop. Dis. 2019, 13, e0007213. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, Y.; Wu, J.; Zheng, P.; Li, Y.; Zheng, X.; Puthiyakunnon, S.; Tu, Z.; Chen, X.-G. The expression profile of Aedes albopictus miRNAs is altered by dengue virus serotype-2 infection. Cell Biosci. 2015, 5, 16. [Google Scholar] [CrossRef]
- Liu, W.; Cheng, P.; Zhang, K.; Gong, M.; Zhang, Z.; Zhang, R. Systematic identification and characterization of long noncoding RNAs (lncRNAs) during Aedes albopictus development. PLoS Negl. Trop. Dis. 2022, 16, e0010245. [Google Scholar] [CrossRef]
- Azlan, A.; Obeidat, S.M.; Das, K.T.; Yunus, M.A.; Azzam, G. Genome-wide identification of Aedes albopictus long noncoding RNAs and their association with dengue and Zika virus infection. PLoS Negl. Trop. Dis. 2021, 15, e0008351. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Wang, G.; Li, C.; Xing, D.; Yan, T.; Zhu, X.; Liu, Q.; Wu, Q.; Guo, X.; Zhao, T. Screening for differentially expressed miRNAs in Aedes albopictus (Diptera: Culicidae) exposed to DENV-2 and their effect on replication of DENV-2 in C6/36 cells. Parasit. Vectors 2019, 12, 44. [Google Scholar] [CrossRef]
- Xu, Y.; Dong, Y.; Xu, Y.; Lai, Z.; Jin, B.; Hao, Y.; Gao, Y.; Sun, Y.; Chen, X.; Gu, J. Differentiation of Long Non-Coding RNA and mRNA Expression Profiles in Male and Female Aedes albopictus. Front. Genet. 2019, 10, 975. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Azlan, A.; Dzaki, N.; Azzam, G. Argonaute: The executor of small RNA function. J. Genet. Genom. 2016, 43, 481–494. [Google Scholar] [CrossRef]
- Kunze-Schumacher, H.; Krueger, A. The Role of MicroRNAs in Development and Function of Regulatory T Cells—Lessons for a Better Understanding of MicroRNA Biology. Front. Immunol. 2020, 11, 2185. [Google Scholar] [CrossRef]
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.D.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020, 11, 1197. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.R.; Abd-Aziz, N.; Affendi, S.; Poh, C.L. Role of microRNAs in antiviral responses to dengue infection. J. Biomed. Sci. 2020, 27, 4. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Li, C.; Zhang, Y.; Yan, T.; Zhu, X.; Zhao, M.; Xing, D.; Dong, Y.; Guo, X.; Zhao, T. Identification of microRNAs expressed in the midgut of Aedes albopictus during dengue infection. Parasit. Vectors 2017, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Avila-Bonilla, R.G.; Yocupicio-Monroy, M.; Marchat, L.A.; De Nova-Ocampo, M.A.; Del Ángel, R.M.; Salas-Benito, J.S. Analysis of the miRNA profile in C6/36 cells persistently infected with dengue virus type 2. Virus Res. 2017, 232, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Batz, Z.A.; Goff, A.C.; Armbruster, P.A. MicroRNAs are differentially abundant during Aedes albopictus diapause maintenance but not diapause induction. Insect Mol. Biol. 2017, 26, 721–733. [Google Scholar] [CrossRef]
- Etebari, K.; Hussain, M.; Asgari, S. Identification of microRNAs from Plutella xylostella larvae associated with parasitization by Diadegma semiclausum. Insect Biochem. Mol. Biol. 2013, 43, 309–318. [Google Scholar] [CrossRef]
