Cmcrf1, a Putative Zn2Cys6 Fungal Transcription Factor, Is Involved in Conidiation, Carotenoid Production, and Fruiting Body Development in Cordyceps militaris
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Cultivation Conditions
2.2. Disruption of Cmcrf1 and Transformation of C. militaris
2.3. Complementation of the Cmcrf1-Null Mutant
2.4. Fungal Development Assays
2.5. Cell Wall Integrity Assays
2.6. Determination of Carotenoids
2.7. Quantitative Real-Time PCR (qRT-PCR)
3. Results
3.1. Identification and Expression Pattern of T-DNA-Tagged Cmcrf1 in C. militaris
3.2. Disruption of Cmcrf1
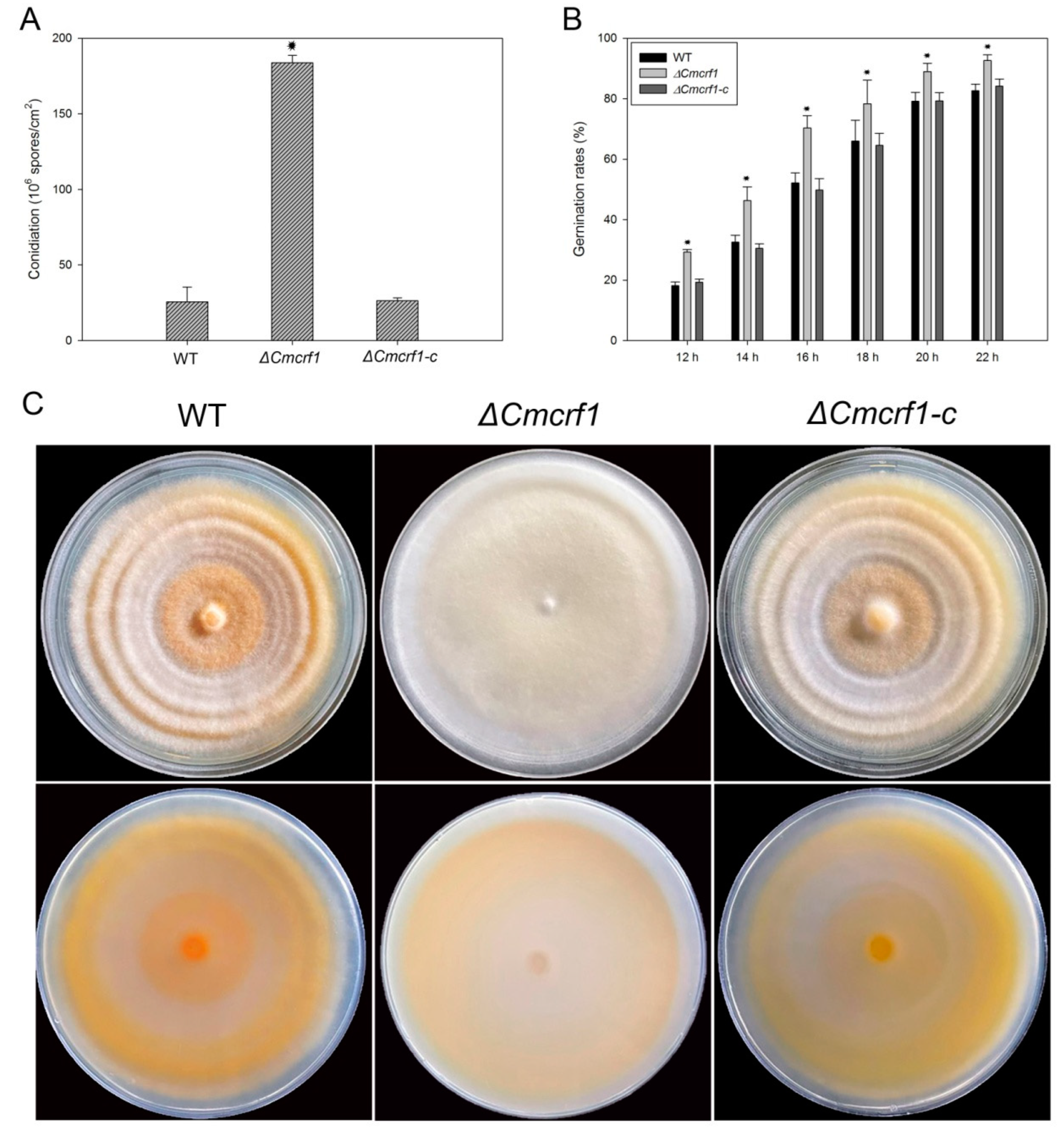
3.3. Deletion of Cmcrf1 Affects the Growth Characteristics of C. militaris
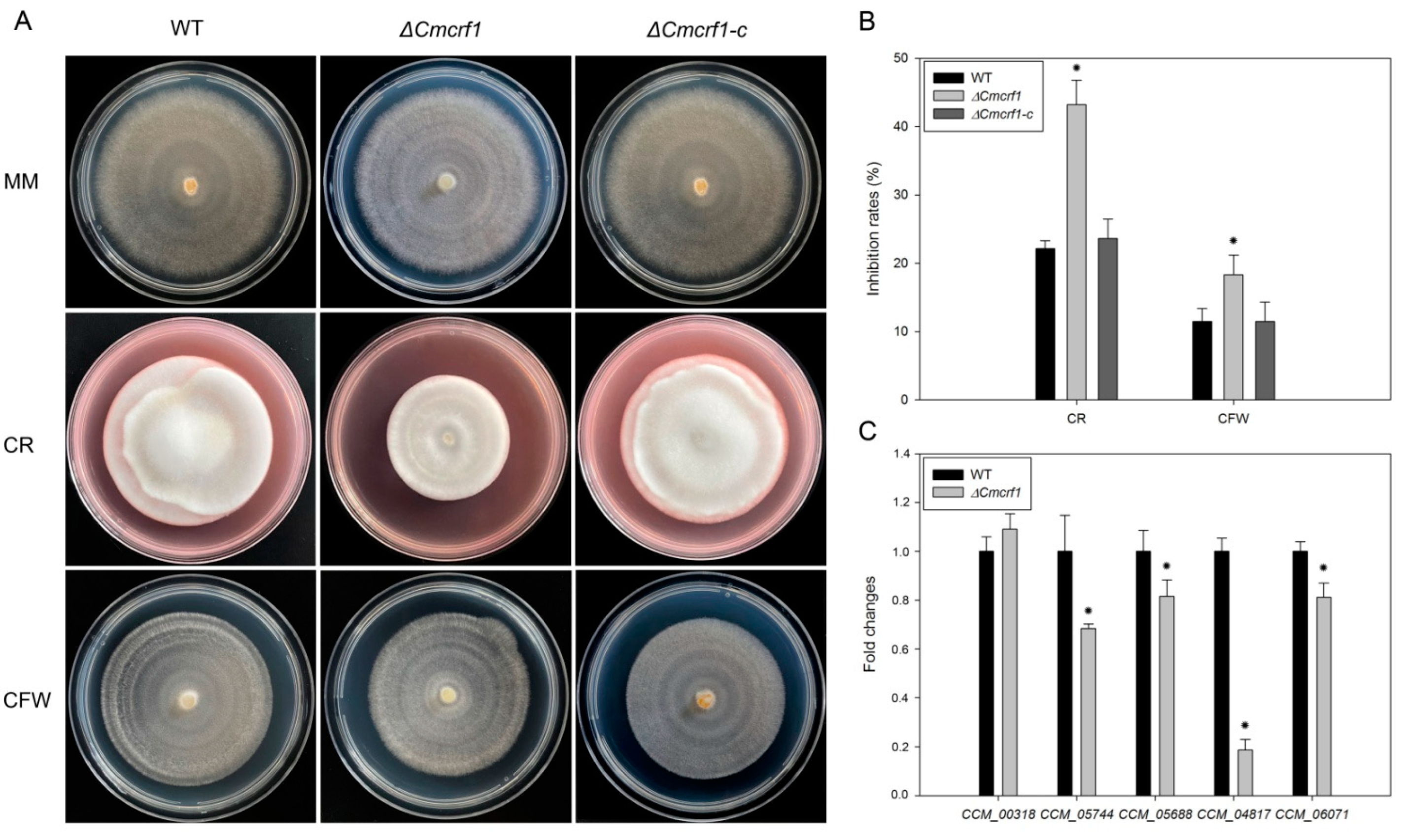
3.4. Effect of Cmcrf1 on Cell-Wall Integrity
3.5. The ΔCmcrf1 Mutant Showed Defective Development of Fruiting Bodies
3.6. Cmcrf1 Affects the Carotenoid Content of C. militaris
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jędrejko, K.J.; Lazur, J.; Muszyńska, B. Cordyceps militaris: An overview of its chemical constituents in relation to biological activity. Foods 2021, 10, 2634. [Google Scholar] [CrossRef] [PubMed]
- Rupa, E.J.; Li, J.F.; Arif, M.H.; Yaxi, H.; Puja, A.M.; Chan, A.J.; Hoang, V.A.; Kaliraj, L.; Yang, D.C.; Kang, S.C. Cordyceps militaris fungus extracts-mediated nanoemulsion for improvement antioxidant, antimicrobial, and anti-inflammatory activities. Molecules 2020, 25, 5733. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, S.L.; Chen, H.Y.; Zou, Y.; Zheng, Q.W.; Guo, L.Q.; Wu, G.H.; Lu, J.; Lin, J.F.; Ye, Z.W. Enhancement of carotenoid production and its regulation in edible mushroom Cordyceps militaris by abiotic stresses. Enzyme Microb. Technol. 2021, 148, 109808. [Google Scholar] [CrossRef] [PubMed]
- Stahl, W.; Sies, H. Bioactivity and protective effects of natural carotenoids. Biochim. Biophys. Acta. 2005, 1740, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Wang, F.F.; Li, K.B.; Liu, Q.; Yang, Y.; Dong, C.H. Heat and light stresses affect metabolite production in the fruit body of the medicinal mushroom Cordyceps militaris. Appl. Microbiol. Biotechnol. 2018, 102, 4523–4533. [Google Scholar]
- Zheng, P.; Xia, Y.; Zhang, S.; Wang, C. Genetics of Cordyceps and related fungi. Appl. Microbiol. Biotechnol. 2013, 97, 2797–2804. [Google Scholar] [CrossRef]
- Gao, Y.L.; Yu, C.; Li, L.J. Heterologous expression of a natural product biosynthetic gene cluster from Cordyceps militaris. Antibiot. 2022, 75, 16–20. [Google Scholar] [CrossRef]
- Meng, G.; Wang, X.; Liu, M.; Wang, F.; Liu, Q.; Dong, C. Efficient CRISPR/Cas9 system based on autonomously replicating plasmid with an AMA1 sequence and precisely targeted gene deletion in the edible fungus, Cordyceps militaris. Microb. Biotechnol. 2022, 15, 2594–2606. [Google Scholar] [CrossRef]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.; Hu, X.; Zhang, S.; Zheng, H.; Huang, Y.; Zhou, Y.; Wang, S.; et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 2011, 12, R116. [Google Scholar] [CrossRef]
- Sung, G.H. Complete mitochondrial DNA genome of the medicinal mushroom Cordyceps militaris (Ascomycota, Cordycipitaceae). Mitochondrial DNA 2015, 26, 789–790. [Google Scholar] [CrossRef]
- Kramer, G.J.; Nodwell, J.R. Chromosome level assembly and secondary metabolite potential of the parasitic fungus Cordyceps militaris. BMC Genomics 2017, 18, 912. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, F.; Liu, M.Q.; Dong, C.H. Hydrophobin CmHYD1 is involved in conidiation, infection and primordium formation, and regulated by GATA transcription factor CmAreA in edible fungus, Cordyceps militaris. J. Fungi 2021, 7, 674. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.W.; Zhao, Y.; Chen, B.X.; Yu, Y.H.; Tang, H.B.; Ye, Z.W.; Lin, J.F.; Guo, L.Q. Cmfhp gene mediates fruiting body development and carotenoid production in Cordyceps militaris. Biomolecules 2020, 10, 410. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.W.; Zhao, Y.; Tang, H.B.; Ye, Z.W.; Wei, T.; Lin, J.F.; Guo, L.Q. Transcriptome analysis of Cordyceps militaris reveals genes associated with carotenoid synthesis and identification of the function of the Cmtns gene. Front. Microbiol. 2019, 10, 2105. [Google Scholar] [CrossRef]