- Etebari, K.; Asgari, S. Accuracy of MicroRNA Discovery Pipelines in Non-Model Organisms Using Closely Related Species Genomes. PLoS ONE 2014, 9, e84747. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Palatini, U.; Masri, R.A.; Cosme, L.V.; Koren, S.; Thibaud-Nissen, F.; Biedler, J.K.; Krsticevic, F.; Johnston, J.S.; Halbach, R.; Crawford, J.E.; et al. Improved reference genome of the arboviral vector Aedes albopictus. Genome Biol. 2020, 21, 215. [Google Scholar] [CrossRef]
- Atieh, T.; Baronti, C.; de Lamballerie, X.; Nougairède, A. Simple reverse genetics systems for Asian and African Zika viruses. Sci. Rep. 2016, 6, 39384. [Google Scholar] [CrossRef]
- Li, J.; Hu, D.; Ding, X.; Chen, Y.; Pan, Y.; Qiu, L.; Che, X. Enzyme-Linked Immunosorbent Assay-Format Tissue Culture Infectious Dose-50 Test for Titrating Dengue Virus. PLoS ONE 2011, 6, e22553. [Google Scholar] [CrossRef]
- Johnson, B.W.; Russell, B.J.; Lanciotti, R.S. Serotype-Specific Detection of Dengue Viruses in a Fourplex Real-Time Reverse Transcriptase PCR Assay. J. Clin. Microbiol. 2005, 43, 4977–4983. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Armisen, J.; Gilchrist, M.J.; Wilczynska, A.; Standart, N.; Miska, E.A. Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome Res. 2009, 19, 1766–1775. [Google Scholar] [CrossRef][Green Version]
- Harding, J.L.; Horswell, S.; Heliot, C.; Armisen, J.; Zimmerman, L.B.; Luscombe, N.M.; Miska, E.A.; Hill, C.S. Small RNA profiling of Xenopus embryos reveals novel miRNAs and a new class of small RNAs derived from intronic transposable elements. Genome Res. 2014, 24, 96–106. [Google Scholar] [CrossRef][Green Version]
- Gu, J.; Hu, W.; Wu, J.; Zheng, P.; Chen, M.; James, A.A.; Chen, X.; Tu, Z. miRNA genes of an invasive vector mosquito, Aedes albopictus. PLoS ONE 2013, 8, e67638. [Google Scholar] [CrossRef]
- Lai, E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002, 30, 363–364. [Google Scholar] [CrossRef] [PubMed]
- Metpally, R.; Nasser, S.; Malenica, I.; Courtright, A.; Carlson, E.; Ghaffari, L.; Villa, S.; Tembe, W.; Van Keuren-Jensen, K. Comparison of Analysis Tools for miRNA High Throughput Sequencing Using Nerve Crush as a Model. Front. Genet. 2013, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-G.; Jiang, X.; Gu, J.; Xu, M.; Wu, Y.; Deng, Y.; Zhang, C.; Bonizzoni, M.; Dermauw, W.; Vontas, J.; et al. Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc. Natl. Acad. Sci. USA 2015, 112, E5907–E5915. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Kim, B.; Kim, V.N. Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E1881–E1889. [Google Scholar] [CrossRef]
- Naganuma, M.; Tadakuma, H.; Tomari, Y. Single-molecule analysis of processive double-stranded RNA cleavage by Drosophila Dicer-2. Nat. Commun. 2021, 12, 4268. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Berezikov, E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011, 12, 846–860. [Google Scholar] [CrossRef]
- Chen, K.; Rajewsky, N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007, 8, 93–103. [Google Scholar] [CrossRef]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Lim, X.-N.; Shan, C.; Marzinek, J.K.; Dong, H.; Ng, T.S.; Ooi, J.S.G.; Fibriansah, G.; Wang, J.; Verma, C.S.; Bond, P.J.; et al. Molecular basis of dengue virus serotype 2 morphological switch from 29 °C to 37 °C. PLoS Pathog. 2019, 15, e1007996. [Google Scholar] [CrossRef]