- Yang, T.; Guo, M.M.; Yang, H.J.; Guo, S.P.; Dong, C.H. The blue-light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl. Microbiol. Biotechnol. 2016, 100, 743–755. [Google Scholar] [CrossRef]
- Zhang, J.J.; Wang, F.F.; Yang, Y.; Wang, Y.; Dong, C.H. CmVVD is involved in fruiting body development and carotenoid production and the transcriptional linkage among three blue-light receptors in edible fungus Cordyceps militaris. Environ. Microbiol. 2020, 22, 466–482. [Google Scholar] [CrossRef]
- Wang, F.; Song, X.H.; Dong, X.M.; Zhang, J.J.; Dong, C.H. DASH-type cryptochromes regulate fruiting body development and secondary metabolism differently than CmWC-1 in the fungus Cordyceps militaris. Appl. Microbiol. Biotechnol. 2017, 101, 4645–4657. [Google Scholar] [CrossRef]
- Thananusak, R.; Laoteng, K.; Raethong, N.; Zhang, Y.; Vongsangnak, W. Metabolic responses of carotenoid and cordycepin biosynthetic pathways in Cordyceps militaris under light-programming exposure through genome-wide transcriptional analysis. Biology 2020, 9, 242. [Google Scholar] [CrossRef]
- In-On, A.; Thananusak, R.; Ruengjitchatchawalya, M.; Vongsangnak, W.; Laomettachit, T. Construction of light-responsive gene regulatory network for growth, development and secondary metabolite production in Cordyceps militaris. Biology 2022, 11, 71. [Google Scholar] [CrossRef]
- Deng, S.; Wang, C.Y.; Zhang, X.; Lin, L. Bidirectional promoter trapping T-DNA for insertional mutagenesis in Verticillium dahliae. Can. J. Microbiol. 2014, 60, 445–454. [Google Scholar] [CrossRef]
- Lv, B.; Zheng, L.; Liu, H.; Tang, J.T.; Hsiang, T.; Huang, J.B. Use of random T-DNA Mutagenesis in identification of gene UvPRO1, a regulator of conidiation, stress response, and virulence in Ustilaginoidea virens. Front. Microbiol. 2016, 7, 2086. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Guo, F.; Wang, Z.; Shen, X.; Deng, Y.; Meng, J.; Jiang, Z.; Chen, B. Genetic dissection of T-DNA insertional mutants reveals uncoupling of dikaryotic filamentation and virulence in sugarcane smut fungus. Phytopathology 2021, 111, 2303–2308. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.H.; Wang, X.L.; Wang, T.H.; Jiang, Q. Agrobacterium-mediated transformation (AMT) of Trichoderma reesei as an efficient tool for random insertional mutagenesis. Appl. Microbiol. Biotechnol. 2007, 73, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Y.; He, C.Y.; Wang, G.L.; Zhang, G.C.; Wang, C.L.; Wang, D.H.; Zou, X.; Wei, G.Y. Improved production of β-glucan by a T-DNA-based mutant of Aureobasidium pullulans. Appl. Microbiol. Biotechnol. 2021, 105, 6887–6898. [Google Scholar] [CrossRef]
- Yu, J.H.; Hamari, Z.; Han, K.H.; Seo, J.A.; Reyes-Domínguez, Y.; Scazzocchio, C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef]
- Liu, X.H.; Lu, J.P.; Zhang, L.; Dong, B.; Min, H.; Lin, F.C. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell 2007, 6, 997–1005. [Google Scholar] [CrossRef]
- de Groot, M.J.A.; Bundock, P.; Hooykaas, P.J.J.; Beijersbergen, A.G.M. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998, 16, 839–842. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual (Fourth Edition): Three-Volume Set; Cold Spring Harbor Laboratory: Long Island, NY, USA, 2012; ISBN 978-7-03-038606-9. [Google Scholar]
- Yang, T.; Sun, J.; Lian, T.T.; Wang, W.Z.; Dong, C.H. Process optimization for extraction of carotenoids from medicinal caterpillar fungus, Cordyceps militaris (Ascomycetes). Int. J. Med. Mushrooms 2014, 16, 125–135. [Google Scholar] [CrossRef]
- He, R.; Ma, L.; Li, C.; Jia, W.; Li, D.; Zhang, D.; Chen, S. Trpac1, a pH response transcription regulator, is involved in cellulase gene expression in Trichoderma reesei. Enzyme Microb. Technol. 2014, 67, 17–26. [Google Scholar]
- Liu, Y.G.; Chen, Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. BioTechniques 2007, 43, 649–650. [Google Scholar] [CrossRef]
- Free, S.J. Fungal cell wall organization and biosynthesis. Adv. Genet. 2013, 81, 33–82. [Google Scholar] [PubMed]
- MacPherson, S.; Larochelle, M.; Turcotte, B. A fungal family of transcriptional regulators: The zinc cluster proteins. Microbiol. Mol. Biol. Rev. 2006, 70, 583–604. [Google Scholar] [CrossRef] [PubMed]