- Hanley, J.P.; Tu, H.A.; Dragon, J.A.; Dickson, D.M.; del Rio-Guerra, R.; Tighe, S.W.; Eckstrom, K.M.; Selig, N.; Scarpino, S.V.; Whitehead, S.S.; et al. Immunotranscriptomic profiling the acute and clearance phases of a human challenge dengue virus serotype 2 infection model. Nat. Commun. 2021, 12, 3054. [Google Scholar] [CrossRef]
- Angleró-Rodríguez, Y.I.; MacLeod, H.J.; Kang, S.; Carlson, J.S.; Jupatanakul, N.; Dimopoulos, G. Aedes aegypti Molecular Responses to Zika Virus: Modulation of Infection by the Toll and Jak/Stat Immune Pathways and Virus Host Factors. Front. Microbiol. 2017, 8, 2050. [Google Scholar] [CrossRef] [PubMed]
- Bonizzoni, M.; Dunn, W.A.; Campbell, C.L.; Olson, K.E.; Marinotti, O.; James, A.A. Complex Modulation of the Aedes aegypti Transcriptome in Response to Dengue Virus Infection. PLoS ONE 2012, 7, e50512. [Google Scholar] [CrossRef] [PubMed]
- Miesen, P.; Ivens, A.; Buck, A.H.; van Rij, R.P. Small RNA Profiling in Dengue Virus 2-Infected Aedes Mosquito Cells Reveals Viral piRNAs and Novel Host miRNAs. PLoS Negl. Trop. Dis. 2016, 10, e0004452. [Google Scholar] [CrossRef]
- Sim, S.; Jupatanakul, N.; Ramirez, J.L.; Kang, S.; Romero-Vivas, C.M.; Mohammed, H.; Dimopoulos, G. Transcriptomic Profiling of Diverse Aedes aegypti Strains Reveals Increased Basal-level Immune Activation in Dengue Virus-refractory Populations and Identifies Novel Virus-vector Molecular Interactions. PLoS Negl. Trop. Dis. 2013, 7, e2295. [Google Scholar] [CrossRef]
- Sim, S.; Dimopoulos, G. Dengue Virus Inhibits Immune Responses in Aedes aegypti Cells. PLoS ONE 2010, 5, e10678. [Google Scholar] [CrossRef]
- Tsujimoto, H.; Hanley, K.A.; Sundararajan, A.; Devitt, N.P.; Schilkey, F.D.; Hansen, I.A. Dengue virus serotype 2 infection alters midgut and carcass gene expression in the Asian tiger mosquito, Aedes albopictus. PLoS ONE 2017, 12, e0171345. [Google Scholar] [CrossRef]
- Zanini, F.; Pu, S.-Y.; Bekerman, E.; Einav, S.; Quake, S.R. Single-cell transcriptional dynamics of flavivirus infection. eLife 2018, 7, e32942. [Google Scholar] [CrossRef]
- Yung, C.-F.; Lee, K.-S.; Thein, T.-L.; Tan, L.-K.; Gan, V.C.; Wong, J.G.X.; Lye, D.C.; Ng, L.-C.; Leo, Y.-S. Dengue serotype-specific differences in clinical manifestation, laboratory parameters and risk of severe disease in adults, singapore. Am. J. Trop. Med. Hyg. 2015, 92, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Avila-Bonilla, R.G.; Yocupicio-Monroy, M.; Marchat, L.A.; Pérez-Ishiwara, D.G.; Cerecedo-Mercado, D.A.; del Ángel, R.M.; Salas-Benito, J.S. miR-927 has pro-viral effects during acute and persistent infection with dengue virus type 2 in C6/36 mosquito cells. J. Gen. Virol. 2020, 101, 825–839. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.P.; Ezzell, R.M.; Arnaout, M.A. Direct interaction of filamin (ABP-280) with the beta 2-integrin subunit CD18. J. Immunol. 1995, 154, 3461–3470. [Google Scholar]
- Edwards, D.N.; Towb, P.; Wasserman, S.A. An activity-dependent network of interactions links the Rel protein Dorsal with its cytoplasmic regulators. Development 1997, 124, 3855–3864. [Google Scholar] [CrossRef]