- Long, N.B.; Orasch, T.; Zhang, S.Z.; Gao, L.; Xu, X.L.; Hortschansky, P.; Ye, J.; Zhang, F.L.; Xu, K.; Gsaller, F.; et al. The Zn2Cys6-type transcription factor LeuB cross-links regulation of leucine biosynthesis and iron acquisition in Aspergillus fumigatus. PLoS Genet. 2018, 14, 10. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.L.; Ye, W.; Zhu, M.Z.; Kong, Y.L.; Li, S.N.; Liu, S.; Zhang, W.M. Interaction of a novel Zn2Cys6 transcription factor DcGliZ with promoters in the gliotoxin biosynthetic gene cluster of the deep-sea-derived fungus Dichotomomyces cejpii. Biomolecules 2019, 10, 56. [Google Scholar] [CrossRef]
- Liao, L.S.; Li, C.X.; Zhang, F.F.; Yan, Y.S.; Luo, X.M.; Zhao, S.; Feng, J.X. How an essential Zn2Cys6 transcription factor PoxCxrA regulates cellulase gene expression in ascomycete fungi? Biotechnol. Biofuels 2019, 12, 105. [Google Scholar] [CrossRef]
- Porquier, A.; Moraga, J.; Morgant, G.; Dalmais, B.; Simon, A.; Sghyer, H.; Collado, I.G.; Viaud, M. Botcinic acid biosynthesis in Botrytis cinerea relies on a subtelomeric gene cluster surrounded by relics of transposons and is regulated by the Zn2Cys6 transcription factor BcBoa13. Curr. Genet. 2019, 65, 965–980. [Google Scholar] [CrossRef]
- Zhang, Z.X.; Jia, H.; Liu, N.; Li, H.X.; Meng, Q.J.; Wu, N.; Cao, Z.Y.; Dong, J.G. The zinc finger protein StMR1 affects the pathogenicity and melanin synthesis of Setosphaeria turcica and directly regulates the expression of DHN melanin synthesis pathway genes. Mol. Microbiol. 2022, 117, 261–273. [Google Scholar] [CrossRef]
- Lu, J.P.; Cao, H.J.; Zhang, L.L.; Huang, P.Y.; Lin, F.C. Systematic analysis of Zn2Cys6 transcription factors required for development and pathogenicity by high-throughput gene knockout in the rice blast fungus. PLoS Pathog. 2014, 10, 10. [Google Scholar] [CrossRef]
- Carrillo, A.J.; Schacht, P.; Cabrera, I.E.; Blahut, J.; Prudhomme, L.; Dietrich, S.; Bekman, T.; Mei, J.; Carrera, C.; Chen, V.; et al. Functional profiling of transcription factor genes in Neurospora crassa. G3 Genes Genomes Genet. 2017, 7, 2945–2956. [Google Scholar]
- Son, H.; Seo, Y.S.; Min, K.; Park, A.R.; Lee, J.; Jin, J.M.; Lin, Y.; Cao, P.Y.; Hong, S.Y.; Kim, E.K.; et al. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog. 2011, 7, e1002310. [Google Scholar] [CrossRef]
- Moise, A.R.; Al-Babili, S.; Wurtzel, E.T. Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 2014, 114, 164–193. [Google Scholar] [CrossRef]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. Bioessays 2006, 28, 799–808. [Google Scholar] [CrossRef]
- Kong, L.A.; Yang, J.; Li, G.T.; Qi, L.L.; Zhang, Y.J.; Wang, C.F.; Zhao, W.S.; Xu, J.R.; Peng, Y.L. Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2012, 8, e1002526. [Google Scholar] [CrossRef] [PubMed]
- Soulie, M.C.; Perino, C.; Piffeteau, A.; Choquer, M.; Malfatti, P.; Cimerman, A.; Kunz, C.; Boccara, M.; Vidal-Cros, A. Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell Microbiol. 2006, 8, 1310–1321. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Sugui, J.A.; Steinberg, G.; Deising, H.B. A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant. Microbe Interact. 2007, 20, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Larson, T.M.; Kendra, D.F.; Busman, M.; Brown, D.W. Fusarium verticillioides chitin synthases CHS5 and CHS7 are required for normal growth and pathogenicity. Curr. Genet. 2011, 57, 177–189. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, R.; Zhang, L.; Lan, J.; Mei, S.; Li, Y. Cmcrf1, a Putative Zn2Cys6 Fungal Transcription Factor, Is Involved in Conidiation, Carotenoid Production, and Fruiting Body Development in Cordyceps militaris. Biology 2022, 11, 1535. https://doi.org/10.3390/biology11101535
He R, Zhang L, Lan J, Mei S, Li Y. Cmcrf1, a Putative Zn2Cys6 Fungal Transcription Factor, Is Involved in Conidiation, Carotenoid Production, and Fruiting Body Development in Cordyceps militaris. Biology. 2022; 11(10):1535. https://doi.org/10.3390/biology11101535
Chicago/Turabian StyleHe, Ronglin, Lin Zhang, Jinling Lan, Shengjie Mei, and Yu Li. 2022. "Cmcrf1, a Putative Zn2Cys6 Fungal Transcription Factor, Is Involved in Conidiation, Carotenoid Production, and Fruiting Body Development in Cordyceps militaris" Biology 11, no. 10: 1535. https://doi.org/10.3390/biology11101535
APA StyleHe, R., Zhang, L., Lan, J., Mei, S., & Li, Y. (2022). Cmcrf1, a Putative Zn2Cys6 Fungal Transcription Factor, Is Involved in Conidiation, Carotenoid Production, and Fruiting Body Development in Cordyceps militaris. Biology, 11(10), 1535. https://doi.org/10.3390/biology11101535