- Loo, D.T.; Kanner, S.B.; Aruffo, A. Filamin binds to the cytoplasmic domain of the beta1-integrin. Identification of amino acids responsible for this interaction. J. Biol. Chem. 1998, 273, 23304–23312. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.; Islam, M.N.; Ye, Y.H.; Chotiwan, N.; Graham, B.; Belisle, J.T.; Kouremenos, K.A.; Dayalan, S.; Tull, D.L.; Klatt, S.; et al. Dengue virus dominates lipid metabolism modulations in Wolbachia-coinfected Aedes aegypti. Commun. Biol. 2020, 3, 518. [Google Scholar] [CrossRef]
- Melo, C.F.O.R.; Delafiori, J.; Dabaja, M.Z.; de Oliveira, D.N.; Guerreiro, T.M.; Colombo, T.E.; Nogueira, M.L.; Proenca-Modena, J.L.; Catharino, R.R. The role of lipids in the inception, maintenance and complications of dengue virus infection. Sci. Rep. 2018, 8, 11826. [Google Scholar] [CrossRef]
- Zaitseva, E.; Yang, S.-T.; Melikov, K.; Pourmal, S.; Chernomordik, L.V. Dengue Virus Ensures Its Fusion in Late Endosomes Using Compartment-Specific Lipids. PLoS Pathog. 2010, 6, e1001131. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Kuhn, R.J.; Rossmann, M.G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005, 3, 13–22. [Google Scholar] [CrossRef]
- Rodenhuis-Zybert, I.A.; Wilschut, J.; Smit, J.M. Dengue virus life cycle: Viral and host factors modulating infectivity. Cell. Mol. Life Sci. 2010, 67, 2773–2786. [Google Scholar] [CrossRef]
- Latanova, A.; Starodubova, E.; Karpov, V. Flaviviridae Nonstructural Proteins: The Role in Molecular Mechanisms of Triggering Inflammation. Viruses 2022, 14, 1808. [Google Scholar] [CrossRef] [PubMed]



| Sample | Raw Reads | Clean Reads | 18–32 bp Reads | Alignment Rate (%) |
|---|---|---|---|---|
| DENV replicate 1 | 78933646 | 76813221 | 51893885 | 95.73 |
| DENV replicate 2 | 77299313 | 74393164 | 50852534 | 96.02 |
| DENV replicate 3 | 55658966 | 54155383 | 42163819 | 95.29 |
| Uninfected replicate 1 | 54757609 | 53082234 | 41476616 | 96.62 |
| Uninfected replicate 2 | 52985076 | 51516847 | 41877287 | 95.78 |
| Uninfected replicate 3 | 58901861 | 57008973 | 41997868 | 95.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azlan, A.; Yunus, M.A.; Abdul Halim, M.; Azzam, G. Revised Annotation and Characterization of Novel Aedes albopictus miRNAs and Their Potential Functions in Dengue Virus Infection. Biology 2022, 11, 1536. https://doi.org/10.3390/biology11101536
Azlan A, Yunus MA, Abdul Halim M, Azzam G. Revised Annotation and Characterization of Novel Aedes albopictus miRNAs and Their Potential Functions in Dengue Virus Infection. Biology. 2022; 11(10):1536. https://doi.org/10.3390/biology11101536
Chicago/Turabian StyleAzlan, Azali, Muhammad Amir Yunus, Mardani Abdul Halim, and Ghows Azzam. 2022. "Revised Annotation and Characterization of Novel Aedes albopictus miRNAs and Their Potential Functions in Dengue Virus Infection" Biology 11, no. 10: 1536. https://doi.org/10.3390/biology11101536
APA StyleAzlan, A., Yunus, M. A., Abdul Halim, M., & Azzam, G. (2022). Revised Annotation and Characterization of Novel Aedes albopictus miRNAs and Their Potential Functions in Dengue Virus Infection. Biology, 11(10), 1536. https://doi.org/10.3390/biology11101536






